Reduction of MS2 bacteriophage and rotavirus in biosand filters†
Hanting
Wang
,
Mingming
Li
,
Kazami
Brockman
and
Thanh H.
Nguyen
*
Department of Civil and Environmental Engineering, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA. E-mail: thn@illinois.edu
First published on 17th March 2016
Abstract
Diarrheal diseases caused by pathogens remain a significant cause of death in developing areas, especially for children under the age of five. Biosand filters (BSFs) are a promising technology implemented worldwide that can effectively reduce levels of bacteria and bacteriophages. However, besides echovirus, the efficacy of enteric virus reduction in BSFs has not been studied. Furthermore, how divalent cation concentrations in the source water used in BSFs influences virus reduction is not clearly understood. In this study, three bench-scale BSFs were fed daily with groundwater containing divalent cations or cation-free buffered solution to determine MS2 or rotavirus reduction as a function of filter depth, residence time, media ripening, and water source. An integrated cell culture and RT-qPCR assay was developed to quantify rotavirus reduction in water samples collected from the filters. Rotavirus reduction obtained by experiments performed in groundwater increased with depth and reached a cumulative average of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 (99.999%) reduction after 31 days. Experiments with 1 mM NaHCO3 spiked with MS2 averaged 1.2
log10 (99.999%) reduction after 31 days. Experiments with 1 mM NaHCO3 spiked with MS2 averaged 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction after 42 days, and there was not an increasing trend of reduction as a function of depth. Finally, MS2 experiments performed in groundwater reached a cumulative average of 5.36
log10 reduction after 42 days, and there was not an increasing trend of reduction as a function of depth. Finally, MS2 experiments performed in groundwater reached a cumulative average of 5.36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction by a BSF that had been in operation for 684 days, but the reduction also did not increase as a function of depth. Overall conclusions include that 1) at the same filter age and using the same water source, rotavirus reduction was higher than what was previously seen with MS2, indicating that MS2 is a conservative surrogate for rotavirus, 2) MS2 reduction efficacy was related to the divalent cation concentrations of the influent water for an unripened filter, and 3) residence time was crucial for increasing virus reduction in all experiments. This is the first study to determine the efficiency of rotavirus reduction in BSFs, which is an essential first step in understanding the extent to which BSFs can reduce human enteric viruses, and hence decrease diarrheal disease incidence.
log10 reduction by a BSF that had been in operation for 684 days, but the reduction also did not increase as a function of depth. Overall conclusions include that 1) at the same filter age and using the same water source, rotavirus reduction was higher than what was previously seen with MS2, indicating that MS2 is a conservative surrogate for rotavirus, 2) MS2 reduction efficacy was related to the divalent cation concentrations of the influent water for an unripened filter, and 3) residence time was crucial for increasing virus reduction in all experiments. This is the first study to determine the efficiency of rotavirus reduction in BSFs, which is an essential first step in understanding the extent to which BSFs can reduce human enteric viruses, and hence decrease diarrheal disease incidence.
Water impactPoor drinking water quality remains an important cause of diarrheal diseases in developing areas, especially for children under the age of five. Biosand filters (BSFs) are a promising technology to reduce the level of pathogens in drinking water. However, there is a lack in understanding of how BSFs work and what factors control BSF efficacy. In this study, we showed, for the first time, that divalent cation concentrations can influence MS2 bacteriophage reduction by BSF and that BSF can effectively reduce 99.99% of rotavirus, a common diarrhea-causing enteric virus in water. The results presented here contribute to the formulation of recommendations on the use of BSFs to reduce diarrheal disease incidence in developing countries. |
1. Introduction
It is estimated that 663 million people worldwide still lack access to improved drinking water sources.1 There are 4 billion cases of diarrheal diseases worldwide annually caused by consumption and use of unimproved drinking water sources and sanitation facilities.2 Diarrheal diseases are a leading cause of child morbidity and mortality in impoverished areas, and this remains a significant public health issue.3 Rotavirus is the leading cause of diarrhea in the world for children under the age of 5, which led to 450![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in 2008.4 It has been shown that household water treatment technologies can help to lower diarrheal disease incidences by 30–40%.5,6 One of the most promising point-of-use technologies currently available is the Centre for Affordable Water and Sanitation Technology (CAWST) V10 concrete biosand filter (BSF), an intermittently-run slow sand filter, which can produce 20–40 L of safe drinking water per day.6 The filter is easy to implement, use, and maintain, and is relatively cheap ($12–50 per filter).7 Twelve liters of source water is poured into the top of the filter and flows through the filter with gravity. The filter is designed and operated so that the 12 L of water sits within the pore spaces for a residence time period of 16–32 hours,8 which is within the 1–48 hour range recommended by CAWST.9 It is possible that small quantities of the 12 L could stay in the filter longer due to mixing within the standing head and storage within the standing head, gravel underdrain, and outlet tube, as shown in a previous study.10 Another 12 L of water is poured into the filter and pushes the filtered water out of an outlet tube into a storage container.
000 deaths in 2008.4 It has been shown that household water treatment technologies can help to lower diarrheal disease incidences by 30–40%.5,6 One of the most promising point-of-use technologies currently available is the Centre for Affordable Water and Sanitation Technology (CAWST) V10 concrete biosand filter (BSF), an intermittently-run slow sand filter, which can produce 20–40 L of safe drinking water per day.6 The filter is easy to implement, use, and maintain, and is relatively cheap ($12–50 per filter).7 Twelve liters of source water is poured into the top of the filter and flows through the filter with gravity. The filter is designed and operated so that the 12 L of water sits within the pore spaces for a residence time period of 16–32 hours,8 which is within the 1–48 hour range recommended by CAWST.9 It is possible that small quantities of the 12 L could stay in the filter longer due to mixing within the standing head and storage within the standing head, gravel underdrain, and outlet tube, as shown in a previous study.10 Another 12 L of water is poured into the filter and pushes the filtered water out of an outlet tube into a storage container.
BSFs have been shown to reduce turbidity levels to under 2 NTU and bacteria by >4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10.11,12 In addition, BSFs that were in operation for 240–300 days showed 4
log10.11,12 In addition, BSFs that were in operation for 240–300 days showed 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction of MS2, a surrogate for enteric viruses.8,13 Even after only 4 weeks of BSF experiments using groundwater, MS2 reduction reached 4
log10 reduction of MS2, a surrogate for enteric viruses.8,13 Even after only 4 weeks of BSF experiments using groundwater, MS2 reduction reached 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10, as long as the sand media depth was at least 44 cm and there was 15.5–31 hours of residence time for the water in the filter.8 This finding contrasts with other studies using surface waters that observed low reduction of less than 1
log10, as long as the sand media depth was at least 44 cm and there was 15.5–31 hours of residence time for the water in the filter.8 This finding contrasts with other studies using surface waters that observed low reduction of less than 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 after four weeks, even with a 55 cm sand depth and 24 hours of residence time14 and a 60 cm sand depth and an average of 15.6 hours of residence time.12 The reasons for the wide range of MS2 reduction in previous studies8,11–15 are not clear and need to be elucidated to better understand BSF operation and efficacy. Some hypotheses for these differences have been suggested previously. The sand source that was used for the two long-term studies8,13 was tested for metal oxides (Al, Fe, Mg, Ca, and Zn), which could enhance MS2 adsorption due to positively charged sand surfaces. Using inductively coupled plasma-optical emission spectrometry (ICP-OES) analysis and concentrated hydrofluoric acid solution to digest the sand media, only low concentrations of metal oxides were detected,8 suggesting that it was not the sand source that caused high MS2 reduction in these studies. Another recent study16 compared MS2 reduction using one water source and two different types of sand (Accusand and granite), both of which had between 1.4–519 times higher concentrations of metal oxides than what was used in the two long-term studies. The granite had 136–423 times higher concentrations of metal oxides than the Accusand, and there was significantly higher MS2 reduction (p < 0.01) shown for the granite (0.62
log10 after four weeks, even with a 55 cm sand depth and 24 hours of residence time14 and a 60 cm sand depth and an average of 15.6 hours of residence time.12 The reasons for the wide range of MS2 reduction in previous studies8,11–15 are not clear and need to be elucidated to better understand BSF operation and efficacy. Some hypotheses for these differences have been suggested previously. The sand source that was used for the two long-term studies8,13 was tested for metal oxides (Al, Fe, Mg, Ca, and Zn), which could enhance MS2 adsorption due to positively charged sand surfaces. Using inductively coupled plasma-optical emission spectrometry (ICP-OES) analysis and concentrated hydrofluoric acid solution to digest the sand media, only low concentrations of metal oxides were detected,8 suggesting that it was not the sand source that caused high MS2 reduction in these studies. Another recent study16 compared MS2 reduction using one water source and two different types of sand (Accusand and granite), both of which had between 1.4–519 times higher concentrations of metal oxides than what was used in the two long-term studies. The granite had 136–423 times higher concentrations of metal oxides than the Accusand, and there was significantly higher MS2 reduction (p < 0.01) shown for the granite (0.62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10) than the Accusand (0.37
log10) than the Accusand (0.37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10). The difference in MS2 reduction levels indicates that high concentrations of metal oxides play a role in increasing MS2 reduction. It has also been hypothesized that sustained filter ripening occurring in long-term BSFs8,13 could have continued to improve MS2 reduction compared to the more typical duration of laboratory BSF experiments of 6–10 weeks.16 However, one of these long-term BSF studies showed that MS2 reduction can meet 4
log10). The difference in MS2 reduction levels indicates that high concentrations of metal oxides play a role in increasing MS2 reduction. It has also been hypothesized that sustained filter ripening occurring in long-term BSFs8,13 could have continued to improve MS2 reduction compared to the more typical duration of laboratory BSF experiments of 6–10 weeks.16 However, one of these long-term BSF studies showed that MS2 reduction can meet 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction in 4 weeks,8 which indicates that other factors, such as microbial communities that develop in the BSF, affect MS2 reduction as well. Because granular filtration also depends on interactions between the particles and the filter media,17 the variability in influent water chemistry used in different studies may affect MS2 reduction. Although most previous BSF studies included some water chemistry data, they did not consider water chemistry as a factor affecting MS2 reduction. Two studies using Newmark groundwater, which has high divalent cation concentrations (0.6–1.5 mM Ca2+ and 1.0–2.3 mM Mg2+) saw MS2 reduction reaching 4
log10 reduction in 4 weeks,8 which indicates that other factors, such as microbial communities that develop in the BSF, affect MS2 reduction as well. Because granular filtration also depends on interactions between the particles and the filter media,17 the variability in influent water chemistry used in different studies may affect MS2 reduction. Although most previous BSF studies included some water chemistry data, they did not consider water chemistry as a factor affecting MS2 reduction. Two studies using Newmark groundwater, which has high divalent cation concentrations (0.6–1.5 mM Ca2+ and 1.0–2.3 mM Mg2+) saw MS2 reduction reaching 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10.8,13 Viruses are uniquely mobile in water, and previous studies have shown that the interactions between different viruses and silica surfaces depend on water chemistry. MS2, ΦX174, and human adenovirus did not significantly adsorb to sand particles when only monovalent cations were present,18,19 but the presence of Ca2+ enhanced adsorption of rotavirus, adenovirus, and poliovirus to silica surfaces.19,20 High divalent cation concentrations have also been shown to enhance MS2 adhesion to organic matter coated sand surfaces.21,22
log10.8,13 Viruses are uniquely mobile in water, and previous studies have shown that the interactions between different viruses and silica surfaces depend on water chemistry. MS2, ΦX174, and human adenovirus did not significantly adsorb to sand particles when only monovalent cations were present,18,19 but the presence of Ca2+ enhanced adsorption of rotavirus, adenovirus, and poliovirus to silica surfaces.19,20 High divalent cation concentrations have also been shown to enhance MS2 adhesion to organic matter coated sand surfaces.21,22
While divalent cation concentrations have been shown to influence virus adsorption to sand, they have not been studied as a factor impacting virus reduction in BSFs, especially for unripened filters where microbial communities have not been developed inside the filter. Previous studies showed that microbial communities influence virus reduction, especially for ripened filters.8,13,15 However, virus reduction levels are low for unripened filters, and this study determined the effect of divalent cation concentrations on virus reduction in unripened filters. Another main limitation in current BSF studies is that besides echovirus, no other enteric viruses have been studied in BSFs, which is a significant constraint to fully understanding BSF efficacy in environmental conditions. This study addressed these two limitations by evaluating the following as a function of filter depth, media aging, and residence time: rotavirus reduction in Newmark groundwater for an unripened filter, MS2 reduction in divalent cation-free water (1 mM NaHCO3) for an unripened filter, and MS2 reduction in Newmark groundwater for a ripened filter of 650–684 days.
2. Materials and methods
2.1 Cell and virus selection
MS2 bacteriophage (ATCC 15597-B1) was obtained from the American Type Culture Collection and was propagated and replicated, as described previously.23 Briefly, Escherichia coli (ATCC 15597) was propagated in tryptic soy broth solution, inoculated with MS2, and incubated at 37 °C. MS2 was then purified by sequential centrifugation (Eppendorf centrifuge 5416) at 5000 rpm (g × 100) for 15 minutes at 20 °C, then filtered through a 0.2 μm low-protein-binding polycarbonate track-etched membrane (Whatman Nucleopore, USA), and finally purified and concentrated using polyethylene glycol (PEG) following a previously described protocol.24 The PEG method for virus concentration is a well-established method and has a recovery of 75–87%.25,26 The purified MS2 stock, concentrated to ∼1012 plaque forming units (PFU) per mL, was stored in 1 mM NaCl at 4 °C. Enumeration of MS2 samples was performed using the double agar layer procedure.23 Dilutions with 30 to 300 plaques were used to calculate the PFU per mL (PFU mL−1).Group A rotavirus OSU strain (ATCC VR-892) was propagated in and extracted from MA-104 Clone 1 (ATCC CRL-2378.1) monkey kidney cells, as described elsewhere.27,28 Rotavirus (RV) purification was performed following the same protocol as for MS2, except the purified RV stock (∼107 focus forming units (FFU) per mL or FFU mL−1) was stored in 1 mM NaCl and 0.1 mM CaCl2 solution at 4 °C to prevent outer capsid protein denaturation. Enumeration of RV samples were performed using an integrated cell culture and reverse transcription quantitative PCR (ICC-RT-qPCR) method developed, as described in detail below.
2.2 Biosand filter experiments
Three 4 inch diameter PVC BSFs with a 55 cm sand height were constructed for MS2 and RV reduction experiments. Seven sampling ports were installed in each filter before packing at the following sand depths (cm): 5.4, 10.9, 16.3, 21.7, 32.6, 43.4, and 54.3. The sand and gravel in the filters were washed, sieved, and packed according to CAWST recommendations, except that the maximum sand size was 0.6 mm, the effective diameter (d10) was 0.35 mm, and the maximum flow rate was 0.51 L min−1.9 The upper and lower layers of gravel (1–6 mm and 6–12 mm diameters, respectively) were used to support the sand layer. The sand had an effective diameter (d10) of 0.35 mm and a uniformity coefficient (UC) of 1.72. A diffuser plate placed 5 cm above the top of the sand layer reduced the speed of water flow. The maximum loading head of the filters was 12.5 cm. One of the filters was constructed and used in a previous study, which had a pore volume of 2.1 L.8 Tracer tests to verify plug flow and determine pore volumes in the two newly constructed filters were performed using four or five pore volumes of 0.1 mM NaCl, as described elsewhere (Fig. 1aS and 1bS†).8,11,13 The dosing volume used for each filter was equal to the pore volume of the sand and gravel for each filter.Feed water for two of the filters was Newmark groundwater (pH 7.8–8.1), which has been well characterized in previous studies.8,13 One of these filters (pore volume 2.2 L) was used to determine RV reduction up to 31 days of use, and the other filter (pore volume 2.1 L) was used to determine MS2 reduction after 650 days of use. A third filter (pore volume 2.0 L) was fed with a cation-free buffered solution (1 mM NaHCO3 in nanopure water, pH 8.1) for up to 42 days. 1 mM NaHCO3 was prepared by adding 168 mg of NaHCO3 into 2.0 L of nanopure water. In order to determine if high divalent cation concentrations of 0.6–1.5 mM Ca2+ and 1.0–2.3 mM Mg2+, as measured by ICP-OES, in the influent water source affects MS2 reduction, MS2 reduction experiments as a function of depth and residence time were performed using 1 mM NaHCO3, which has similar pH as the groundwater. The high hardness of this groundwater source (116 mg L−1 as calculated using 1.5 mM Ca2+ and 2.3 mM Mg2+) comes from exposure to limestone in the aquifer, not exposure to the sand media used to pack the filters. MS2 and RV were spiked into the feed waters at concentrations of 108 PFU mL−1 and 106 FFU mL−1, respectively. Due to the difficulty in propagating RV, experiments on RV reduction were only conducted with an unripened BSF fed with groundwater for 31 days. Maximum filter flow rates were taken during each feed. Short residence time (average of ∼10 min) samples were collected once the feed water finished flowing through the filter. Long residence time samples were collected immediately prior to another filter feed, ∼20–29 hours after short residence time samples were collected. Seven port samples were collected from the sampling ports at both short and long residence times. An effluent sample was collected for long residence time through the outlet tube. All sample volumes were 1 mL, collected in 1.7 mL autoclaved centrifuge tubes, and stored in 4 °C until further processing, but no longer than 48 hours after sample collection. Control tests to determine natural degradation of 108 PFU mL−1 of MS2 and 106 FFU mL−1 of RV in groundwater and 1 mM NaHCO3 placed at room temperature were performed. Negligible natural degradation of RV in groundwater and MS2 in 1 mM NaHCO3, and 1 log10 of natural degradation of MS2 in groundwater were observed. Replicate filters for different treatments were not constructed due to the large physical space required for these filters, especially for those that were operated for years. This is a limitation of the study and future work should include replicate columns when feasible. In this study, experiments were repeated as frequently as possible, and dependent samples t-tests were used to statistically analyze the effects of different solutions on virus reduction trends.
2.3 ICC-RT-qPCR for rotavirus quantification
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 copy number of replicated RV genomes per cell, which allows for quantification of only infectious RV.
log10 copy number of replicated RV genomes per cell, which allows for quantification of only infectious RV.
Two control tests were performed to test the feasibility of the ICC-RT-qPCR method. First, the Newmark groundwater used in this study causes the MA-104 cells to detach when using the FFU method, making it impossible to accurately infect the cells with RV. Hence, the first control test was to ensure that cell detachment would not occur when using the ICC-RT-qPCR method. The second control test was to determine if any components in Newmark groundwater inhibit qPCR. A stock of cDNA containing the NSP3 gene used for RV quantification was diluted into two sets, one with nuclease-free water and one with groundwater, from the stock solution (1011 copies per μL). The dilutions for 101–109 copies per μL for both sets were used to create calibration curves and to compare the efficiencies and Ct values.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 g for 60 minutes. The supernatant for each sample was pipetted out while being careful not to disrupt the pellets. Finally, 100 μL of 1 mM NaCl + 0.1 mM CaCl2 was added to the centrifuge tubes and the samples were rotated at 15 rpm in 4 °C overnight.
500 g for 60 minutes. The supernatant for each sample was pipetted out while being careful not to disrupt the pellets. Finally, 100 μL of 1 mM NaCl + 0.1 mM CaCl2 was added to the centrifuge tubes and the samples were rotated at 15 rpm in 4 °C overnight.
All samples were incubated at 37 °C and 5% CO2 for 30 minutes to allow for activation of RV. After this incubation, 100 μL of serum-free Eagle's minimum essential medium (MEM) was added to each sample. This gave a total volume of 200 μL per sample for a total of 21 samples per set of data. 21 wells out of a 24 well plate of confluent MA-104 cells were rinsed twice with 500 μL phosphate buffered saline (PBS) per well to remove unbound cells. 150 μL of serum-free MEM + RV + trypsin solution was added to each of the 21 wells. The samples were incubated at 37 °C for 30 minutes to allow RV binding and penetration. The cells were then rinsed twice with 500 μL serum-free MEM per well to remove unbound viruses. 500 μL of serum-free MEM was added into each well and the samples were incubated at 37 °C and 5% CO2 for 18 hours for RV replication.
The number of MA-104 cells per well could not be assumed to be the same in each well. Hence, in order to accurately quantify the amount of RV infecting the MA-104 cells per well, the number of cells per well also needed to be quantified. Counting the number of cells for hundreds of wells per experiment through the hemocytometer would have been too labor-intensive. Instead the GAPDH housekeeping gene was tracked through qPCR based on the fact that there are two GAPDH housekeeping genes per cell.30 The GAPDH primers adapted from a previous study31 were GAPDHF (5′-AATCCCATCACCATCTTCCAG-3′) and GAPDHR (5′-AAATGAGCCCCAGCCTTC-3′). Using the cell counts from the hemocytometer as a basis of the cell concentration per well (∼107 copies per μL), a calibration curve to quantify the GAPDH gene was made consisting of ten-fold dilutions (concentrations between 103 and 107 copies per μL) of the extracted RNA from the cells-only well (Fig. 3S†). For each sample, the NSP3 and GAPDH genes were quantified in the same thermal cycle. Dilutions of the four primers and cDNA standard were stored in −20 °C and were thawed on ice prior to each RT-qPCR run.
2.4 Data analysis
Ct values were obtained in triplicates for all samples: influent, effluent, ports, cDNA calibration curve, cell calibration curve, RV stock calibration curve, and negative control. The average Ct value between the triplicates was used for quantification of NSP3 and GAPDH genes.3. Results and discussion
3.1 ICC-RT-qPCR for rotavirus
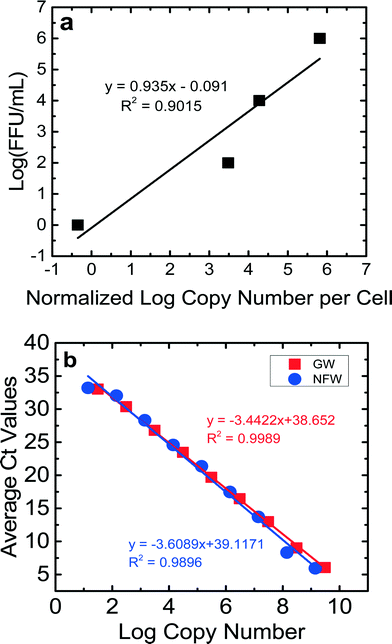
For the calibration curves obtained for the NSP3 and GAPDH genes (Fig. 2S and 3S†), the ranges of efficiencies were 86–101% for JVK and 88–118% for GAPDH. The detection limit for a Ct value of 35 was between 20 copies μL−1 and 80 copies μL−1 for JVK and GAPDH primers, respectively. The R2 values of the calibration curves were consistently between 0.98–0.99 for both JVK and GAPDH primers. Using these JVK and GAPDH primer calibration curves and the FFU concentrations of the RV stock, a calibration curve comparing infectious RV (log10 FFU mL−1) vs.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 normalized copy number of replicated RV genomes per cell is produced for each experiment (Fig. 1a). This calibration curve was used to quantify only infectious RV for BSF virus reduction experiments. By determining the normalized log copy number of replicated RV genomes per cell for each of the 16 samples collected per experiment, we used the calibration curve (Fig. 1a) to determine the log (FFU mL−1) of each sample. Eight RV experiments were performed in this study, and each experiment produced a calibration curve that showed that 1 FFU mL−1 could be achieved. The R2 values for the calibration curves ranged from 0.84–0.99. Fig. 1a is an example of a calibration curve from one experiment. Based on this calibration curve, the detection limit of this ICC-RT-qPCR was 1 FFU mL−1, which is lower than that of the traditional FFU method (100 FFU mL−1). This lower detection limit allowed us to quantify RV reduction using the bench-scale BSF.
log10 normalized copy number of replicated RV genomes per cell is produced for each experiment (Fig. 1a). This calibration curve was used to quantify only infectious RV for BSF virus reduction experiments. By determining the normalized log copy number of replicated RV genomes per cell for each of the 16 samples collected per experiment, we used the calibration curve (Fig. 1a) to determine the log (FFU mL−1) of each sample. Eight RV experiments were performed in this study, and each experiment produced a calibration curve that showed that 1 FFU mL−1 could be achieved. The R2 values for the calibration curves ranged from 0.84–0.99. Fig. 1a is an example of a calibration curve from one experiment. Based on this calibration curve, the detection limit of this ICC-RT-qPCR was 1 FFU mL−1, which is lower than that of the traditional FFU method (100 FFU mL−1). This lower detection limit allowed us to quantify RV reduction using the bench-scale BSF.
The results of the control experiments to check for PCR inhibition by Newmark groundwater are shown in Fig. 1b. The efficiencies and values for the dilutions for both nuclease-free water and groundwater were similar, suggesting that components in the groundwater did not inhibit qPCR. The Ct values for both sets of experiments were correlated with the copy numbers. The slopes of these correlation lines were statistically similar (p = 0.41). The efficiencies for the qPCR runs were 98% for nuclease-free water and 95% for groundwater. These results showed the lack of qPCR inhibition by groundwater.
3.2 Rotavirus reduction in Newmark groundwater
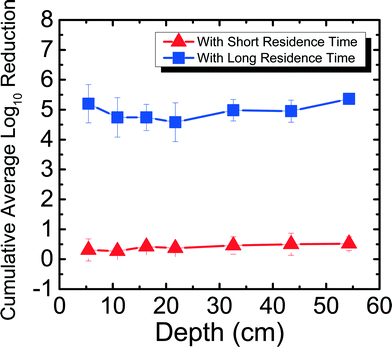
The reduction of RV spiked in Newmark groundwater was observed as a function of depth and residence time. Fig. 2 shows RV reduction in Newmark groundwater as a function of filter depth from days 1–31. Samples were taken from seven ports for both SRT (∼10 min) and LRT (24 hours). RV reduction reached a cumulative average of 3.54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for SRT and 4.92
log10 for SRT and 4.92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for LRT, which meets U.S. EPA and WHO standards for virus reduction. Similar to the MS2 reduction shown in Newmark groundwater for the PVC port column in a previous study,8 RV reduction followed an exponential increase of reduction as a function of depth for SRT (R2 = 0.97). Although there was not an exponential increase of reduction as a function of depth for LRT (R2 = 0.40), there was still significantly higher reduction seen during LRT than SRT. These patterns are demonstrated through dependent samples t-tests performed, showing that RV reduction was significantly different (p < 0.05) for all ports except between ports 1 and 2, 1 and 3, 2 and 3, 2 and 4, and 5 and 6 for SRT. For LRT, RV reduction was only significantly different (p < 0.05) between ports 1 and 4, 1 and 7, and 3 and 7. Overall, increased residence time and increased sand media depth improved RV reduction due to longer contact time between the water and the sand media. This observation suggests the important role of physical factors on virus reduction in agreement with previous studies.8,13,16
log10 for LRT, which meets U.S. EPA and WHO standards for virus reduction. Similar to the MS2 reduction shown in Newmark groundwater for the PVC port column in a previous study,8 RV reduction followed an exponential increase of reduction as a function of depth for SRT (R2 = 0.97). Although there was not an exponential increase of reduction as a function of depth for LRT (R2 = 0.40), there was still significantly higher reduction seen during LRT than SRT. These patterns are demonstrated through dependent samples t-tests performed, showing that RV reduction was significantly different (p < 0.05) for all ports except between ports 1 and 2, 1 and 3, 2 and 3, 2 and 4, and 5 and 6 for SRT. For LRT, RV reduction was only significantly different (p < 0.05) between ports 1 and 4, 1 and 7, and 3 and 7. Overall, increased residence time and increased sand media depth improved RV reduction due to longer contact time between the water and the sand media. This observation suggests the important role of physical factors on virus reduction in agreement with previous studies.8,13,16
 | ||
| Fig. 2 Cumulative average log10 reduction of rotavirus in Newmark groundwater as a function of filter depth and short residence time (∼10 min) and long residence time (24 hours) for days 1–31. | ||
Even without ripening, RV reduction reached 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 in the first 5 cm of the filter (port 1) alone for LRT, which is 1
log10 in the first 5 cm of the filter (port 1) alone for LRT, which is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 more than what was seen for MS2 reduction in port 1 for LRT.8 The extent of RV reduction in groundwater after 31 days for port 1 is similar compared to the cumulative average MS2 reduction in groundwater after 240 days for port 1. This suggests that high levels of virus reduction may not be entirely dependent on filter ripening, as shown in a previous study for MS2,8 but could occur for certain viruses and filter usage conditions. For example, it has been shown previously that RV aggregates and adsorbs to silica surfaces in high concentrations of divalent cations.20 Thus, different factors, such as depth of the filter and the hardness of the source water, are important for virus reduction for an unripened filter.
log10 more than what was seen for MS2 reduction in port 1 for LRT.8 The extent of RV reduction in groundwater after 31 days for port 1 is similar compared to the cumulative average MS2 reduction in groundwater after 240 days for port 1. This suggests that high levels of virus reduction may not be entirely dependent on filter ripening, as shown in a previous study for MS2,8 but could occur for certain viruses and filter usage conditions. For example, it has been shown previously that RV aggregates and adsorbs to silica surfaces in high concentrations of divalent cations.20 Thus, different factors, such as depth of the filter and the hardness of the source water, are important for virus reduction for an unripened filter.
There are limited studies looking at enteric virus reduction in slow sand filtration. Although the results from the studies vary, important conclusions can be drawn. In one study, reovirus reduction reached at least 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 after 7, 91, and 147 days when using river water as a water source in a 15 cm diameter filter with a 90 cm sand depth and up to 8 hours of hydraulic residence time.32 The study also showed insignificant differences in reduction between ripened and unripened filters. The long filter depth could have played a role in the high reduction even before ripening, especially since it was shown that the majority of the reduction took place within the top 42 cm of the filter. In another study, poliovirus reduction varied between 0.1
log10 after 7, 91, and 147 days when using river water as a water source in a 15 cm diameter filter with a 90 cm sand depth and up to 8 hours of hydraulic residence time.32 The study also showed insignificant differences in reduction between ripened and unripened filters. The long filter depth could have played a role in the high reduction even before ripening, especially since it was shown that the majority of the reduction took place within the top 42 cm of the filter. In another study, poliovirus reduction varied between 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 and 4
log10 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 depending on water source, sand type, and flow rate.33 Through this current study and previous studies, it can be concluded that certain filter characteristics such as water source and filter depth influence enteric virus reduction efficacy, especially for an unripened filter. The higher RV reduction observed in an unripened filter in this study compared to the lower MS2 reduction observed in an unripened filter8 using the same setup shown previously suggests that MS2 is a conservative surrogate for RV. Because of this and the limited ability to propagate RV, we next determined the role of divalent cations on MS2 reduction in an unripened filter.
log10 depending on water source, sand type, and flow rate.33 Through this current study and previous studies, it can be concluded that certain filter characteristics such as water source and filter depth influence enteric virus reduction efficacy, especially for an unripened filter. The higher RV reduction observed in an unripened filter in this study compared to the lower MS2 reduction observed in an unripened filter8 using the same setup shown previously suggests that MS2 is a conservative surrogate for RV. Because of this and the limited ability to propagate RV, we next determined the role of divalent cations on MS2 reduction in an unripened filter.
3.3 Short term MS2 reduction in 1 mM NaHCO3
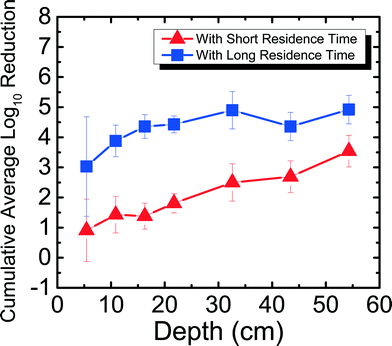
Fig. 3 shows the reduction of MS2 in 1 mM NaHCO3 as a function of filter depth from days 11–42, which encompasses an unripened filter and the start of the ripened period for the filter. Samples were taken from seven ports for both short residence time (SRT) of ∼10 min and long residence time (LRT) of 20–25 hours. MS2 reduction reached a cumulative average of 0.35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for SRT and 1.2
log10 for SRT and 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for LRT. Performing dependent samples t-tests showed that MS2 reduction was not statistically different (p > 0.05) between most ports for both SRT and LRT except between ports 2 and 3 and ports 2 and 4 for SRT and between ports 1 and all other ports for LRT. This low level of MS2 reduction is consistent with the results obtained by a BSF that was run with Cane Creek Reservoir water for 42–52 days.15 Similarly, in another study using Lac St Louis lake water in Quebec, MS2 reduction observed as a function of depth showed reduction averaging between 1
log10 for LRT. Performing dependent samples t-tests showed that MS2 reduction was not statistically different (p > 0.05) between most ports for both SRT and LRT except between ports 2 and 3 and ports 2 and 4 for SRT and between ports 1 and all other ports for LRT. This low level of MS2 reduction is consistent with the results obtained by a BSF that was run with Cane Creek Reservoir water for 42–52 days.15 Similarly, in another study using Lac St Louis lake water in Quebec, MS2 reduction observed as a function of depth showed reduction averaging between 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 and 2
log10 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 throughout the filter.14 The MS2 reduction level obtained with 1 mM NaHCO3 shown in Fig. 3 and with surface water sources14,15 was much lower than what we found using groundwater with the same setup as this study.8 It is noted that MS2 reduction in 1 mM NaHCO3 was higher than in previous studies using surface waters. A possible reason is because surface water has organic matter, which can compete with MS2 for available adsorption sites on the sand media. This phenomenon was shown in a previous studies comparing MS2 and natural organic matter adsorption onto hematite23 and interactions between MS2 and silica surface under environmentally relevant conditions.34
log10 throughout the filter.14 The MS2 reduction level obtained with 1 mM NaHCO3 shown in Fig. 3 and with surface water sources14,15 was much lower than what we found using groundwater with the same setup as this study.8 It is noted that MS2 reduction in 1 mM NaHCO3 was higher than in previous studies using surface waters. A possible reason is because surface water has organic matter, which can compete with MS2 for available adsorption sites on the sand media. This phenomenon was shown in a previous studies comparing MS2 and natural organic matter adsorption onto hematite23 and interactions between MS2 and silica surface under environmentally relevant conditions.34
 | ||
| Fig. 3 Cumulative average log10 reduction of MS2 in 1 mM NaHCO3 as a function of filter depth and short residence time (∼10 min) and long residence time (20–25 hours) for days 11–42. | ||
These varying levels of MS2 reduction using different water sources suggest that the divalent cation concentration present at up to 1.5 mM Ca2+ and 2.3 mg Mg2+ in the groundwater influenced MS2 reduction efficacy for an unripened filter. MS2 has an isoelectric point of 3.6,23 and the quartz sand packed in BSFs has an isoelectric point of 2.44.35 If the source water has a pH of 7.5–8.0, there is electrostatic repulsion between negatively charged MS2 and negatively charged sand. However, charge neutralization of MS2 by divalent cations21 can reduce the repulsion between MS2 and the sand surface, allowing higher MS2 reduction in groundwater compared to reduction in 1 mM NaHCO3 and surface waters, which are assumed to be relatively soft compared to groundwater.36 Finally, it is important to note that although the MS2 reduction for SRT is negligible, observing over 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction with 20–25 hours of residence time shows the significance of physical reduction that occurs in the filter with sufficient residence time, similar to what was shown in a previous study.8
log10 reduction with 20–25 hours of residence time shows the significance of physical reduction that occurs in the filter with sufficient residence time, similar to what was shown in a previous study.8
3.4 Long-term MS2 reduction in Newmark groundwater
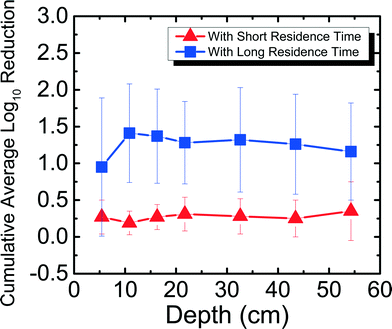
Field studies 10–12 years after project implementation have found BSFs still in use.37 While it is encouraging to see that people continue to use their filters, no laboratory studies have determined pathogen reduction efficacy in a filter that has been used for more than 300 days. It is important to determine how pathogen reduction efficacy changes with time to better understand the mechanism of pathogen reduction in the filter. This is the first study to determine the efficacy and trend of MS2 reduction for a BSF that has been run for more than 650 days. Fig. 4 shows MS2 reduction in Newmark groundwater as a function of filter depth from days 650–684. Samples were taken from seven ports for both SRT (∼10 min) and LRT (24–29 hours). MS2 reduction reached a cumulative average of 0.52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for SRT and 5.36
log10 for SRT and 5.36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for LRT. MS2 reduction was only statistically different between ports 2 and 3, 4, 5, 6, and 7 for SRT (p < 0.05), and between ports 7 and 2, 3, 4, 5, and 6 for LRT (p < 0.05).
log10 for LRT. MS2 reduction was only statistically different between ports 2 and 3, 4, 5, 6, and 7 for SRT (p < 0.05), and between ports 7 and 2, 3, 4, 5, and 6 for LRT (p < 0.05).
In this study, the same PVC port filter was used as in a previous study8 for MS2 reduction experiments for days 9–28, which has a shorter SRT (∼10 min) compared to a full-scale BSF (∼45 min). Also reported in the previous study was the concrete port filter that ran for 240 days and showed an average cumulative MS2 reduction of 5.16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for SRT and 5.64
log10 for SRT and 5.64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for LRT.8 Although both the PVC port filter used in this study and the concrete port filter used in the previous study8 were fully ripened, there was significantly lower MS2 reduction for SRT for the PVC port filter. This observation could suggest that a SRT period of 10 min is not enough time for significant MS2 reduction to occur when the filter has ripened, even when using water that has high divalent cations. The filter's flow rates that were slightly higher (average 0.44 L min−1, maximum 0.51 L min−1) than what CAWST recommends (0.4 L min−1) could also explain why the MS2 reduction was low for SRT. The higher flow rates indicate that the results in this study are conservative compared to what could be seen at lower flow rates. The experiments in this study did not compare BSF performance in the lab with performance in the field, but were instead performed to better understand factors that affect virus reduction. In another study that used Lac St Louis, similar reduction levels (∼0.5
log10 for LRT.8 Although both the PVC port filter used in this study and the concrete port filter used in the previous study8 were fully ripened, there was significantly lower MS2 reduction for SRT for the PVC port filter. This observation could suggest that a SRT period of 10 min is not enough time for significant MS2 reduction to occur when the filter has ripened, even when using water that has high divalent cations. The filter's flow rates that were slightly higher (average 0.44 L min−1, maximum 0.51 L min−1) than what CAWST recommends (0.4 L min−1) could also explain why the MS2 reduction was low for SRT. The higher flow rates indicate that the results in this study are conservative compared to what could be seen at lower flow rates. The experiments in this study did not compare BSF performance in the lab with performance in the field, but were instead performed to better understand factors that affect virus reduction. In another study that used Lac St Louis, similar reduction levels (∼0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10) were observed at different depths throughout a 10 cm diameter acrylic filter on day 60 with residence time of 4 hours, but reduction levels reached around 2
log10) were observed at different depths throughout a 10 cm diameter acrylic filter on day 60 with residence time of 4 hours, but reduction levels reached around 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 with 24 hours of residence time,11 once again showing how residence time can impact reduction regardless of the water source used.
log10 with 24 hours of residence time,11 once again showing how residence time can impact reduction regardless of the water source used.
Furthermore, for the concrete port filter, there was increasing cumulative MS2 reduction as a function of depth for both SRT and LRT, where the most significant reduction occurred in the first 5 cm of the filter.8 However, as shown in Fig. 4, for the PVC port filter, there was no significant increasing or decreasing trend for both SRT and LRT. One possible reason why MS2 reduction did not increase with depth could be because of a shift in microbial community structure throughout the filter. Previously, it was shown that the highest abundance and diversity of species populated the first 5 cm of the filter, allowing for the most reduction in the biolayer.8 Perhaps with time (>650 days), the microbial communities shifted and evened out throughout the filter. This hypothesis could be tested by studying the microbial communities throughout the filter. While this was not done within this study because it would destroy the experimental filters, future work should focus on understanding further how microbial communities influence long-term virus reduction, which is important for communities where BSFs are the main technology for drinking water treatment. Nevertheless, based on the results presented in this study that MS2 reduction can reach U.S. EPA and WHO standards only with long residence times, BSF users should operate the filters with the typical 16–32 hours of residence time8 to ensure that more effective virus reduction takes place.
4. Conclusions
Overall, this study shed light on two major limitations in previous BSF studies. First, this was the first study to determine the efficacy of enteric virus reduction in BSFs. After 31 days, RV reduction reached 4.92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10, which exceeds U.S. EPA and WHO standards. This level of reduction was seen when using Newmark groundwater as the source water, and is comparable to previous studies looking at reovirus and poliovirus reduction in slow sand filtration. Based on the results presented here that RV reduction by BSF can approach 5
log10, which exceeds U.S. EPA and WHO standards. This level of reduction was seen when using Newmark groundwater as the source water, and is comparable to previous studies looking at reovirus and poliovirus reduction in slow sand filtration. Based on the results presented here that RV reduction by BSF can approach 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10, it is recommended that filter users use the CAWST V10 BSF, especially with groundwater used as the source water. In addition, an ICC-RT-qPCR method was developed to more efficiently and effectively quantify RV reduction in the BSF samples. Second, this study showed that the water chemistry of the influent source waters affects MS2 reduction efficacy for an unripened filter. While MS2 reduction using Newmark groundwater that has high divalent cation concentrations exceeded U.S. EPA and WHO standards of 4
log10, it is recommended that filter users use the CAWST V10 BSF, especially with groundwater used as the source water. In addition, an ICC-RT-qPCR method was developed to more efficiently and effectively quantify RV reduction in the BSF samples. Second, this study showed that the water chemistry of the influent source waters affects MS2 reduction efficacy for an unripened filter. While MS2 reduction using Newmark groundwater that has high divalent cation concentrations exceeded U.S. EPA and WHO standards of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction in a previous study,8 MS2 reduction using 1 mM NaHCO3, which has no cations, showed only a low reduction of 1.2
log10 reduction in a previous study,8 MS2 reduction using 1 mM NaHCO3, which has no cations, showed only a low reduction of 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10. Although there are other differences between the groundwater and the buffered solution, the main factor that controls virus adsorption to silica surfaces in an unripened filter is the presence of divalent cations in the groundwater, as shown in our previous studies.20–22 For a ripened filter, divalent cations may not play as large of a role as microbial communities would in increasing virus reduction.
log10. Although there are other differences between the groundwater and the buffered solution, the main factor that controls virus adsorption to silica surfaces in an unripened filter is the presence of divalent cations in the groundwater, as shown in our previous studies.20–22 For a ripened filter, divalent cations may not play as large of a role as microbial communities would in increasing virus reduction.
Future work should focus on the following experiments to more comprehensibly understand how viruses are reduced in BSFs. First, RV reduction efficacy was only studied for the BSF's unripened period. While this is invaluable data, showing that enteric viruses can be reduced effectively by BSFs, long term studies of RV and short and long term studies of other enteric viruses are needed to fully understand the efficacy of enteric virus reduction in BSFs. Furthermore, these experiments should explore different water sources, especially those that mimic field conditions, and water chemistry should be systematically characterized. Only two water sources have been characterized in BSF studies8 and a few water sources have been characterized for slow sand filtration.32,33 In addition, studies on how sand media composition containing other minerals such as calcites or dolomites found in limestone should also be conducted to determine whether other sand media serve as a more favorable media for virus reduction. Finally, microbial community analysis of filters operated under field conditions around the world, similar to what was done for the first time on one BSF in a previous study,8 and pathogen reduction efficacy experiments of these filters should be conducted to better understand the mechanism of virus reduction in the filter. Results from these experiments would help to improve the BSF and quality of life of BSF users.
References
- UNICEF/WHO, Progress on drinking water and sanitation, 2015 Search PubMed.
- UNICEF, Global water supply and sanitation assessment 2000 report, OMS, 2000 Search PubMed.
- WHO, Diarrhoeal disease, http://www.who.int/mediacentre/factsheets/fs330/en/) Search PubMed.
- WHO, Estimated rotavirus deaths for children under 5 years of age, 2008, p. 453000, http://www.who.int/immunization/monitoring_surveillance/burden/estimates/rotavirus/en/ Search PubMed.
- L. Fewtrell, R. B. Kaufmann, D. Kay, W. Enanoria, L. Haller and J. M. Colford, Lancet Infect. Dis., 2005, 5, 42–52 CrossRef PubMed.
- M. D. Sobsey, C. E. Stauber, L. M. Casanova, J. M. Brown and M. A. Elliott, Environ. Sci. Technol., 2008, 42, 4261–4267 CrossRef CAS PubMed.
- CAWST, Biosand Filters Knowledge Base. Fact Sheet: Biosand Filter, http://biosandfilters.info/technical/fact-sheet-biosand-filter) Search PubMed.
- H. Wang, T. Narihiro, A. P. Straub, C. R. Pugh, H. Tamaki, J. F. Moor, I. M. Bradley, Y. Kamagata, W.-T. Liu and T. H. Nguyen, Environ. Sci. Technol., 2014, 48, 6702–6709 CrossRef CAS PubMed.
- CAWST, Biosand Filter Construction Manual, http://resources.cawst.org/node/6150 Search PubMed.
- C. Young-Rojanschi and C. Madramootoo, J. Water Supply: Res. Technol.--AQUA, 2015, 64, 157–167 CrossRef.
- M. Elliott, C. Stauber, F. Koksal, F. DiGiano and M. Sobsey, Water Res., 2008, 42, 2662–2670 CrossRef CAS PubMed.
- M. W. Jenkins, S. K. Tiwari and J. Darby, Water Res., 2011, 45, 6227–6239 CrossRef CAS PubMed.
- I. Bradley, A. Straub, P. Maraccini, S. Markazi and T. H. Nguyen, Water Res., 2011, 45, 4501–4510 CrossRef CAS PubMed.
- C. Young-Rojanschi and C. Madramootoo, Water Res., 2014, 49, 1–10 CrossRef CAS PubMed.
- M. Elliott, F. DiGiano and M. Sobsey, Water Res., 2011, 45, 4092–4102 CrossRef CAS PubMed.
- M. Elliott, C. E. Stauber, F. A. DiGiano, A. F. de Aceituno and M. D. Sobsey, Int. J. Environ. Res. Public Health, 2015, 12, 10276–10299 CrossRef CAS PubMed.
- M. Elimelech, J. Gregory and X. Jia, Particle deposition and aggregation: measurement, modelling and simulation, Butterworth-Heinemann, 2013 Search PubMed.
- Y. Jin, M. V. Yates, S. S. Thompson and W. A. Jury, Environ. Sci. Technol., 1997, 31, 548–555 CrossRef CAS.
- K. Wong, B. Mukherjee, A. M. Kahler, R. Zepp and M. Molina, Environ. Sci. Technol., 2012, 46, 11145–11153 CrossRef CAS PubMed.
- L. Gutierrez, S. E. Mylon, B. Nash and T. H. Nguyen, Environ. Sci. Technol., 2010, 44, 4552–4557 CrossRef CAS PubMed.
- M. Pham, E. A. Mintz and T. H. Nguyen, J. Colloid Interface Sci., 2009, 338, 1–9 CrossRef CAS PubMed.
- B. Yuan, M. Pham and T. H. Nguyen, Environ. Sci. Technol., 2008, 42, 7628–7633 CrossRef CAS PubMed.
- L. Gutierrez, X. Li, J. Wang, G. Nangmenyi, J. Economy, T. B. Kuhlenschmidt, M. S. Kuhlenschmidt and T. H. Nguyen, Water Res., 2009, 43, 5198–5208 CrossRef CAS PubMed.
- T. H. Nguyen, N. Easter, L. Gutierrez, L. Huyett, E. Defnet, S. E. Mylon, J. K. Ferri and N. A. Viet, Soft Matter, 2011, 7, 10449–10456 RSC.
- G. D. Lewis and T. G. Metcalf, Appl. Environ. Microbiol., 1988, 54, 1983–1988 CAS.
- P. Vilaginès, A. Suarez, B. Sarrette and R. Vilagines, Water Sci. Technol., 1997, 35, 455–459 CrossRef.
- M. D. Rolsma, H. B. Gelberg and M. S. Kuhlenschmidt, J. Virol., 1994, 68, 258–268 CAS.
- O. C. Romero-Maraccini, N. J. Sadik, S. L. Rosado-Lausell, C. R. Pugh, X.-Z. Niu, J.-P. Croué and T. H. Nguyen, