Impact of acclimation methods on microbial communities and performance of anaerobic fluidized bed membrane bioreactors†
Nicole
LaBarge
 a,
Yaoli
Ye
a,
Kyoung-Yeol
Kim
a,
Yasemin Dilsad
Yilmazel
a,
Pascal E.
Saikaly
b,
Pei-Ying
Hong
b and
Bruce E.
Logan
*a
a,
Yaoli
Ye
a,
Kyoung-Yeol
Kim
a,
Yasemin Dilsad
Yilmazel
a,
Pascal E.
Saikaly
b,
Pei-Ying
Hong
b and
Bruce E.
Logan
*a
aDepartment of Civil and Environmental Engineering, The Pennsylvania State University, University Park, PA 16802, USA. E-mail: nicole.a.labarge@gmail.com; yeyaoli007@gmail.com; kuk32@psu.edu; yyilmazel@engr.psu.edu; blogan@psu.edu
bBiological and Environmental Sciences and Engineering Division, Water Desalination and Reuse Center, King Abdullah University of Science and Technology (KAUST), 4700 King Abdullah Boulevard, Thuwal 23955-6900, Saudi Arabia. E-mail: Pascal.Saikaly@kaust.edu.sa; Peiying.Hong@kaust.edu.sa
First published on 17th October 2016
Abstract
An anaerobic fluidized bed membrane bioreactor (AFMBR) is a new and effective method for energy-efficient treatment of low strength wastewater, but the factors that affect performance are not well known. Different inocula and acclimation methods of the granular activated carbon (GAC) used in the reactor were examined here to determine their impact on chemical oxygen demand (COD) removal and microbial community composition of domestic wastewater-fed AFMBRs. AFMBRs inoculated with anaerobic digester sludge (D) or domestic wastewater (W) and fed domestic wastewater, or inoculated with a microbiologically diverse anaerobic bog sediment and acclimated using methanol (M), all produced the same COD removal of 63 ± 12% using a diluted wastewater feed (100 ± 21 mg L−1 COD). However, an AFMBR with GAC inoculated with anaerobic digester sludge and acclimated using acetate (A) showed significantly increased wastewater COD removal to 84 ± 6%. In addition, feeding the AFMBR with the M-acclimated GAC with an acetate medium for one week subsequently increased COD removal to 70 ± 6%. Microbial communities enriched on the GAC included Geobacter, sulfur-reducing bacteria, Syntrophaceae, and Chlorobiaceae, with reactor A having the highest relative abundance of Geobacter. These results showed that acetate was the most useful substrate for acclimation of GAC communities, and GAC harbors unique communities relative to those in the AFMBR influent and recirculated solution.
Water impactAnaerobic membrane bioreactors are being used to treat wastewater as they avoid the high energy demands of aerobic membrane bioreactors and activated sludge. Fluidizing granular activated carbon (GAC) can minimize membrane fouling and achieve good levels of COD removal from wastewater. We examined different GAC acclimation methods and determined that GAC enrichment on acetate could subsequently improve bioreactor performance. |
1 Introduction
There is great interest in the development of new technologies to make the operation of wastewater treatment plants energy neutral, or even net energy producers. Microbial fuel cells (MFCs) are being examined as a technology for achieving net electricity production by directly generating electricity from organic matter in wastewater,1–3 but current production in MFCs rapidly declines when the chemical oxygen demand (COD) decreases to less than ∼100–150 mg L−1.4 To treat wastewater to levels needed for discharge, and maintain high power production, a post-treatment system is required. One new treatment technology that has been shown to be effective when treating effluents from MFCs and other anaerobic processes, or low-strength wastewaters is an anaerobic fluidized bed membrane bioreactor (AFMBR). In an AFMBR, granular activated carbon (GAC) is used as a surface for organic matter adsorption and biofilm growth. Recirculation of the wastewater fluidizes the GAC, which scours the membranes to minimize fouling. Membrane relaxation, accomplished by cycling the effluent pump on and off, is also used to help minimize fouling and maintain low transmembrane pressures (TMPs).5AFMBRs were originally developed as an energy-efficient method for treating effluent from an anaerobic fluidized bed reactor (AFBR).5,6 The effluent COD of a lab-scale AFMBR treating municipal wastewater was 25 ± 10 mg L−1, with a feed of 68 ± 18 mg COD L−1 (64% removal).5 A pilot-scale AFMBR produced an effluent COD of 14 ± 8 mg L−1, with a higher influent COD of 152 ± 36 mg L−1 (91% removal).6 AFMBRs have also been used to treat effluent from MFCs, producing low effluent COD concentrations of 16 ± 3 mg L−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 and 36 ± 6 mg L−1 of COD.8 All of these effluent CODs would be sufficient to meet discharge limitations of <30 mg L−1 of BOD5, assuming a 2
7 and 36 ± 6 mg L−1 of COD.8 All of these effluent CODs would be sufficient to meet discharge limitations of <30 mg L−1 of BOD5, assuming a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 COD
