Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Evaluation of electron donors for biological perchlorate removal highlights the importance of diverse perchlorate-reducing populations†
Nadine
Kotlarz
a,
Giridhar
Upadhyaya
b,
Paul
Togna‡
c and
Lutgarde
Raskin
*a
aDepartment of Civil and Environmental Engineering, University of Michigan, Ann Arbor, MI, USA. E-mail: raskin@umich.edu
bCarollo Engineers, Orange County, CA, USA
cEnvironmental Operating Solutions Inc., Bourne, MA, USA
First published on 10th October 2016
Abstract
This research investigated the treatment of a synthetic groundwater with approximately 100 mg L−1 perchlorate (ClO4−) and 15 mg L−1 nitrate (NO3−-N) using a bench-scale, fluidized-bed bioreactor (FBR). The groundwater was amended sequentially with acetate and MicroC2000™, a proprietary, glycerol-based electron donor. Nitrate reduction to less than 0.05 mg L−1 NO3−-N and perchlorate removal to less than 0.3 mg L−1 ClO4− occurred under both electron donor regimes, although a higher biomass yield was observed and a higher influent COD concentration was required to maintain the same effluent quality when MicroC2000™ was used as the electron donor. High-throughput sequencing of partial 16S rRNA genes from biomass collected at several time points revealed that a single Dechloromonas population dominated the perchlorate-reducing community under both electron donors and Dechloromonas species comprised greater than 30% relative abundance of the bacterial community by the end of reactor operation. The same Dechloromonas population was abundant in two bench-scale systems fed lower perchlorate concentrations, although several other perchlorate-reducing bacteria, presumably with higher affinities for perchlorate, were also abundant in those systems. The results suggest that to reduce perchlorate to levels that allow groundwater to serve as a drinking water source, distinct environments for diverse perchlorate-reducing bacteria with high and low affinities for perchlorate are needed. Such conditions can be created by using two bioreactors in series.
Water impactBiological removal of perchlorate and nitrate from a highly contaminated groundwater can be accomplished by feeding acetate or a proprietary electron donor to a bioreactor system. To reduce perchlorate to levels that allow groundwater to serve as a drinking water source, distinct environments for perchlorate-reducing bacteria with high and low affinities for perchlorate are needed in the bioreactor system. |
1. Introduction
Perchlorate (ClO4−) has been used as an oxidant in various industrial and defense-related applications, including the manufacturing of solid missile and rocket fuel, air bags, and road flares.1–3 Uncontained disposal of perchlorate-contaminated wastewater, made possible by the lack of a federal drinking water standard or cleanup requirement, has resulted in widespread environmental contamination across the United States.4 A 2007 study reported that 395 sites across 35 states, the District of Columbia, and two commonwealths had measurable levels of perchlorate in drinking water, groundwater, surface water, and sediment or soil.4 In 11 of these sites, perchlorate concentrations exceeded 500 mg L−1 and the highest level reported (3.7 g L−1) was in groundwater. The highest perchlorate concentrations were found in Arkansas, California, Nevada, Texas and Utah. Outside of the United States, reported perchlorate concentrations of contaminated groundwater and surface waters generally have been less than 1 mg L−1.5–8Human exposure to perchlorate is a health concern for reproductive-age women because perchlorate decreases thyroid hormone production and adversely affects fetal development.9 Exposure to perchlorate occurs primarily through ingestion of contaminated tap water and food.10 Ingestion of contaminated tap water was estimated to increase total perchlorate intake levels in reproductive-age women in the United States by 3–24%.11 These estimates were based on measurements of perchlorate concentrations in drinking water from the United States Environmental Protection Agency (U.S. EPA) UCMR1 database and data from other regional studies of perchlorate contamination. Additionally, the use of perchlorate-contaminated groundwater for irrigation increases the risk of human ingestion of perchlorate because several crops can absorb and accumulate perchlorate from irrigation waters.12 Certain food items, such as dairy products, fruits and vegetables, have been found to significantly contribute to perchlorate exposure.13 For these reasons, remediation of perchlorate-contaminated waters is important to limit human exposure to perchlorate and protect public health.
Biological reduction is an established approach for remediation of perchlorate-contaminated waters.14 Dissimilatory perchlorate-reducing bacteria are facultative anaerobes that can use perchlorate or chlorate as terminal electron acceptors for anaerobic respiration.15 Through this metabolism, perchlorate is reduced by the perchlorate reductase enzyme to chlorate (ClO3−), which is subsequently reduced by perchlorate reductase to chlorite (ClO2−).1 Chlorite dismutase detoxifies chlorite by converting it to chloride (Cl−) and molecular oxygen. The innocuous nature of the products of biological perchlorate reduction and the low chemical input needed to support biological treatment16 make biological remediation of groundwater containing high concentrations of perchlorate more attractive than technologies that employ chemical reducing agents.3,17,18
Biological water treatment often takes place in biofilm systems, such as fixed-bed bioreactor and fluidized-bed bioreactor (FBR) systems.19 In an FBR, biofilm develops on a support medium (e.g., granular activated carbon [GAC]) that is suspended or fluidized within the bioreactor. Biological perchlorate removal requires the addition of an electron donor to provide sufficient reducing equivalents to reduce perchlorate. An electron donor is also needed to reduce oxygen, the energetically more favorable electron acceptor, and the commonly co-occurring contaminant nitrate (NO3−).20,21 Suitable electron donors in such applications include acetate, ethanol, lactate and hydrogen.1,22 A variety of alternative electron donors are also available, including proprietary electron donors such as MicroC products (Environmental Operating Solutions, Bourne, MA). MicroC products have been used as electron donors for denitrification in wastewater treatment.23–26 In particular, several municipal and industrial denitrifying wastewater treatment plants are using MicroC2000™, a glycerol-based electron donor with an undisclosed composition. However, MicroC2000™ has not been studied for perchlorate removal from groundwater. Feasibility studies of diverse electron donors are important for providing the water treatment industry with flexibility for electron donor choice.
We recently reported using two bench-scale bioreactors (one fixed-bed bioreactor and one FBR) for treatment of a synthetic groundwater containing 200 μg L−1 ClO4− and 15 mg L−1 NO3− as N.27 Perchlorate reduction was established in both bioreactors with acetate as the electron donor and then acetate was replaced with MicroC4000™, a commercial, carbohydrate-based product. Biomass yields increased during operation with MicroC4000™ and more reducing equivalents (or chemical oxygen demand (COD)) were required than acetate COD to achieve comparable effluent quality. In the present study, we tested the feasibility of replacing acetate with a glycerol-based MicroC product (MicroC2000™) for treatment of a synthetic perchlorate-contaminated groundwater in a bench-scale FBR. In this case, the influent perchlorate levels were much higher (100 mg L−1) than in our previous study.27 First, contaminant removals and estimates of biomass yield under the two electron donor regimes were determined. Then the bacterial community structure in this “high-perchlorate” FBR was characterized by pyrosequencing of 16S ribosomal RNA (rRNA) genes from samples collected during operation with acetate and MicroC2000™. Additionally, biomass was collected from the “low-perchlorate” bench-scale systems reported in our previous study27 to compare perchlorate-reducing populations across three bioreactors that were inoculated in the same way but were operated with different perchlorate loadings and using different MicroC products. While nitrate and perchlorate removals in the low-perchlorate systems were described previously,27 their bacterial community structures are reported here for the first time.
2. Materials and methods
2.1 Reactor setup and operation
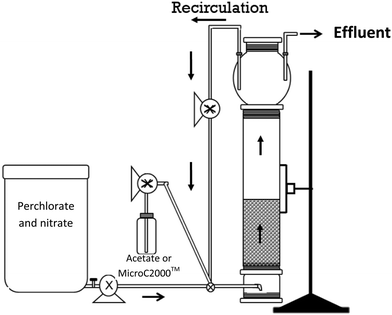
An FBR consisting of a glass column, 4.9 cm inner diameter and 56 cm height, fitted with a 3 L bulb at the top, was operated to treat a simulated groundwater containing 100 mg L−1 ClO4− and 15 mg L−1 NO3−-N. The column was packed with GAC particles (bituminous F816, 0.9 mm effective diameter, 1.25 g cm−3 true density, and 0.69 g cm−3 bulk density) collected from a pilot-scale nitrate- and perchlorate-removing bioreactor that used acetate as the electron donor20 (stored at 4 °C under aerobic conditions for several years) to achieve a settled bed height of 34 cm. The settled and fluidized bed volumes were 640 and 1075 cm3, resulting in empty bed contact times (EBCT) of 42.7 and 80.5 minutes, respectively. Excluding a few brief interruptions due to mechanical problems, the FBR was operated continuously for 222 days at 20.5 ± 0.4 °C in a temperature-controlled room. A second, identical FBR and a fixed-bed bioreactor system, which were described previously,27 were operated in parallel to treat a simulated groundwater containing approximately 200 μg L−1 ClO4− and 15 mg L−1 NO3−-N.Synthetic groundwater simulating a groundwater in Rialto (CA) (except for a higher perchlorate concentration) was prepared (Table 1). The groundwater recipe was amended with ammonium and phosphate to provide sufficient nitrogen and phosphorus for microbial growth and a trace metal solution, SL-10 (Table S1†), which included molybdenum, a trace element required for perchlorate reduction.28 The groundwater was prepared using deionized water and chemical stocks that were sterilized by filtration or autoclaving. The influent tank was covered to limit deposition of microbes from the air. The synthetic groundwater was pumped into the bottom of the FBR at a flow rate of 15 mL min−1 using a peristaltic pump. The majority of the water that passed through the FBR was recirculated from the top to the bottom at a flow rate of 1.8 L min−1 using a peristaltic pump to allow expansion of the GAC from approximately 34 cm to 44 cm (i.e., 29% expansion before biomass growth). The bed height fluctuated over time and attained a final expansion of 88% (expanded bed height was 64 cm at the end of the study). During reactor operation with acetate, the liquid in the bulb of the FBR was stirred periodically with a brush to resuspend the solids deposited on the sides and to remove excess biomass deposited in the bulb. After the electron donor was changed from acetate to a glycerol product, the reactor maintenance protocol was changed to include daily mixing and wasting of the liquid in the bulb.
| Chemicals used | Target concentration (mg L−1) | Expressed as |
|---|---|---|
| NaNO3 | 15 | NO3−-N |
| NaCl, MgCl2·6H2O and CaCl2 | 36.3 | Cl− |
| K2CO3 | 6 | CO32− |
| NaHCO3 | 213.5 | HCO3− |
| Na2SO4 | 12.5 | SO42− |
| NH4Cl | 12 | N |
| NaClO4 | 100 | ClO4− |
| KH2PO4 | 2.60 | P |
| Trace element solution SL-10 | See Table S4 |
2.2 Reactor inoculation
The GAC was inoculated with 70 mL of a suspended culture of Azospira suillum JPLRND (formerly called Dechlorosoma suillum JPLRND), a strain capable of using oxygen, nitrate, and perchlorate as terminal electron acceptors.29 The strain was isolated from a groundwater sample collected from an aquifer located near the Jet Propulsion Laboratory in Pasadena, California, USA.29 Additionally, the FBR was seeded with approximately 7 g of biologically active carbon (BAC) particles collected from a bench-scale fixed-bed bioreactor operated for nitrate and arsenic removal.30 The bench-scale nitrate and arsenic removing system had originally been seeded with BAC from a system operated for nitrate and perchlorate removal.20 To establish the microbial communities in the filter beds, synthetic groundwater containing nitrate, sulfate, perchlorate (approximately 200 μg L−1), salts, and trace metals was recirculated through the reactors for two days.2.3 Electron donor
The electron donor was pumped into the influent line using a peristaltic pump to achieve target influent chemical oxygen demand (COD) levels. Acetate was supplied as the electron donor during the first 114 days of operation. Assuming a net biomass yield of 0.4 g CODbiomass g−1 CODacetate (ref. 31) for all electron acceptors and ammonium-N as the nitrogen source, 190 mg L−1 acetate COD is required to remove 15 mg L−1 NO3−-N, 100 mg L−1 ClO4−, and 39 mg L−1 DO (including 7 mg L−1 DO in the influent and 32 mg L−1 DO resulting from the complete reduction of 100 mg L−1 ClO4−). On day 114, acetate was replaced with MicroC2000™, a glycerol product with a COD of 1086 g L−1 in the originally delivered aqueous product. This glycerol product was dosed at a target COD level equivalent to the acetate COD fed during days 81–114, although the measured influent COD concentration was somewhat higher (216.2 mg L−1 C on day 142). Over time, electron donor limitation in the reactor was suspected and the influent concentration of the glycerol product was increased to 491.1 mg L−1 C (average of two data points) on day 142. To minimize the sulfate reduction observed after increasing the influent COD, the influent COD was lowered on day 197 to approximately 442 mg L−1 C.2.4 Chemical analyses
The reactors were operated for 222 days and the performance was monitored for 217 days. Influent and effluent pH, effluent oxidation reduction potential (ORP), and influent DO concentrations were monitored regularly. DO levels were measured in the influent tank using a WTW multi340 meter with CellOx325 sensors (Weilheim, Germany) (detection limit 0.01 mg L−1). pH was measured using a Seven Easy pH meter (Mettler Toledo, Columbus, OH) and ORP was measured in flow-through cells using an Oakton pH meter (Oakton Instruments, Vernon Hills, IL) fitted with an ORP electrode.Samples were collected from the influent when a new influent batch was prepared (every 3–4 days) and samples were collected from the effluent approximately every other day. Influent samples were taken from the influent tank and effluent samples were collected from the top of the reactor through a tube positioned at the liquid control point from the top of the bulb (Fig. 1). The samples were filtered through 0.22 μm filters (Fisher, Pittsburgh, PA) and stored at 4 °C until analysis.
Two ion chromatography systems were used to measure concentrations of anions in the FBR influent and effluent. Influent acetate, nitrate, chloride, and sulfate concentrations were measured using a Dionex DX 100 ion chromatograph (Dionex, Sunnyvale, CA) with an AS-14 column (4 × 250 mm) equipped with a conductivity detector, as previously described,30 and carbonate/bicarbonate eluent. Influent and effluent perchlorate concentrations and effluent concentrations of acetate, nitrate, chloride and sulfate were determined using a second ion chromatograph, a Dionex ICS-2100, with a conductivity detector. The anions were chromatographically separated using a Dionex AS-16 column (2 × 250 mm) and a gradient flow of potassium hydroxide (KOH) was used to elute the anions through the column. The second ion chromatography system had a longer run time and better sensitivity for perchlorate analysis. The method detection limits for acetate, chloride, nitrate as N, sulfate and perchlorate were determined to be 100 μg L−1, 40 μg L−1, 50 μg L−1, 40 μg L−1 and 3 μg L−1, respectively.
Soluble non-purgeable organic carbon (sNPOC) was measured in effluent samples using a Total Organic Carbon Analyzer TOC-V CSH (Shimadzu, Columbia, MD) (method detection limit 0.3 mg L−1 C). All samples were sparged and acidified in the instrument to remove inorganic carbon.
Effluent samples were occasionally analyzed for soluble chemical oxygen demand (sCOD). The HACH COD micro digestion method (HACH, Loveland, CO) for low range COD (0–150 mg L−1 C) was used for sample digestion and COD concentrations were measured colorimetrically using a HACH DR/4000 spectrophotometer at 420 nm (method detection limit 3.7 mg L−1 C). For time periods when few effluent sCOD measurements were taken, sCOD values were estimated using sNPOC measurements and the relationship between sNPOC and sCOD for effluent samples (Fig. S2†).
2.5 Biomass yield calculations
Biomass yields (g CODbiomass g−1 CODelectron donor) were estimated for four different electron donor operating conditions distinguished by composition and/or influent COD concentration and corresponding to data collected during days 102–114, 130–142, 178–189 and 200–210. The time periods chosen to estimate yields for conditions 1 and 2 corresponded with the final 12 days of operation for each condition. Earlier periods were chosen for conditions 3 and 4 because of limited effluent sCOD or sNPOC measurements (Fig. S2†). Data from the first days after a change in condition were not included in any yield estimates to avoid transition periods. Yield was estimated based on COD consumed (i.e., influent COD − effluent COD) and the calculated COD requirement for the amount of perchlorate, nitrate and DO removal observed (sulfate reduction was considered if it occurred). The COD consumed beyond the amount required to reduce perchlorate, nitrate, and DO (and sulfate for days 178–189) was considered to be used for biomass growth. Yield was estimated as the ratio of COD used for biomass to total COD consumed. COD equivalents of 2.86 mg COD mg−1 NO3−-N, 0.67 mg COD mg−1 ClO4−, and 0.322 mg COD mg−1 SO42− were used in the calculations. Estimates of yield for other time periods for comparison are presented in Table S2.†2.6 Biomass sampling, DNA extraction and 16S rRNA gene sequence analysis
Biomass samples were collected from the FBR and a low-perchlorate FBR27 on days 86, 96, 189, 196, and 222 by removing BAC particles from the top of each column. Biomass samples were collected from a low-perchlorate fixed-bed bioreactor27 on days 86 and 189. A composite sample was prepared for each sampling day by removing BAC particles from each of the sampling ports along the depth of the fixed-bed reactor and mixing the BAC particles. Samples were flash-frozen and stored at −80 °C. Total DNA was extracted from biomass samples using a phenol–chloroform–isoamyl (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 at pH 8) extraction protocol.32 The V3–V5 region of bacterial 16S rRNA genes was amplified using primers 563F-modified and 909R.33 PCR products were pooled based on equal DNA concentrations and submitted for titanium 454-pyrosequencing at the University of Illinois Keck Center.
1 at pH 8) extraction protocol.32 The V3–V5 region of bacterial 16S rRNA genes was amplified using primers 563F-modified and 909R.33 PCR products were pooled based on equal DNA concentrations and submitted for titanium 454-pyrosequencing at the University of Illinois Keck Center.
Sequences were analyzed using mothur version 1.36.1.34 The mothur implementation of AmpliconNoise35 was used to reduce sequencing errors and PCR single base errors. Each sequence was trimmed after the first 450 flows and sequences with any mismatches to the barcode or more than one mismatch to the primer were removed. Sequences with homopolymers longer than 8 bp and sequences less than 200 bp long were also removed. Alignment to the SILVA SSU database version 123 was performed using the Needleman–Wunsch algorithm (ksize = 8) and chimeras were removed using UCHIME.36 This resulted in 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 524 sequences that were approximately 200 bp long for further analysis.
524 sequences that were approximately 200 bp long for further analysis.
Taxonomic assignment of individual sequences was based on the naïve Bayesian method and the reference Greengenes taxonomy database with a confidence score threshold of 80%. Sequences were grouped into operational taxonomic units (OTUs) using the average neighbor approach at three percent sequence divergence. The consensus taxonomy for each OTU was determined using the mothur classify.otu command and the Greengenes taxonomy database.
3. Results and discussion
3.1 Perchlorate and nitrate removal
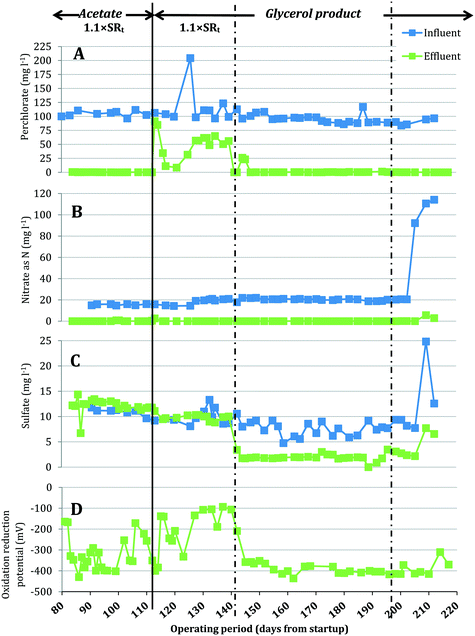
A bench-scale FBR was fed synthetic groundwater containing 100 mg L−1 ClO4− and 15 mg L−1 NO3−-N and operated under two electron donor regimes. After demonstrating consistent contaminant removal with acetate as the electron donor, acetate was replaced with MicroC2000™, a proprietary, glycerol-based product. Fig. 2 presents the influent and effluent perchlorate, nitrate and sulfate concentrations after start-up and optimization of operating conditions (days 81–217). As expected, the effluent pH did not change significantly during reactor operation (two tailed t-test, p = 0.07) and averaged 6.9 ± 0.3 for days 81–217. When acetate was used as the electron donor (days 81–114), the average influent NO3−-N concentration was 15.4 ± 0.6 mg L−1 and effluent NO3−-N concentrations were below the method detection limit (0.05 mg L−1 N), excluding three data points (days 101, 102 and 114) (Fig. 2B). For the same period, the influent perchlorate concentration averaged 104.4 ± 4.9 mg L−1 and perchlorate was reduced to 0.22 ± 0.13 mg L−1 in the effluent (Fig. 2A). The redox potential in the effluent averaged −326 ± 79 mV (Fig. 2D), indicating that redox conditions favorable for biological perchlorate reduction were established.37 Influent COD concentrations were maintained at 199.8 ± 11.9 mg L−1 during operation with acetate. Acetate COD concentrations in the effluent averaged 18.09 ± 8.02 mg L−1, which closely matched effluent sCOD concentrations (17.8 ± 10.2 mg L−1).On day 114, acetate was replaced with the glycerol product dosed at a target COD value equal to the acetate COD dose applied for days 81–114. This change in electron donor composition briefly impacted nitrate removal (Fig. 3), but effluent nitrate concentrations fell below detection again during days 116–142. Perchlorate removal also was impacted by the change in electron donor and effluent perchlorate concentrations first increased (91.5 mg L−1 and 84.8 mg L−1 ClO4− on days 115 and 116, respectively), decreased briefly (11.2 mg L−1 and 8.5 mg L−1 on days 119 and 123) and then increased gradually (54.3 ± 10.0 mg L−1 during days 127–142). The higher effluent perchlorate levels for days 127–142 corresponded with higher ORP (Fig. 2D) and lower effluent sCOD (4.7 mg L−1 ± 2.2 mg L−1) (Fig. 3A).
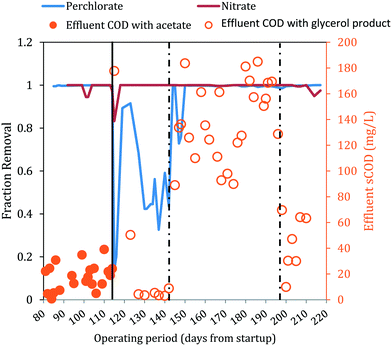 | ||
| Fig. 3 Fraction removal of perchlorate and nitrate and effluent sCOD concentrations. The solid line indicates when acetate was changed to a glycerol electron donor. Influent COD levels were adjusted twice (represented by dashed lines). Closed circles are effluent acetate COD. Open circles are effluent sCOD measured either directly by COD analysis or indirectly through a correlation with effluent sNPOC measurements (ESI†). | ||
After the influent concentration of the glycerol-based electron donor was increased on day 142 (influent COD was measured to be 484.19 mg L−1 on day 142), the effluent perchlorate concentrations decreased to an average of 0.28 ± 0.20 mg L−1 for days 145–193, excluding two measurements (26.1 mg L−1 and 23.2 mg L−1 on days 147 and 148, respectively). The effluent nitrate concentrations were below the detection limit during this period except on days 186, 189 and 193 when the effluent had 0.11, 0.13 and 5.65 mg L−1 NO3−-N, respectively. While the higher influent COD helped to restore perchlorate removal, significant sulfate reduction (73 ± 9% sulfate removed) was observed during days 145–193 (Fig. 2C). Lowering the influent COD on day 197 to approximately 442 mg L−1 helped to lessen sulfate reduction and the effluent sulfate concentration increased from 1.94 ± 0.62 mg L−1 SO42− (days 145–193) to 2.77 ± 0.53 mg L−1 (days 200–210), but sulfate reduction was not completely eliminated. Lowering the influent COD did not affect perchlorate removal (i.e., effluent perchlorate concentrations averaged 0.26 ± 0.10 mg L−1 for days 200–210).
3.2 Biomass yield with the glycerol-based electron donor
More biomass production was observed visually within 24 hours after changing from acetate to the glycerol-based electron donor and the reactor required more frequent wasting of biomass to prevent clogging and control bed expansion. Estimates of biomass yield indicated that the yield was higher with the glycerol product (0.54 g CODbiomass g−1 CODsubstrate) than with acetate (0.34 g CODbiomass g−1 CODsubstrate) even when influent COD levels were similar (Table 2). The estimated biomass yield increased further (0.62 g CODbiomass g−1 CODsubstrate) after the influent concentration of the glycerol product was increased on day 142. This increased yield observed with the glycerol product is consistent with previous reports of higher biomass yields with glycerol compared with other electron donors. In a bench-scale study with denitrifying, moving bed biofilm reactors, biomass growth was observed to be thicker and heavier with glycerol as the electron donor compared with methanol or sulfide.38 This study also reported that reactor operation with glycerol was associated with higher levels of effluent total suspended solids and greater plugging of media void spaces.38 Their estimate of the observed biomass yield with a glycerol-based electron donor (0.5 g CODbiomass g−1 CODsubstrate [based on average effluent volatile suspended solids data]) is close to the estimated yield with the glycerol product tested in the present study (0.54 g CODbiomass g−1 CODsubstrate, Table 2). Similarly, in a study with bench-scale, denitrifying sequencing batch reactors, the biomass yield increased over time as biomass grew on glycerol or a glycerol by-product from biodiesel fuel production.25| Operating condition | Period reported (days) | Electron donor | Measured influent COD (mg L−1) | Period included for yield calculations (days) | Estimated yielda (mg CODbiomass mg−1 CODelectron donor) |
|---|---|---|---|---|---|
| a COD consumed beyond the amount required for contaminant removal was considered to be used for biomass growth. Yield was estimated as the ratio of COD used for biomass growth to total COD consumed. COD equivalents of 2.86 mg COD mg−1 NO3−-N, 0.67 mg COD mg−1 ClO4−, and 0.322 mg COD mg−1 SO42− were used in the calculations. | |||||
| 1 | 81 to 114 | Acetate | 200 | 102 to 114 | 0.34 |
| 2 | 115 to 142 | Glycerol product | 216 | 130 to 142 | 0.55 |
| 3 | 143 to 197 | Glycerol product | 484 | 178 to 189 | 0.61 |
| 4 | 197 to 222 | Glycerol product | 442 | 200 to 210 | 0.69 |
Microbial populations that use glycerol with higher yields likely increased their relative abundance in the FBR after the change in electron donor, as suggested by Al-Omari et al.39 With more electrons channeled to biomass production, fewer electrons would have been available for the reduction of the electron acceptors. Indeed, replacing acetate with the glycerol product at an equivalent COD negatively affected perchlorate removal from days 130 to 142 (Fig. 3), during which time relatively low levels of soluble COD were also observed in the effluent (4.8 ± 2.4 mg L−1 sCOD) (Fig. 3A). As discussed above, after the influent concentration of the glycerol-based electron donor was increased on day 142, the effluent perchlorate concentrations decreased. Therefore, the changing effluent perchlorate levels observed between days 115 and 143 after converting the electron donor from acetate to the glycerol product was likely caused by substrate-limitation brought on by a higher biomass yield with the glycerol product.
3.3 Bacterial community structure
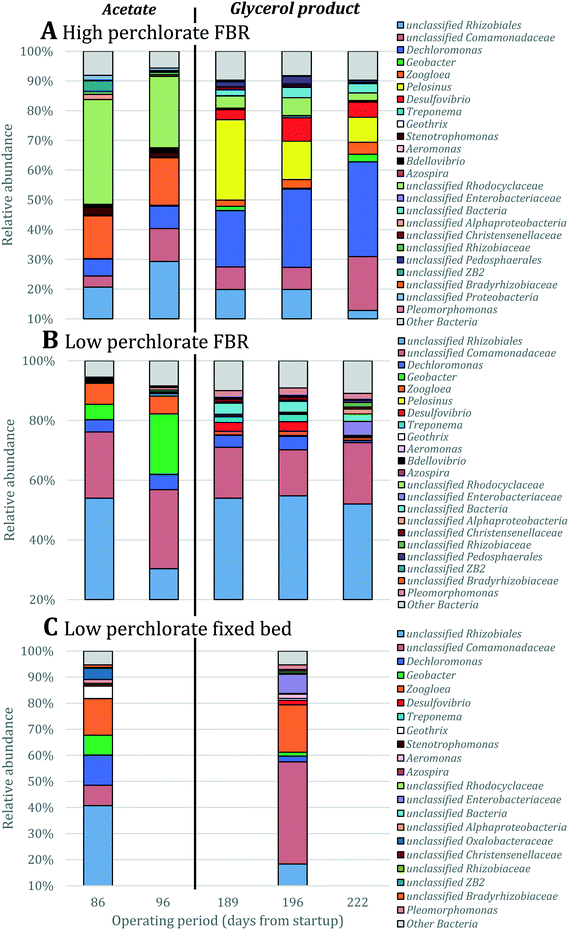
To determine the microbial community structures in the FBR under two electron donor regimes, 16S rRNA genes extracted from biomass collected during operation with acetate or MicroC2000™ were sequenced using 454 pyrosequencing. Additionally, 16S rRNA genes from biomass collected from another bench-scale FBR and a fixed-bed system (Fig. S1†) fed 200 μg L−1 ClO4− and 15 mg L−1 NO3−-N were sequenced. These “low-perchlorate” systems were operated using acetate as the electron donor for 100 days and then acetate was replaced with MicroC4000™, a carbohydrate-based, proprietary product with unknown composition. We compared the community structures in the low-perchlorate systems with the community structure in the high-perchlorate FBR to discuss the impact of bulk electron acceptor concentrations and reactor configurations.Bacterial members of 15 phyla were present in the three reactors, and Proteobacteria were most abundant (Fig. S3†). Perchlorate-reducing bacteria that perform canonical perchlorate respiration have been isolated exclusively from Proteobacteria.40Dechloromonas and Azospira, two genera within the Betaproteobacteria, encompass the (per)chlorate-reducing species most commonly isolated from the environment.40 The majority of the sequences associated with Dechloromonas in the high-perchlorate FBR were from a single OTU (Dechloromonas_OTU0003 in Fig. 5), which was also the dominant Dechloromonas OTU in the low-perchlorate FBR and fixed-bed systems, but to a lesser extent (Fig. 5). Many perchlorate-reducing bacteria can respire nitrate and Dechloromonas spp. have been found to be abundant in nitrate-reducing systems even in the absence of perchlorate,41 including in the nitrate- and arsenic-removing bioreactor,30 which provided biomass to inoculate the FBR and the low-perchlorate reactors. Therefore, the presence of nitrate was likely important for the initial selection of Dechloromonas OTU0003, but the higher bulk perchlorate concentrations in the high-perchlorate FBR (e.g., approximately 250 μg L−1 compared with 6 μg L−1 ClO4− in the low-perchlorate FBR) apparently enriched for this population. In the high-perchlorate FBR, the relative abundance of Dechloromonas increased from 5.8% to 32.1% over the reactor operating period (Fig. 5). Enrichment of Dechloromonas species in communities fed high concentrations of perchlorate is consistent with findings from other studies. For example, Dechloromonas represented 69% of clones from a mixed culture acclimated to 50–1500 mg L−1 ClO4−.42 In addition, the dominant (per)chlorate-reducing bacteria in a sequence batch reactor treating 1200 mg L−1 ClO4− included Dechloromonas (19% of clones), although Dechlorosoma (53%) and Ideonella (28%) species were more abundant.43 The enrichment for a single Dechloromonas OTU in the high-perchlorate FBR, compared with the many more Dechloromonas populations identified in the low-perchlorate systems (Fig. 5), implies that higher perchlorate concentrations select for a single perchlorate-reducing population. The increase in relative abundance of OTU0003 from 7.6% (day 96) to 19.2% (day 189) after the change from acetate to MicroC2000™ suggests that this population can utilize glycerol efficiently.
The perchlorate- and nitrate-reducing Azospira suillum JPLRND29 was used as one of the inocula in this study. OTU0051 matched the 16S rRNA gene sequence of Azospira suillum PS44 (Fig. S4†) and, therefore, it is possible that this Azospira population is the Azospira suillum JPLRND in the inoculum. This OTU was present in the first set of biomass samples collected from all reactors (day 86). However, it was not abundant in the FBR relative to other community members during operation with either acetate or the glycerol product (Fig. 5). The strain was originally isolated using medium amended with 1000 mg L−1 ClO4−, although the groundwater from which the strain was isolated contained only 300 μg L−1 ClO4−.45 Apparently this strain was not able to compete with other perchlorate-reducing bacteria in the FBR. Further evidence of its competitive exclusion from the FBR is that the putative inoculum had higher relative abundances in fixed-bed and FBR systems with lower bulk perchlorate levels (Fig. 5) but inoculated in the same way.27
Operation of the FBR with acetate for 114 days before converting to the glycerol-based product means that the final bacterial community structure was derived from the community that developed first with acetate. This sequence of electron donors was chosen to represent a practical scenario in which an electron donor commonly used for biological, inorganic contaminant removal (e.g., acetate) is replaced by an alternative, proprietary product. Thus, certain populations that were present during operation with the glycerol-based electron donor may have been present because they were initially established during operation with acetate. The abundance of Zoogloea spp., which can utilize nitrate as an electron acceptor46 and have been found to be abundant in bioreactors fed acetate,47 while the glycerol-based product was used may have been contingent on the use of acetate as the electron donor first. Zoogloea spp. were abundant in all three perchlorate reactors (16.0%, 6.1%, and 14.3% relative abundance on day 96 in the high- and low-perchlorate FBRs and low-perchlorate fixed-bed reactor, respectively) but decreased markedly in the FBRs after replacing acetate with the glycerol-based product. In contrast, the relative abundance of Zoogloea spp. increased slightly to 18.3% in the fixed-bed reactor after converting to the glycerol product. Compared with biofilms in a fixed-bed reactor, biofilms in an FBR are expected to have higher detachment rates considering the particle-to-particle contact and turbulence that occurs with fluidization of the media,48 and therefore there may be less retention of populations in biofilms in an FBR compared with biofilms in a fixed-bed system.
3.4 Nitrate utilization
Nitrate removal was impacted only briefly and to a lesser extent than perchlorate removal after the change in electron donor (Fig. 3A), suggesting that nitrate reduction was favored before perchlorate reduction. The preferential use of nitrate before perchlorate is unexpected from a thermodynamic perspective because (per)chlorate reduction yields slightly more energy than nitrate reduction at standard conditions (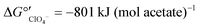 and
and 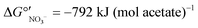 ).15,49 However, the preferential use of nitrate has been reported for some perchlorate-reducing cultures,28 including Azospira suillum JPLRND, which was used to inoculate the FBR, and several undefined or environmental communities.27,50–56 Indeed, nitrate inhibition of biological perchlorate reduction has been reported in several studies.28,51,54,57–59 Understanding the utilization of these competing electron acceptors is important to optimize bioreactor operation and predict perchlorate reduction rates.
).15,49 However, the preferential use of nitrate has been reported for some perchlorate-reducing cultures,28 including Azospira suillum JPLRND, which was used to inoculate the FBR, and several undefined or environmental communities.27,50–56 Indeed, nitrate inhibition of biological perchlorate reduction has been reported in several studies.28,51,54,57–59 Understanding the utilization of these competing electron acceptors is important to optimize bioreactor operation and predict perchlorate reduction rates.
A few possibilities may explain why nitrate reduction was favored over perchlorate reduction in the FBR. Firstly, a single Dechloromonas population (Dechloromonas_OTU0003) increased in relative abundance over time and became the most abundant population in the reactor by the end of the study (Fig. 5A). This population may have been involved in reducing nitrate as well as perchlorate. The mixed community inoculum originated from a nitrate- and arsenic-removing bioreactor,30 which was itself inoculated with biomass from a perchlorate- and nitrate-reducing bioreactor.20 This history of nitrate-reducing conditions may have increased the selective pressure for Dechloromonas populations capable of respiring both nitrate and perchlorate. Nitrate reduction has also been attributed to Dechloromonas in other mixed community studies. For example, in a bench-scale, two-stage membrane biofilm reactor system, Dechloromonas dominated the bacterial community in the first reactor where the majority of nitrate reduction occurred.60 If the dominating Dechloromonas (OTU0003) population in the FBR was responsible for the majority of nitrate and perchlorate reduction, it is unclear why this population would prefer nitrate reduction when the electron donor substrate was limited. While there are some examples of purified perchlorate reductase enzymes that reduce nitrate,61 many (per)chlorate-reducing bacteria that respire nitrate contain a separate nitrate reductase for nitrate reduction.62–65 It is therefore unlikely that nitrate competes with perchlorate for a single active binding site in perchlorate reductase, but without further experiments on the dominating Dechloromonas population in the FBR, it cannot be ruled out that the preferential use of nitrate before perchlorate observed was not related to competitive inhibition.
The presence of nitrate has been shown to significantly alter the community structure of perchlorate-reducing communities. In long-term, perchlorate-reducing enrichment cultures receiving acetate as the sole electron donor and seeded with marine sediment, Rhodobacteraceae were the prevailing populations based on shotgun metagenomics and 16S rRNA gene sequencing.66 When nitrate was added in addition to perchlorate, the abundance of Rhodocyclaceae increased and Rhodobacteraceae decreased.66Rhodocyclaceae were well represented in the high-perchlorate FBR, particularly during operation with acetate (Fig. 4), whereas Rhodobacteraceae were rare. More research is needed to understand if Rhodocyclaceae prefer nitrate over perchlorate. A selective pressure by the presence of nitrate could explain why nitrate removal was favored over perchlorate removal when acetate became limiting in low-perchlorate fixed-bed and recirculating bioreactors fed 1 mg L−1 ClO4− and 10–16 mg L−1 NO3−-N, even though reactor biofilms had been enriched under perchlorate-reducing conditions.51 The presence of nitrate, irrespective of inoculation strategy, may have selected for microbial populations in the FBR that prefer nitrate reduction and/or compete effectively against perchlorate-reducing bacteria for space in the biofilm.67
 | ||
| Fig. 4 Relative abundance of genera of the top 20 most abundant OTUs. Data were obtained for biomass collected from (A) the FBR and (B) an identical FBR and (C) a fixed-bed bioreactor fed 200 μg L−1 ClO4− as described previously.27 The low-perchlorate systems were fed much lower concentrations of perchlorate and were operated using acetate or a proprietary carbohydrate-based electron donor. Truncated y axes are shown to accentuate changes in abundance. | ||
3.5 Sulfate reduction
While the higher influent COD after day 142 helped to restore perchlorate removal, significant sulfate reduction (73 ± 9% sulfate removed) was observed during days 145–193. The highest relative abundance of the sulfate-reducing bacteria Desulfovibrio observed in the high-perchlorate FBR (8.0% on day 196) corresponded with a period when sulfate reduction was high (88.0% of influent sulfate was reduced on day 196). Lowering the influent COD on day 197 to approximately 442 mg L−1 helped minimize sulfate reduction and the effluent sulfate concentration increased from 1.94 ± 0.62 mg L−1 SO42− (days 145–193) to 2.77 ± 0.53 mg L−1 (days 200–210), but sulfate reduction was not completely eliminated. Lowering the influent COD did not affect perchlorate removal (i.e., effluent perchlorate concentrations averaged 0.26 ± 0.10 mg L−1 for days 200–210), and as the relative abundance of perchlorate-reducing Dechloromonas increased, so did the relative abundance of sulfate-reducing Desulfovibrio (Fig. 4). Similarly, in a membrane biofilm reactor, the greatest perchlorate removal was achieved when the greatest sulfate reduction occurred.50 When influent COD from the glycerol product was decreased on day 197 and sulfate reduction subsequently declined, perchlorate removal did not change dramatically. This is in contrast with observations in previous studies27,50 that reported a trade-off between perchlorate removal and the control of sulfate reduction. In the study by Ontiveros-Valencia and colleagues (2013), reducing the electron donor concentration in the influent fed to a membrane biofilm reactor not only helped control sulfate reduction but also resulted in slightly higher effluent perchlorate concentrations.50 They suggested that sulfate reducers compete with perchlorate reducers for space in the biofilm.503.6 Incomplete perchlorate reduction
Even with greater than 99.5 percent removal of perchlorate in the FBR, average effluent perchlorate concentrations during steady-state operation with acetate and the glycerol product were still well above the U.S. EPA's recommended preliminary remediation goal of 15 μg L−1 ClO4− for Superfund sites with a drinking water exposure pathway. Given the concentrations of sCOD in the effluent during the same time periods (Fig. 3), the incomplete removal of perchlorate in the FBR was probably not caused by electron donor substrate limitation. We speculate that even within the biofilm, the electron donor substrate concentration was not limiting perchlorate reduction as the biofilm in an FBR is typically thin due to increased detachment caused by particle-to-particle attrition and turbulence48 and thus did not promote concentration differences between the bulk liquid and the areas within the biofilm close to the substratum. Biofilm modeling could be used to evaluate possible substrate limitations but is beyond the scope of the current study. In addition, the relative abundance of Dechloromonas species increased during the reactor operating period and, by the end of the study, comprised greater than 30% of the bacterial community (Fig. 4 and 5), suggesting that incomplete perchlorate reduction was not caused by low abundance of perchlorate-reducing populations in the system. We recognize that the relative abundance of perchlorate-reducing bacteria based on sequencing of 16S rRNA genes may not provide an accurate representation of perchlorate-reducing activity. Quantifying the expression of functional genes related to perchlorate reduction (e.g., chlorite dismutase or perchlorate reductase68,69) or sequencing of 16S rRNA70 would have provided additional information on the activity of perchlorate-reducing species and their involvement in perchlorate removal.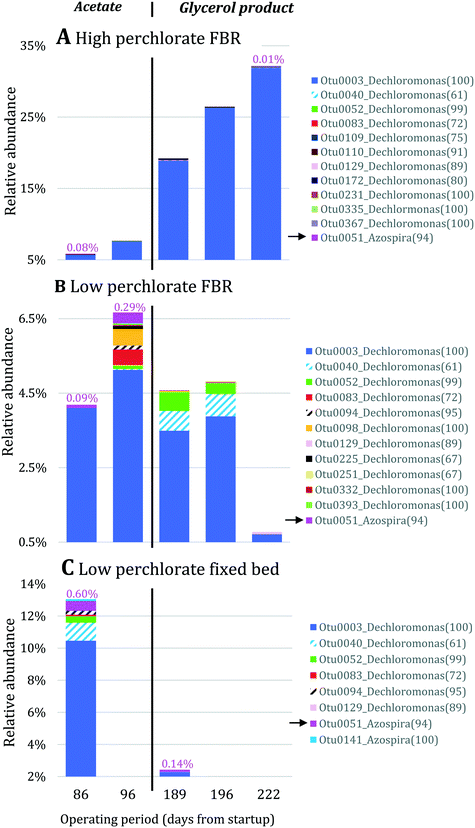 | ||
| Fig. 5 Relative abundance of OTUs classified as Dechloromonas and Azospira based on 16S rRNA gene sequencing. Singleton OTUs are not included. Data were obtained for biomass collected from (A) the FBR, (B) a low-perchlorate FBR and (C) a low-perchlorate fixed-bed reactor. The low-perchlorate systems treated much lower concentrations of perchlorate (approximately 200 μg L−1) and were operated using acetate or a proprietary carbohydrate-based electron donor (Fig. S1†).27 Arrows point to the putative Azospira inoculum seeded into the three reactors. Numbers above bars represent the percent relative abundance of the Azospira inoculum. Data were normalized by total number of 16S rRNA gene sequences. Samples from the FBRs were collected twice during reactor operation with acetate (days 86 and 96) and three times after converting acetate to the proprietary products (days 189, 196 and 222). Samples from the fixed-bed reactor were collected once before and once after converting to the carbohydrate-based product. | ||
One reason the FBR did not remove perchlorate to lower levels in the effluent may be explained by considering characteristics of steady-state biofilms. To sustain a steady-state biofilm, the bulk electron acceptor substrate concentration must be above a minimum concentration (Smin, eqn (1)), so that growth occurs rapidly enough to replace biofilm losses due to cell decay (bdecay is a decay rate, d−1) and detachment (bdetach is a detachment rate, d−1).71
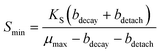 | (1) |
A perchlorate concentration below Smin is expected to lead to diminishing biofilm thickness and wash-out of perchlorate-reducing populations. Smin changes proportionally with the half-saturation constant (Ks), the concentration of electron acceptor substrate at which a population grows at half its maximum growth rate (μmax), and Ks varies inversely with a population's affinity for the electron acceptor substrate. Systems with low concentrations of perchlorate are expected to select for perchlorate-reducing populations with higher affinity (and lower Ks values) for perchlorate and/or populations that utilize perchlorate as a secondary growth substrate,72 whereas systems with high perchlorate concentrations would select for populations which use perchlorate as a primary substrate and have lower affinity for perchlorate.73 Interestingly, despite the two orders of magnitude difference in bulk perchlorate levels between the high and low perchlorate systems, the same Dechloromonas population (OTU0003) dominated the bacterial community in all three bioreactors (Fig. 5). While several more Dechloromonas populations were abundant in the FBR with low perchlorate (Fig. 4 and 5), OTU0003 Dechloromonas was particularly dominant in the high-perchlorate FBR and presumably outcompeted the other Dechloromonas populations in this system. It is possible that higher concentrations of perchlorate selected for a Dechloromonas population with a lower affinity and higher Ks, thereby driving up the lowest perchlorate concentration attainable in the high-perchlorate FBR.
Detachment rate (bdetach) also may have limited the lowest perchlorate concentration achievable in the FBR. As discussed above, biofilms in an FBR are expected to have higher detachment rates than biofilms in fixed-bed systems considering the particle-to-particle contact and turbulence that occurs with fluidization of the media.48 A higher detachment rate would result in higher effluent perchlorate concentrations (eqn (1)). In agreement with this, effluent perchlorate concentrations in the low-perchlorate FBR were higher (approximately 6 μg L−1 ClO4− for the first 100 days of reactor operation) than effluent concentrations in the fixed-bed reactor (less than 3 μg L−1 ClO4−) treating the same influent for the same time period.27
In a fixed-bed system, the influent moves through the bed in a plug-flow manner and concentration gradients occur along the depth of the bed.47 Changes in electron acceptor concentrations along the depth of the fixed bed may allow greater spatial separation and selection for perchlorate-reducing bacterial populations with a wider range of affinities (Ks) for perchlorate. Consistent with this, a comparison of Dechloromonas populations in the fixed-bed system and in the low-perchlorate FBR (Fig. 5C) for the first biomass sampling event (day 86) revealed that several more Dechloromonas populations were present in the fixed-bed system. It is therefore possible that lower effluent perchlorate concentrations would have been achieved if a fixed-bed system would have been used to treat synthetic groundwater with the high perchlorate concentration. In practice, the mixing of microorganisms that occurs during backwashing reduces the spatial separation achieved during operation of a fixed-bed system.74 Therefore, the use of a two-stage system with two bioreactors in series30 may be a more promising approach as different microbial communities are expected to be selected for in the “lead” and “lag” reactors.47,60,75
4. Conclusions and future directions
This study demonstrated the feasibility of replacing acetate with MicroC2000™, a proprietary glycerol-based electron donor, for biological reduction of 100 mg L−1 perchlorate from groundwater. Electron donor choice by the water treatment industry is determined primarily by cost and operational requirements. The need for higher COD doses and more frequent wasting of biomass with MicroC2000™ compared with acetate is consistent with our results of an earlier study testing MicroC4000™ for biological treatment of lower concentrations of perchlorate.27 These observations with MicroC products require follow-up investigation through pilot and full-scale applications. More feasibility studies of diverse electron donors will provide the water treatment industry with greater flexibility for electron donor choice. A recent lifecycle assessment reported that the environmental impacts associated with the production of acetate contributed 87–98% of the overall environmental impacts of biological perchlorate reduction.76 Future research investigating environmental impacts and life cycle costs associated with electron donors used for perchlorate reduction would provide additional information to guide electron donor choice. However, using a product with undisclosed composition makes it difficult to fully assess the product's environmental impacts. As several full-scale wastewater treatment plants are already using MicroC2000™ for denitrification, it appears that the use of MicroC2000™ has been accepted by the water treatment industry in spite of its undisclosed composition. As treated wastewater is increasingly being considered for water reuse applications, the use of proprietary products without full disclosure may need to be reconsidered.Effluent perchlorate concentrations did not satisfy the U.S. EPA's recommended preliminary remediation goal for Superfund sites. We hypothesize that the incomplete perchlorate removal observed was partly because high perchlorate loadings enriched for a single Dechloromonas population with low affinity for perchlorate. Utilizing two FBRs in series may achieve better effluent quality by supporting the growth of perchlorate-reducing populations with higher affinity for perchlorate in the second reactor. However, this two-stage approach would increase the physical and energy footprints of treatment. More research on community structure along concentration gradients that develop in fixed-bed systems, and the effects of routine backwashing on the positioning of microbial populations in fixed-bed biofilms, is needed to evaluate whether a single fixed-bed bioreactor can be competitive with a two-stage FBR in terms of effluent quality and operational convenience.
Acknowledgements
This work was partially funded by Environmental Operating Solutions, Inc (EOSi). While EOSi was a partner in this research, University of Michigan researchers executed the work, analyzed and interpreted all results and wrote the manuscript. NK was partially supported by a fellowship from the Susan E. Stutz-McDonald Scholarship Foundation. The authors would like to thank Ashley Hammerbeck and Xinsheng Chu for their help with reactor maintenance and chemical analyses, Paul Hatzinger and Shaw Environmental for providing the Azospira suillum JPLRND culture, and Sam Ledwell for helpful discussions.References
- J. D. Coates and L. A. Achenbach, Microbial perchlorate reduction: rocket-fuelled metabolism, Nat. Rev. Microbiol., 2004, 2, 569–580 CrossRef CAS PubMed.
- C. W. Trumpolt, M. Crain, G. D. Cullison, S. J. Flanagan, L. Siegel and S. Lathrop, Perchlorate: sources, uses, and occurrences in the environment, Remediation, 2005, 16, 65–89 CrossRef.
- E. T. Urbansky, Perchlorate chemistry: implications for analysis and remediation, Biorem. J., 1998, 2, 81–95 CrossRef CAS.
- United States Government Accountability Office Testimony Before the Subcommittee on Environmental and Hazardous Materials, House Committee on Energy and Commerce, Perchlorate; EPA Does Not Systematically Track Incidents of Contamination, 2007.
- K. Kannan, M. L. Praamsma, J. F. Oldi, T. Kunisue and R. K. Sinha, Occurrence of perchlorate in drinking water, groundwater, surface water and human saliva from India, Chemosphere, 2009, 76, 22–26 CrossRef CAS PubMed.
- K. Kosaka, M. Asami, Y. Matsuoka, M. Kamoshita and S. Kunikane, Occurrence of perchlorate in drinking water sources of metropolitan area in Japan, Water Res., 2007, 41, 3474–3482 CrossRef CAS PubMed.
- O. Quiñones, J. E. Oh, B. Vanderford, J. H. Kim, J. Cho and S. A. Snyder, Perchlorate assessment of the Nakdong and Yeongsan watersheds, Republic of Korea, Environ. Toxicol. Chem., 2007, 26, 1349–1354 CrossRef.
- Q. Wu, T. Zhang, H. Sun and K. Kannan, Perchlorate in tap water, groundwater, surface waters, and bottled water from China and its association with other inorganic anions and with disinfection byproducts, Arch. Environ. Contam. Toxicol., 2010, 58, 543–550 CrossRef CAS PubMed.
- R. J. Brechner, G. D. Parkhurst, W. O. Humble, M. B. Brown and W. H. Herman, Ammonium perchlorate contamination of Colorado River drinking water is associated with abnormal thyroid function in newborns in Arizona, J. Occup. Environ. Med., 2000, 42, 777–782 CrossRef CAS.
- B. Gu and J. D. Coates, Perchlorate: environmental occurrence, interactions and treatment, Springer Science & Business Media, 2006 Search PubMed.
- W. Mendez, E. Dederick and J. Cohen, Drinking water contribution to aggregate perchlorate intake of reproductive-age women in the United States estimated by dietary intake simulation and analysis of urinary excretion data, J. Exposure Sci. Environ. Epidemiol., 2010, 20, 288–297 CrossRef CAS PubMed.
- W. A. Jackson, P. Joseph, P. Laxman, K. Tan, P. N. Smith, L. Yu and T. A. Anderson, Perchlorate accumulation in forage and edible vegetation, J. Agric. Food Chem., 2005, 53, 369–373 CrossRef CAS PubMed.
- F. K. Lau, B. Rey deCastro, L. Mills-Herring, L. Tao, L. Valentin-Blasini, K. U. Alwis and B. C. Blount, Urinary perchlorate as a measure of dietary and drinking water exposure in a representative sample of the United States population 2001–2008, J. Exposure Sci. Environ. Epidemiol., 2013, 23, 207–214 CrossRef CAS PubMed.
- P. B. Hatzinger, Perchlorate biodegradation for water treatment, Environ. Sci. Technol., 2005, 39, 239A–247A CrossRef CAS PubMed.
- G. Rikken, A. Kroon and C. Van Ginkel, Transformation of (per) chlorate into chloride by a newly isolated bacterium: reduction and dismutation, Appl. Microbiol. Biotechnol., 1996, 45, 420–426 CrossRef CAS.
- J. C. Brown, V. L. Snoeyink, L. Raskin and R. Lin, The sensitivity of fixed-bed biological perchlorate removal to changes in operating conditions and water quality characteristics, Water Res., 2003, 37, 206–214 CrossRef CAS PubMed.
- B. E. Logan, Peer reviewed: assessing the outlook for perchlorate remediation, Environ. Sci. Technol., 2001, 35, 482A–487A CrossRef CAS PubMed.
- L. Ye, H. You, J. Yao and H. Su, Water treatment technologies for perchlorate: a review, Desalination, 2012, 298, 1–12 CrossRef CAS.
- J. Brown, R. S. Summers, M. LeChevallier, H. Collins, J. A. Roberson, S. Hubbs and E. Dickenson, Biological Drinking Water Treatment? Naturally, J. - Am. Water Works Assoc., 2015, 107, 20–30 CrossRef.
- X. Li, G. Upadhyaya, W. Yuen, J. Brown, E. Morgenroth and L. Raskin, Changes in the structure and function of microbial communities in drinking water treatment bioreactors upon addition of phosphorus, Appl. Environ. Microbiol., 2010, 76, 7473–7481 CrossRef CAS PubMed.
- J. C. Brown, R. D. Anderson, J. H. Min, L. Boulos, D. Prasifka and G. J. Juby, Fixed-bed biological treatment of perchlorate-contaminated drinking water, J. - Am. Water Works Assoc., 2005, 97, 70–81 CAS.
- J. Xu, Y. Song, B. Min, L. Steinberg and B. E. Logan, Microbial degradation of perchlorate: principles and applications, Environ. Eng. Sci., 2003, 20, 405–422 CrossRef CAS.
- A. Grabińska-Ńoniewska, T. Słomczyński and Z. Kańska, Denitrification studies with glycerol as a carbon source, Water Res., 1985, 19, 1471–1477 CrossRef.
- J. C. Akunna, C. Bizeau and R. Moletta, Nitrate and nitrite reductions with anaerobic sludge using various carbon sources: glucose, glycerol, acetic acid, lactic acid and methanol, Water Res., 1993, 27, 1303–1312 CrossRef.
- K. Uprety, Evaluation of glycerol and waste alcohol as supplemental carbon sources for denitrification: Virginia Tech, 2013 Search PubMed.
- K. Selock, W. Burton, C. Bott, N. Cutting, B. Wimmer, J. Neethling, S. Amad, J. Hinojosa and S. Murthy, Glycerin 101-Lessons Learned While Pilot Testing Glycerin to Enhance Denitrification, Water Environment Federation, 2008 Search PubMed.
- G. Upadhyaya, N. Kotlarz, P. Togna and L. Raskin, Carbohydrate-Based Electron Donor for Biological Nitrate and Perchlorate Removal From Drinking Water, J. - Am. Water Works Assoc., 2015, 107, 12 CrossRef.
- S. K. Chaudhuri, S. M. O'Connor, R. L. Gustavson, L. A. Achenbach and J. D. Coates, Environmental factors that control microbial perchlorate reduction, Appl. Environ. Microbiol., 2002, 68, 4425–4430 CrossRef CAS PubMed.
- N. C. Sturchio, P. B. Hatzinger, M. D. Arkins, C. Suh and L. J. Heraty, Chlorine isotope fractionation during microbial reduction of perchlorate, Environ. Sci. Technol., 2003, 37, 3859–3863 CrossRef CAS PubMed.
- G. Upadhyaya, J. Jackson, T. M. Clancy, S. P. Hyun, J. Brown, K. F. Hayes and L. Raskin, Simultaneous removal of nitrate and arsenic from drinking water sources utilizing a fixed-bed bioreactor system, Water Res., 2010, 44, 4958–4969 CrossRef CAS PubMed.
- B. E. Logan, Microbial fuel cells, John Wiley & Sons, 2008 Search PubMed.
- H. Urakawa, W. Martens-Habbena and D. A. Stahl, High abundance of ammonia-oxidizing Archaea in coastal waters, determined using a modified DNA extraction method, Appl. Environ. Microbiol., 2010, 76, 2129–2135 CrossRef CAS PubMed.
- T.-H. Chiao, T. M. Clancy, A. Pinto, C. Xi and L. Raskin, Differential resistance of drinking water bacterial populations to monochloramine disinfection, Environ. Sci. Technol., 2014, 48, 4038–4047 CrossRef CAS PubMed.
- P. D. Schloss, S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn and C. F. Weber, Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities, Appl. Environ. Microbiol., 2009, 75, 7537 CrossRef CAS PubMed.
- C. Quince, A. Lanzen, R. J. Davenport and P. J. Turnbaugh, Removing noise from pyrosequenced amplicons, BMC Bioinf., 2011, 12, 1 CrossRef PubMed.
- R. C. Edgar, B. J. Haas, J. C. Clemente, C. Quince and R. Knight, UCHIME improves sensitivity and speed of chimera detection, Bioinformatics, 2011, 27, 2194–2200 CrossRef CAS PubMed.
- J. D. Shrout and G. F. Parkin, Influence of electron donor, oxygen, and redox potential on bacterial perchlorate degradation, Water Res., 2006, 40, 1191–1199 CrossRef CAS PubMed.
- K. A. Bill, C. B. Bott, P. H. Yi, C. Ziobro and S. N. Murthy, Evaluation of alternative electron donors in anoxic moving bed biofilm reactors (MBBRs) configured for post-denitrification, Water Environment Federation, 2008 Search PubMed.
- A. Al-Omari, S. Murthy, I. Takacs, Y. Mokhayeri, R. Riffat and I. Nopens, Modeling the use of external carbon substrate for denitrification by generalists and specialists, Water Environment Federation, 2011 Search PubMed.
- M. D. Youngblut, O. Wang, T. P. Barnum and J. D. Coates, (Per) chlorate in Biology on Earth and Beyond, Annu. Rev. Microbiol., 2016, 70, 435–457 CrossRef CAS PubMed.
- R. Nerenberg, Y. Kawagoshi and B. E. Rittmann, Microbial ecology of a perchlorate-reducing, hydrogen-based membrane biofilm reactor, Water Res., 2008, 42, 1151–1159 CrossRef CAS PubMed.
- Y. Zhu, N. Gao, W. Chu, S. Wang and J. Xu, Bacterial reduction of highly concentrated perchlorate: Kinetics and influence of co-existing electron acceptors, temperature, pH and electron donors, Chemosphere, 2016, 148, 188–194 CrossRef CAS PubMed.
- S. J. Nor, S. H. Lee, K.-S. Cho, D. K. Cha, K. I. Lee and H. W. Ryu, Microbial treatment of high-strength perchlorate wastewater, Bioresour. Technol., 2011, 102, 835–841 CrossRef CAS PubMed.
- V. M. Markowitz, I.-M. A. Chen, K. Palaniappan, K. Chu, E. Szeto, Y. Grechkin, A. Ratner, B. Jacob, J. Huang and P. Williams, IMG: the integrated microbial genomes database and comparative analysis system, Nucleic Acids Res., 2012, 40, D115–D122 CrossRef CAS PubMed.
- SERDP Project CU-1163 Final Report: In Situ Bioremediation of Perchlorate, Envirogen, Inc., 2002 Search PubMed.
- C.-H. Xie and A. Yokota, Zoogloea oryzae sp. nov., a nitrogen-fixing bacterium isolated from rice paddy soil, and reclassification of the strain ATCC 19623 as Crabtreella saccharophila gen. nov., sp. nov., Int. J. Syst. Evol. Microbiol., 2006, 56, 619–624 CrossRef CAS PubMed.
- G. Upadhyaya, T. M. Clancy, J. Brown, K. F. Hayes and L. Raskin, Optimization of arsenic removal water treatment system through characterization of terminal electron accepting processes, Environ. Sci. Technol., 2012, 46, 11702–11709 CrossRef CAS PubMed.
- H. T. Chang, B. E. Rittmann, D. Amar, R. Heim, O. Ehlinger and Y. Lesty, Biofilm detachment mechanisms in a liquid-fluidized bed, Biotechnol. Bioeng., 1991, 38, 499–506 CrossRef CAS PubMed.
- N. Bardiya and J.-H. Bae, Dissimilatory perchlorate reduction: a review, Microbiol. Res., 2011, 166, 237–254 CrossRef CAS PubMed.
- A. Ontiveros-Valencia, Y. Tang, R. Krajmalnik-Brown and B. E. Rittmann, Perchlorate reduction from a highly contaminated groundwater in the presence of sulfate-reducing bacteria in a hydrogen-fed biofilm, Biotechnol. Bioeng., 2013, 110, 3139–3147 CrossRef CAS PubMed.
- H. Choi and J. Silverstein, Inhibition of perchlorate reduction by nitrate in a fixed biofilm reactor, J. Hazard. Mater., 2008, 159, 440–445 CrossRef CAS PubMed.
- M. Nozawa-Inoue, M. Jien, K. Yang, D. E. Rolston, K. R. Hristova and K. M. Scow, Effect of nitrate, acetate, and hydrogen on native perchlorate-reducing microbial communities and their activity in vadose soil, FEMS Microbiol. Ecol., 2011, 76, 278–288 CrossRef CAS PubMed.
- R. Nerenberg, B. E. Rittmann and I. Najm, Perchlorate reduction in a hydrogen-based membrane-biofilm reactor, J. - Am. Water Works Assoc., 2002, 94, 103 CAS.
- D. Xie, H. Yu, C. Li, Y. Ren, C. Wei and C. Feng, Competitive microbial reduction of perchlorate and nitrate with a cathode directly serving as the electron donor, Electrochim. Acta, 2014, 133, 217–223 CrossRef CAS.
- M. C. Ziv-El and B. E. Rittmann, Systematic evaluation of nitrate and perchlorate bioreduction kinetics in groundwater using a hydrogen-based membrane biofilm reactor, Water Res., 2009, 43, 173–181 CrossRef CAS PubMed.
- H. P. Zhao, S. Van Ginkel, Y. Tang, D.-W. Kang, B. Rittmann and R. Krajmalnik-Brown, Interactions between perchlorate and nitrate reductions in the biofilm of a hydrogen-based membrane biofilm reactor, Environ. Sci. Technol., 2011, 45, 10155–10162 CrossRef CAS PubMed.
- D. C. Herman and W. T. Frankenberger, Bacterial reduction of perchlorate and nitrate in water, J. Environ. Qual., 1999, 28, 1018–1024 CrossRef CAS.
- A. R. Ricardo, G. Carvalho, S. Velizarov, J. G. Crespo and M. A. Reis, Kinetics of nitrate and perchlorate removal and biofilm stratification in an ion exchange membrane bioreactor, Water Res., 2012, 46, 4556–4568 CrossRef CAS PubMed.
- R. Wang, L. Chen, F. Liu, H. H. Chen, J. W. Zhang and M. Chen, Inhibition of nitrate and the accumulated denitrification intermediate (nitrite) on perchlorate bioreduction, Water Qual. Res. J. Can., 2014, 49, 346–353 CrossRef CAS.
- H. P. Zhao, A. Ontiveros-Valencia, Y. Tang, B. O. Kim, Z. E. Ilhan, R. Krajmalnik-Brown and B. Rittmann, Using a two-stage hydrogen-based membrane biofilm reactor (MBfR) to achieve complete perchlorate reduction in the presence of nitrate and sulfate, Environ. Sci. Technol., 2013, 47, 1565–1572 CAS.
- S. W. Kengen, G. B. Rikken, W. R. Hagen, C. G. Van Ginkel and A. J. Stams, Purification and characterization of (per) chlorate reductase from the chlorate-respiring strain GR-1, J. Bacteriol., 1999, 181, 6706–6711 CAS.
- Å. Malmqvist, T. Welander, E. Moore, A. Ternström, G. Molin and I.-M. Stenström, Ideonella dechloratans gen. nov., sp. nov., a new bacterium capable of growing anaerobically with chlorate as an electron acceptor, Syst. Appl. Microbiol., 1994, 17, 58–64 CrossRef.
- A. Wolterink, S. Kim, M. Muusse, I. S. Kim, P. J. Roholl, C. G. van Ginkel, A. J. Stams and S. W. Kengen, Dechloromonas hortensis sp. nov. and strain ASK-1, two novel (per) chlorate-reducing bacteria, and taxonomic description of strain GR-1, Int. J. Syst. Evol. Microbiol., 2005, 55, 2063–2068 CrossRef CAS PubMed.
- A. F. Wolterink, E. Schiltz, P.-L. Hagedoorn, W. R. Hagen, S. W. Kengen and A. J. Stams, Characterization of the chlorate reductase from Pseudomonas chloritidismutans, J. Bacteriol., 2003, 185, 3210–3213 CrossRef CAS PubMed.
- J. Xu, J. J. Trimble, L. Steinberg and B. E. Logan, Chlorate and nitrate reduction pathways are separately induced in the perchlorate-respiring bacterium Dechlorosoma sp. KJ and the chlorate-respiring bacterium Pseudomonas sp. PDA, Water Res., 2004, 38, 673–680 CrossRef CAS PubMed.
- V. G. Stepanov, Y. Xiao, Q. Tran, M. Rojas, R. C. Willson, Y. Fofanov, G. E. Fox and D. J. Roberts, The presence of nitrate dramatically changed the predominant microbial community in perchlorate degrading cultures under saline conditions, BMC Microbiol., 2014, 14, 1 CrossRef PubMed.
- Y. Tang, H. Zhao, A. K. Marcus, R. Krajmalnik-Brown and B. E. Rittmann, A steady-state biofilm model for simultaneous reduction of nitrate and perchlorate, part 2: parameter optimization and results and discussion, Environ. Sci. Technol., 2012, 46, 1608–1615 CrossRef CAS PubMed.
- S. K. De Long, K. A. Kinney and M. J. Kirisits, qPCR assays to quantify genes and gene expression associated with microbial perchlorate reduction, J. Microbiol. Methods, 2010, 83, 270–274 CrossRef CAS PubMed.
- M. Nozawa-Inoue, M. Jien, N. S. Hamilton, V. Stewart, K. M. Scow and K. R. Hristova, Quantitative detection of perchlorate-reducing bacteria by real-time PCR targeting the perchlorate reductase gene, Appl. Environ. Microbiol., 2008, 74, 1941–1944 CrossRef CAS PubMed.
- A. L. Smith, S. J. Skerlos and L. Raskin, Membrane biofilm development improves COD removal in anaerobic membrane bioreactor wastewater treatment, Microb. Biotechnol., 2015, 8, 883–894 CrossRef CAS PubMed.
- C. L. Grady Jr, G. T. Daigger, N. G. Love and C. D. Filipe, Biological wastewater treatment, CRC press, 2011 Search PubMed.
- R. Nerenberg and B. E. Rittmann, Perchlorate as a secondary substrate in a denitrifying, hollow-fiber membrane biofilm reactor, Water Sci. Technol.: Water Supply, 2002, 2, 259–265 CAS.
- B. E. Rittmann and P. L. McCarty, Environmental biotechnology: principles and applications, McGraw Hill, New York, 2001 Search PubMed.
- X. Li, W. Yuen, E. Morgenroth and L. Raskin, Backwash intensity and frequency impact the microbial community structure and function in a fixed-bed biofilm reactor, Appl. Microbiol. Biotechnol., 2012, 96, 815–827 CrossRef CAS PubMed.
- H. P. Zhao, A. Ontiveros-Valencia, Y. Tang, B. O. Kim, S. VanGinkel, D. Friese, R. Overstreet, J. Smith, P. Evans and R. Krajmalnik-Brown, Removal of multiple electron acceptors by pilot-scale, two-stage membrane biofilm reactors, Water Res., 2014, 54, 115–122 CrossRef CAS PubMed.
- J. K. Choe, M. H. Mehnert, J. S. Guest, T. J. Strathmann and C. J. Werth, Comparative assessment of the environmental sustainability of existing and emerging perchlorate treatment technologies for drinking water, Environ. Sci. Technol., 2013, 47, 4644–4652 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ew00181e |
| ‡ Current affiliation: Envirogen Technologies Inc., Kingwood, TX, USA. |
| This journal is © The Royal Society of Chemistry 2016 |