1 COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BOD5 ratio.9,10 Energy requirements for AFBR-AFMBR and MFC-AFMBR systems are significantly lower than conventional aerobic or other anaerobic membrane processes, and their energy costs could potentially be offset by electricity generation from methane recovery5,6 or electricity generated by MFCs.7 Fluidizing the GAC to minimize membrane fouling through scouring11 uses less energy than other methods to control membrane fouling, such as biogas sparging,12 and AFMBRs can have relatively short hydraulic retention times (HRTs) of 2–4 h,13 1 h,7 and 1.2–3.8 h.8 The percentage of COD removed by an AFMBR was found to be constant when the influent COD was varied between 150 to 300 mg L−1, but membrane fouling was best controlled with an influent COD of ≤200 mg L−1.14
BOD5 ratio.9,10 Energy requirements for AFBR-AFMBR and MFC-AFMBR systems are significantly lower than conventional aerobic or other anaerobic membrane processes, and their energy costs could potentially be offset by electricity generation from methane recovery5,6 or electricity generated by MFCs.7 Fluidizing the GAC to minimize membrane fouling through scouring11 uses less energy than other methods to control membrane fouling, such as biogas sparging,12 and AFMBRs can have relatively short hydraulic retention times (HRTs) of 2–4 h,13 1 h,7 and 1.2–3.8 h.8 The percentage of COD removed by an AFMBR was found to be constant when the influent COD was varied between 150 to 300 mg L−1, but membrane fouling was best controlled with an influent COD of ≤200 mg L−1.14
The AFMBR is a relatively new treatment process, and thus there are few studies that have examined the impact of GAC acclimation or operational conditions on performance. AFMBRs have been inoculated in different ways, for example, by using GAC from another anaerobic reactor7,8,13 or by microorganisms in the effluent of the AFBR.5,6,13,15 COD removal efficiencies for AFMBRs operated as a post-treatment step for wastewater treatment have varied from over a wide range of 64%5 to 91%, and the reactors have been operated under different conditions which precludes direct comparisons.6 It was therefore not known if the differences in performance were due to the composition of the feed, slight variations in reactor operation, or reactor acclimation methods. To better understand the impact of reactor acclimation on subsequent performance, different acclimation procedures were examined here for AFMBRs using various microbial inocula and substrates, with all reactors fed domestic wastewater that was diluted to match the COD of a typical upstream reactor effluent from an AFBR or MFC. DNA samples were obtained from the solution and the GAC to examine whether different microbial communities developed based on the acclimation methods and inoculum sources, and to assess which microbes could be important for AFMBR function based on their relative abundance in the samples.
2 Materials and methods
2.1 Reactor setup and operation
The AFMBR body (65 mL) was made of a clear PVC tube (30 cm tall, 16 mm diameter; U.S. Plastic Corp.) as previously described7 (Fig. S1, ESI†). The reactor contained a submerged membrane module consisting of eight, 24 cm-long polyvinylidene-fluoride (PVDF) hollow fiber membranes each 2.0 mm in outer diameter and 0.8 mm inner diameter, with a 0.1 μm pore size (Kolon Inc., South Korea). For biogas collection, a Hungate tube (10 mL, Bellco Glass Inc., Vineland, NJ) with the bottom cut off was epoxied onto the top of the reactor and sealed with a butyl rubber stopper (20 mm diameter, Chemglass Inc., Vineland, NJ). The reactor body was covered with aluminum foil to minimize algae growth. Plastic mesh (27 × 27, McMaster-Carr, NJ) was used at the bottom of the reactor body to retain the GAC (DARCO MRX, 10 × 30 mesh, Norit Activated Carbon, Cabot, GA). The GAC used was manufactured from lignite coal, and contained 0.22 mL g−1 micropores (<2 nm), 0.35 mL g−1 mesopores (2–50 nm), and 0.45 mL g−1 macropores (>50 nm), according to the manufacturer. Each reactor included 10 g wet weight of GAC, equivalent to 3.4–3.8 g dry weight, for a GAC concentration of 153 g L−1 in the reactor.Domestic wastewater used as the feed in all AFMBR experiments was obtained from the primary clarifier effluent at the Penn State Wastewater Treatment Plant. The wastewater was diluted to 100 mg L−1 COD using distilled water, in order to simulate the effluent concentration from an MFC or AFBR. Sodium bicarbonate was added to increase the wastewater conductivity to 1.2 mS cm−1, similar to that of a typical domestic wastewater. If the pH approached 8.0 by sodium bicarbonate addition, sodium chloride was instead added to complete the conductivity increase. The AFMBRs were operated at room temperature (22 ± 10 °C). The feed reservoir was placed on ice, and the wastewater warmed to room temperature in the feed tube prior to entering the reactor.
2.2 Acclimation methods
Four different GAC acclimation procedures were examined to evaluate their impact on subsequent AFMBR performance in terms of COD removal (Table 1). For two of these procedures, GAC was acclimated while being recirculated in the same column used for AFMBR tests, but without membranes, in order to produce acclimation conditions similar to those used in previous studies to acclimate GAC in anaerobic fluidized beds.6 This operational mode simulated operation of an AFBR and avoided any membrane fouling during acclimation. For these two tests, the inoculum to the column was either anaerobic digester sludge (D) or wastewater (W). GAC was fluidized to cover half of the reactor height by recirculating liquid inside the reactor at a flowrate of 190 mL min−1. Reactor D acclimation procedures achieved a high concentration of inoculum biomass by adding anaerobic digester sludge (6.5 mL, 10% v/v) into raw domestic wastewater, with closed-loop recirculation to keep the GAC (3.8 g) suspended, as done by Shin et al. for AFBR reactor inoculation.6 Membranes were then added, and the reactor was fed raw wastewater (257 ± 60 mg COD L−1), with an initial HRT of 11 h (1.5 L m−2 h−1 membrane flux) for three weeks, consistent with Shin et al.6 and another anaerobic membrane bioreactor.16 During installation of the membranes, the headspace in the open reactor body was flushed with N2/CO2 to maintain an anaerobic environment. GAC and reactor fluid was temporarily stored in a separate bottle flushed with N2/CO2, and replaced after installation was complete. After three weeks, influent was then switched to diluted wastewater (100 mg COD L−1), and the HRT was gradually reduced over the course of a week to 1 h (16 L m−2 h−1 membrane flux), resulting in an organic loading rate of 2.5 g L−1 d−1. A settling chamber was installed between the top of the reactor body and the Hungate tube to prevent overflow of GAC into the recirculation line. The reactor body was covered after day 10 of normal operation. The digester sludge was from a mesophilic anaerobic digester at the Penn State Wastewater Treatment Plant, which has a very high concentration of methanogens, but a relatively low diversity as it was primarily composed of acetoclastic Methanosaeta.17 To maintain anaerobic feed conditions, the feed to reactor D was bubbled with N2/CO2 (20% CO2, 80% N2), and gas bags containing N2/CO2 were attached to the closed feed bottle.| Reactor | Inoculum | Acclimation substrate | Acclimation procedure | Feed (mg L−1 COD) | HRT (h) |
|---|---|---|---|---|---|
| D | Anaerobic digester sludge | Wastewater | GAC acclimated in column for 1 week, then membranes added, and HRT gradually decreased from 11 h HRT to 1 h | 92 ± 14 | 1 h |
| W | Wastewater | Wastewater | GAC acclimated in column for 1 month, then membranes added | 112 ± 21 | 1 h |
| A | Anaerobic digester sludge | Wastewater + acetate + methanol | GAC acclimated in a serum bottle, and then transferred to AFMBR | 153 ± 19 | 1.3 h |
| M | Bog sediment | Methanol | GAC acclimated in a serum bottle, and then transferred to AFMBR | 106 ± 25 | 1 h |
For the W reactor, only diluted wastewater was used for inoculation, and therefore a simpler startup procedure was used compared to that used for reactor D that was inoculated with a high concentration of solids. GAC (3.4 g) was added into the column (no membranes), and the reactor was run with continuous flow of wastewater diluted to 100 mg L−1 COD at 1 h HRT for one month, and then the membranes were added into the system.
Two additional acclimation procedures were used based on GAC acclimation in serum vials, which avoids the need for pumping during acclimation, and with added organic matter (acetate and methanol). This serum bottle approach was used for AFMBR experiments by Ren et al.,7 with GAC inoculated using anaerobic sludge but acclimated to only wastewater. Here, we examined whether specific substrates could be used to avoid the need for diluted wastewater. Because methanogens in anaerobic digesters are primarily acetoclastic, it was reasoned that pre-acclimation using acetate might improve acclimation as it would provide the main growth substrate for acetoclastic methanogens. In addition, methanol is also known to stimulate the rate of methanogenesis, and has been added to the feed during the startup of anaerobic reactors.18 Therefore, GAC used for reactor A was acclimated in serum bottles (165 mL) using domestic wastewater (100 mL) amended with acetate (1 g L−1) and methanol (1 g L−1), anaerobic digester sludge (20 mL), and GAC (10 g wet weight).14 Approximately once per week, 70% of the medium was removed and replaced with fresh medium. GAC (10 g wet weight) was then used in the AFMBR with the membranes directly installed.
For the second set of serum bottle acclimation tests, only methanol was used as the substrate in order to enrich a wider diversity of methanogens than those that might develop in acetate-fed reactors. Therefore, for reactor M, the GAC was inoculated using sediment of an anaerobic bog, and only methanol (no acetate) was used. Anaerobic sediment was obtained from the Black Moshannon bog (40°54′20.6′′N, 78°03′11.1′′W), as this site is known to contain a highly diverse methanogen community compared to those in anaerobic digesters.19 Solids and fibers were removed from the bog sample by sieving under anaerobic conditions, followed by centrifugation for 5 min. at 7650 × g (Sorvall Evolution RC Centrifuge, Thermo Scientific, MA). Samples were decanted and mixed with bicarbonate-buffered methanogen medium20 to create a 50/50 (v/v) slurry. GAC was acclimated in a serum bottle (3.4 g), with the headspace flushed with nitrogen gas, with 10 mL of the bog sediment slurry, 40 mL bicarbonate medium, and 20 μL methanol (24 mg COD equivalent). GAC was then directly added to the AFMBR with membranes already installed.
2.3 AFMBR operation
AFMBRs inoculated with the D, W, and M acclimated GAC were operated at an HRT of 1 h (Table 1) by cycling the pump for 10 minutes on at 1.2 mL min−1, and then 1 minute off. On/off cycling helps to reduce fouling through membrane relaxation.5 GAC was fluidized by recirculating liquid inside the reactor at a flowrate of 210–250 mL min−1, so that the fluidized GAC covered the height of the membranes. Peristaltic pumps were used for influent, effluent, and recirculation lines (FH100M, Cole Parmer, Barrington, IL). The AFMBR inoculated with acetate-acclimated GAC (A), was operated at a slightly longer HRT (1.3 h), due to conditions needed for this reactor as a control reactor in other tests. However, it was expected that this slightly different HRT would not impact COD removal, as previous AFMBR tests have shown that HRTs of up to 3.8 h do not significantly impact the percentage of COD removal.8To study the COD removal from a reactor fed only soluble COD (sCOD), and determine if performance of an operating AFMBR could be improved by enrichment on an acetate feed, the AFMBR with the M acclimated GAC was switched to an acetate feed for one week, and then a feed of filtered diluted wastewater for one week. The acetate feed was made using 0.147 g L−1 of acetate (∼100 mg L−1 COD) in bicarbonate-buffered methanogen medium. The diluted wastewater (∼100 mg COD L−1), with the conductivity adjusted to 1.2 mS cm−1, was filtered through 0.45 μm pore diameter nylon filters (Micron Separations, Inc, Westborough, MA) for a final feed of 46 ± 10 mg COD L−1. When this AFMBR was then switched back to dilute wastewater feed, its operation was designated MA to distinguish performance from the previous tests.
2.4 Analytical methods
The performance of the AFMBRs was assessed on the basis of COD removal. COD was measured using a chemical kit (method 5220, HACH Company, Loveland, CO) and standard methods.21 Soluble COD was measured after filtering samples with 0.45 μm pore size syringe filters (PVDF, 25 mm size, Restek Corporation, PA). Gas produced by the reactor was collected in a gas bag attached to the top of the Hungate tube using a syringe needle to pierce the septum. The gas composition was measured using gas chromatography, (SRI 310C, SRI Instruments, Torrance, CA), and total gas volume was measured using a gastight syringe. Methane production in the gas phase was calculated by measuring the methane volume in the headspace over one to two days, with the reactor remaining sealed between measurements, as: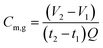 | (1) |
| Cm,l = 1.24 × 103 kHyPTVmρw | (2) |
2.5 Microbial community analysis
Microbial communities were examined for biomass extracted from GAC and from the solution in the AFMBRs inoculated with the D, W and A GAC. For the reactors inoculated with GAC that was originally enriched on methanol, samples were taken after the reactor was fed acetate, and thus these samples are indicated to be from reactor MA. No previous tests had been conducted on microbial communities in AFMBRs. Since we were particularly interested in using AFMBRs used to treat effluent from MFCs, we also examined microbial communities in an AFMBR fed with MFC effluent to see if they were similar to those that had evolved in the AFMBRs fed with diluted wastewater. For these tests, we used the AFMBR in our laboratory that were previously examined by Kim et al.,8 with these samples designated as reactor MFC. Details on reactor acclimation and COD removal and operational conditions of the AFMBR fed with MFC effluent was already described by Kim et al.,8 but no microbial community analysis was done as a part of that study. That AFMBR had also used GAC acclimated to anaerobic digester sludge, similar to that done here for the reactor D tests. Prior to DNA sampling, the MFC-fed AFMBR had been operated an HRT of 1.2 h.Microbial communities were examined using DNA extracted from GAC (0.25 g wet), or from the reactor fluid (13 mL) or the reactor influent (52 mL). Samples were obtained at the end of operation of the D, W, and MA reactors. DNA sampling was conducted on day 45 for reactor A, and day 38 for reactor MFC. All COD values reported here were obtained before DNA sampling. All liquid samples were centrifuged at 4500 × g for 1 h (Eppendorf 5804, NY) to generate a pellet, and the supernatant was decanted. DNA was extracted from the pellets or GAC using the MO Bio PowerSoil DNA extraction kit with the following modifications. Bead tubes with 0.1 mm glass beads were used instead of the garnet bead-beating tube included in the kit to reduce shear. The sample (GAC or pellet) and 750 μL bead solution were added to the bead-beating tube, and instead of vortexing for cell lysis, tubes were put to a bead mill (Bead Ruptor 12 Homogenizer, Kennesaw, GA) for 45 s on the medium setting. Tubes were centrifuged for 1 minute, and incubated at 4 °C for 10 minutes.
PCR was performed on the isolated DNA using the primer set 515F/805R because of its coverage of the domains bacteria and archaea. Amplicon sequences were obtained using Illumina MiSeq and were classified using the Ribosomal Database Project (RDP) at a 95% confidence interval. Relative abundance of each genus was estimated by normalizing the number of reads assigned to each genus against the total reads obtained for that sample. To assess similarities among samples, Bray–Curtis similarities were computed based on square-root transformed relative abundances. Bootstrapped multidimensional scaling (MDS) plots were generated from these Bray–Curtis similarities using Primer-E software, version 7.22 All high-throughput sequencing files were deposited in the Short Read Archive (SRA) of the European Nucleotide Archive (ENA) under study accession number PRJEB14201.
3 Results and discussion
3.1 COD removal efficiency
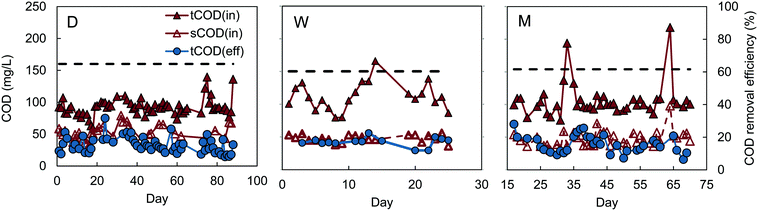
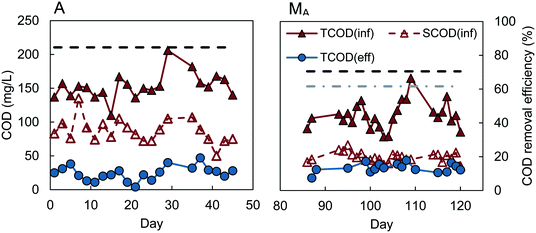
Reactors D, W, and M had similar COD removals of 62–64% (t-test, P > 0.4), despite different inocula and acclimation procedures (Fig. 1). The feed to reactors M and W were not significantly different (P = 0.29), with average influent CODs of 106 ± 25 mg L−1 for reactor M, and 112 ± 21 mg L−1 for W. Reactor D had a slightly lower average influent COD of 92 ± 14 mg L−1, but the effluent COD was also lower (33 ± 13 mg L−1), producing the same percentage COD removal (64 ± 13%) as reactors M and W. A constant percentage of COD removal, with different organic loading rates, was also observed in the MFC-fed AFMBR by Kim et al.8 Although the average COD removal for the MFC-fed reactor was slightly higher (67 ± 7%) on average than that obtained for reactors M, W, and D, our subsequent analysis of the data showed that the COD removals were not significantly different (P > 0.06). This similarity in COD removals for the D and W inoculated GAC suggests that the use of digester sludge versus wastewater as the GAC inoculum was not a significant factor in subsequent AFMBR performance. In addition, the finding of similar COD removals using a different inoculum (anaerobic bog sediment) suggested that the more diverse bog inoculum was not a factor. The use of methanol under these conditions did also not appear to be a factor due to the similar performance of all three of these approaches. Acclimation of GAC using a digester sludge that was amended with additional acetate (A), however, showed a significantly higher percentage of COD removal of 84 ± 6% (Fig. 2) (P < 5 × 10−9). These results suggested acetate acclimation was the key factor for the improved COD removal.The AFMBR inoculated with the M procedure was further examined for soluble COD removal and acetate removal. COD removal using the filtered wastewater was 54 ± 3%, indicating conversion and removal of particulate COD in the previous tests, as there was a higher percentage of COD with the non-filtered wastewater. The COD removal of acetate in these tests was inconclusive, due to interferences of chloride and ammonia in the bicarbonate medium which prohibited accurate measurements.23,24 When this reactor was subsequently used for treatment of diluted wastewater (results denoted MA), COD removal was significantly improved (P = 0.001) to 70 ± 6% (MA). These results, coupled with the A reactor tests showing improved COD removals, suggests that performance of an AFMBR can be improved either through the initial acclimation of the GAC to acetate prior to AFMBR tests, or through subsequent acetate feeding to the AFMBR acclimated by a different method. However, the impact of acetate feeding on AFMBRs on subsequent performance should be further explored to verify this finding.
All reactors reduced COD to levels permissible for discharge, assuming a typical COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BOD5 ratio of 2
BOD5 ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which has also been previously observed for wastewater samples from the same treatment plant used in this study.9 Effluent of lower-performing reactors M, W, and D averaged 36 ± 13 mg COD L−1, equivalent to 18 ± 6 mg BOD5 L−1, which is below the Environmental Protection Agency (EPA) secondary treatment standard of 30 mg BOD5 L−1 for a 30 day average.25 Effluent COD concentrations for other reactors were even lower, with estimated effluent BOD concentrations of 18 ± 3 mg L−1 for the MFC-fed AFMBR, 17 ± 3 mg L−1 for reactor MA, and 12 ± 5 mg L−1 for reactor A.
1, which has also been previously observed for wastewater samples from the same treatment plant used in this study.9 Effluent of lower-performing reactors M, W, and D averaged 36 ± 13 mg COD L−1, equivalent to 18 ± 6 mg BOD5 L−1, which is below the Environmental Protection Agency (EPA) secondary treatment standard of 30 mg BOD5 L−1 for a 30 day average.25 Effluent COD concentrations for other reactors were even lower, with estimated effluent BOD concentrations of 18 ± 3 mg L−1 for the MFC-fed AFMBR, 17 ± 3 mg L−1 for reactor MA, and 12 ± 5 mg L−1 for reactor A.
3.2 Methane generation
As COD removals were generally similar among the M, W and D reactors, methane gas was collected from a single reactor (D), at two points during operation when gas production over 1–2 days was sufficient to be measured. Both gas samples were taken after the reactor body was covered to prevent light entry. For the first measurements (days 16–18), 0.54 mL methane was collected from the headspace per liter of wastewater treated, and 1.53 mL L−1 was calculated for dissolved methane, for a total of 2.1 mL L−1. This recovery was much lower than that calculated for complete conversion of the COD removed into methane (23 mL L−1, assuming 382 mL methane per gram COD at 25 °C and 1 atm).26 However, the low amount of methane recovered is similar to that previously obtained for the same reactor configuration by Ren et al.7 of 1.67 mL L−1. The methane produced here during the second set of tests (days 62–63) was much higher, at 13.5 mL L−1 total (1.15 mL L−1 headspace gas, 12.4 mL L−1 dissolved). These measurements indicated highly variable methane production rates throughout the operation of an AFMBR, for reasons which are not well understood. However, there was likely COD removal supported by alternate electron acceptors, such as oxygen leaking into the reactor through the recirculation tubing or fittings.3.3 Microbial communities
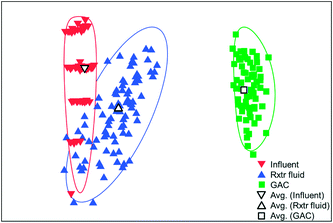
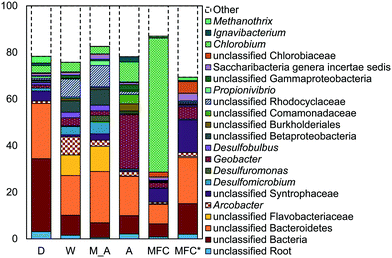
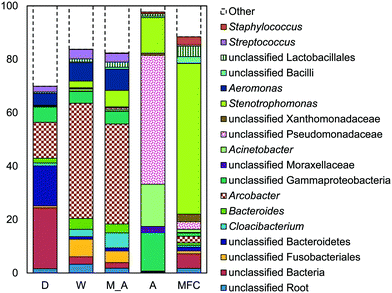
The microbial communities on the GAC in D, W, MA, A, and MFC-fed AFMBRs were compared to those in suspension within the reactor body and the AFMBR influent. DNA recovered from the GAC ranged from 2.3 to 3.7 μg DNA/g-GAC (see ESI,† Table S2). Based on bootstrapping DNA sequencing results, microbial communities on all GAC particles were significantly different than those in the influent (ANOSIM, R = 0.68, P = 0.008) or reactor fluid (ANOSIM, R = 0.34, P = 0.02) (Fig. 3). Microbial communities in the influent were not significantly different from the cells in suspension in the reactors (ANOSIM, R = −0.02, P = 0.47).Some of these microorganisms enriched on the GAC (Fig. 4) have been identified as those capable of exocellular electron transfer. Geobacter abundance on the GAC, ranging from 1.2% to 23%, was higher than that seen in the reactor fluid (<0.5%). In particular, reactor A showed high abundance of Geobacter (23%), over seven times that of the other GAC samples. Sulfate reducing bacteria (SRB), as assessed by the three most prevalent genera detected (Desulfobulbus, Desulfuromonas, and Desulfomicrobium) were enriched in reactors MA (10%), W (6%), and D (2%). Geobacter and some SRB (Desulfobulbus and Desulforomonas) can transfer electrons exocellularly without mediators.27–30 Additionally, methanogenesis by a co-culture of Geobacter metallireducens and Methanosarcina barkeri was enhanced by addition of GAC as a conductive surface for attached growth.31Desulfomicrobium may play a role in microbiologically influenced corrosion.32 The family Syntrophaceae showed strong correlation with GAC, as compared with reactor fluid and influent (Pearson >0.85). Members of Syntrophaceae are strict anaerobes, and grow in conjunction with microbes that utilize H2 or formate, and some members of this family are SRB.33 The close association of microbes provided by a biofilm environment may have selected for these types of syntrophic bacteria on the GAC.
While addition of GAC to co-cultures of G. metallireducens and G. sulfurreducens, as well as G. sulfurreducens and M. barkeri, has been shown to promote direct interspecies electron transfer (DIET),31 a specific reason for enriched populations of Geobacter or SRB in mixed-culture studies on GAC has not been reported. Liu et al.31 reported that the conductivity of GAC was substantially higher than that of microbial aggregates from anaerobic digesters, so the greater conductivity of GAC may select for microbes capable of exocellular electron transfer to other microorganisms on the GAC.
Reactor D contained the most archaea, with 3% Methanothrix and 5% archaea overall on the GAC (Fig. 4). All other GAC samples contained <1% total archaea. The fluid in reactor D was enriched in archaea (1.6%) and Methanothrix (0.8%), while other reactor fluid samples contained an order of magnitude less archaea (0.1% maximum). The influent for all samples contained low archaeal abundance (0.2% maximum). Methanothrix is an acetoclastic methanogen with a high affinity for acetate and a low growth rate, and it is therefore found in environments with low concentrations of acetate.34
A high abundance of Chlorobium, within the phylum Chlorobia, was found in the AFMBR treating MFC effluent (Fig. 4). Chlorobia are phototrophic obligate anaerobes. Instead of undergoing oxygenic photosynthesis and producing oxygen, Chlorobium undergo anoxygenic phototrophy using hydrogen or reduced sulfur as the electron donor.35 Although this process does not release oxygen and compromise the anaerobic environment of the reactor, sulfate-reduction from the products of anoxic phototrophy can compete with methanogenesis, which is undesirable for methods relying on methane production for energy capture. Reactors M, W, D and A were covered with aluminum foil during operation to prevent algae growth, but the MFC-fed AFMBR was operated without a cover, likely leading to the proliferation of phototrophic microbes.
Reactors MA and W showed very similar GAC communities (79% similarity, Bray Curtis, square root transformed), and both reactors showed slightly lower similarity (Fig. S3†) with reactor D (60 ± 1%). Reactor A showed a high similarity to MA and W (63 ± 1%), but it had less SRBs and Syntrophaceae, and more Geobacter than MA, W, and D. The high abundance of Chlorobium in the MEC-fed reactor likely produced a lower similarity to reactors M (54%), W (53%), D (54%), and A (55%). Removing Chlorobium from the relative abundance plot for better visualization (Fig. 4) showed that the MFC reactor also contained the distinguishing groups of Syntrophaceae, Geobacter, and SRBs.
There was less consistency of microbial populations among reactor fluid samples than among GAC samples (Fig. 5). As reactors MA and W each received wastewater from the same bottle, this may be the main reason they showed very similar microbial communities (79% similarity), if most of the selective microbial growth occurred on the GAC. Reactors MA and W showed lower similarity to reactors D (58 ± 2%) and A (33 ± 3%), although they all received diluted wastewater influent from the same treatment plant. The AFMBR that received MFC effluent, rather than diluted wastewater, showed moderate similarity to reactors MA (55%) and W (54%). Greater differences among reactor fluid for the five reactors tested suggest that the suspended environment was less selective for the growth of specific microbes.
 | ||
| Fig. 5 DNA sequencing results (>2% relative abundance) for reactor fluid samples from D, W, M, A, and MFC AFMBRs, displaying both bacteria and archaea. | ||
4 Conclusions
Acclimation of GAC to acetate significantly improved subsequent COD removal (84 ± 6%) in AFMBRs compared to GAC acclimated using other methods (63 ± 12% COD removal). Exposing a methanol-acclimated reactor to acetate for one week also resulted in a subsequent increase in COD removal from 62% to 71%, suggesting that temporary acetate amendment could be used to improve AFMBR performance, but this finding would need to be further studied for its long-term impact on AFMBR performance. Geobacter and SRB, which have previously been associated with conditions involving exocellular electron transfer, were found in higher abundance on the GAC than in the reactor fluid and influent. GAC was a more selective environment for microbes (Pearson >0.85) than the reactor fluid. These findings show that acclimation of GAC communities to an acetate substrate, or subsequent additional acclimation of a reactor using an acetate amendment, are useful methods for improving AFMBR performance.Acknowledgements
The authors would like to thank Dr. Hiroyuki Kashima for help with community analysis. This work was supported by Strategic Environmental Research and Development Program (SERDP), and Award OSR-2015-SEED-2450-01 from the King Abdullah University of Science and Technology (KAUST).References
- H. Liu, R. Ramnarayanan and B. E. Logan, Environ. Sci. Technol., 2004, 38, 2281–2285 CrossRef CAS PubMed.
- B. E. Logan, M. J. Wallack, K.-Y. Kim, W. He, Y. Feng and P. E. Saikaly, Environ. Sci. Technol. Lett., 2015, 2, 206–214 CrossRef CAS.
- B. E. Logan and K. Rabaey, Science, 2012, 337, 686–690 CrossRef CAS PubMed.
- X. Zhang, W. He, L. Ren, J. Stager, P. J. Evans and B. E. Logan, Bioresour. Technol., 2015, 176, 23–31 CrossRef CAS PubMed.
- R. Yoo, J. Kim, P. L. McCarty and J. Bae, Bioresour. Technol., 2012, 120, 133–139 CrossRef CAS PubMed.
- C. Shin, P. L. McCarty, J. Kim and J. Bae, Bioresour. Technol., 2014, 159, 95–103 CrossRef CAS PubMed.
- L. Ren, Y. Ahn and B. E. Logan, Environ. Sci. Technol., 2014, 48, 4199–4206 CrossRef CAS PubMed.
- K. Y. Kim, W. Yang, Y. Ye, N. LaBarge and B. E. Logan, Bioresour. Technol., 2016, 208, 58–63 CrossRef CAS PubMed.
- S. Hays, F. Zhang and B. E. Logan, J. Power Sources, 2011, 196, 8293–8300 CrossRef CAS.
- D. Mara and N. Horan, Water and Wastewater Microbiology, 2003 Search PubMed.
- M. Aslam, P. L. McCarty, J. Bae and J. Kim, Sep. Purif. Technol., 2014, 132, 10–15 CrossRef CAS.
- J. K. Kim, K. Kim, H. Ye, E. Lee, C. Shin, P. L. McCarty and J. Bae, Environ. Sci. Technol., 2011, 45, 576–581 CrossRef CAS PubMed.
- J. Bae, C. Shin, E. Lee, J. Kim and P. L. McCarty, Bioresour. Technol., 2014, 165, 75–80 CrossRef CAS PubMed.
- Y. Ye, N. LaBarge, H. Kashima, K.-Y. Kim, P. Hong, P. E. Saikaly and B. E. Logan, Environ. Sci.: Water Res. Technol., 2016 10.1039/c6ew00203j.
- K. Dutta, M. Y. Lee, W. W. Lai, C. H. Lee, A. Y. Lin, C. F. Lin and J. G. Lin, Bioresour. Technol., 2014, 165, 42–49 CrossRef CAS PubMed.
- A. Akram and D. C. Stuckey, Environ. Technol., 2008, 29, 1053–1065 CrossRef CAS PubMed.
- M. Siegert, X. F. Li, M. D. Yates and B. E. Logan, Front. Microbiol., 2014, 5, 778 Search PubMed.
- C. Shin, J. Bae and P. L. McCarty, Bioresour. Technol., 2012, 109, 13–20 CrossRef CAS PubMed.
- L. M. Steinberg and J. M. Regan, Appl. Environ. Microbiol., 2008, 74, 6663–6671 CrossRef CAS PubMed.
- M. Siegert, M. D. Yates, A. M. Spormann and B. E. Logan, ACS Sustainable Chem. Eng., 2015, 3, 1668–1676 CrossRef CAS.
- Standard Methods for the Examination of Water and Wastewater: Centennial, ed. A. D. Eaton, L. S. Clesceri, E. W. Rice, A. E. Greenberg and M. A. H. Franson, 21st edn, 2005, p. 1368 Search PubMed.
- M. Harb, Y. Xiong, J. Guest, G. Amy and P.-Y. Hong, Environ. Sci.: Water Res. Technol., 2015, 1, 800–813 Search PubMed.
- W. Boyles, Sci. Chem. Oxyg. Demand, 1997 Search PubMed.
- B. R. Kim, J. - Water Pollut. Control Fed., 1989, 61, 614–617 Search PubMed.
- NPDES, U.S. Environmental Protection Agency NPDES Permit Writers' Manual, 2010 Search PubMed.
- R. E. Speece, Anaerobic Biotechnology for Industrial Wastewaters, 1996 Search PubMed.
- D. R. Bond, D. E. Holmes, L. M. Tender and D. R. Lovely, Science, 2002, 295, 483–485 CrossRef CAS PubMed.
- D. R. Bond and D. R. Lovely, Appl. Environ. Microbiol., 2003, 69, 1548–1555 CrossRef CAS PubMed.
- D. E. Holmes, D. R. Bond and D. R. Lovely, Appl. Environ. Microbiol., 2004, 70, 1234–1237 CrossRef CAS PubMed.
- B. E. Logan, Nat. Rev. Microbiol., 2009, 7, 375–381 CrossRef CAS PubMed.
- F. Liu, A.-E. Rotaru, P. M. Shrestha, N. S. Malvankar, K. P. Nevin and D. R. Lovley, Energy Environ. Sci., 2012, 5, 8982 CAS.
- K. E. Duncan, B. M. Perez-Ibarra, G. Jenneman, J. B. Harris, R. Webb and K. Sublette, Appl. Microbiol. Biotechnol., 2014, 98, 907–918 CrossRef CAS PubMed.
- J. Kuever, in The Prokaryotes, ed. E. F. Rosenberg, S. Lory, E. Sackenbrandt and F. Thompson, 2014, pp. 281–288 Search PubMed.
- M. S. M. Jetten, A. J. M. Stams and A. J. B. Zehnder, FEMS Microbiol. Rev., 1992, 88, 181–198 CrossRef CAS.
- J. A. Eisen, K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant and C. M. Fraser, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9509–9514 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ew00237d |
| This journal is © The Royal Society of Chemistry 2016 |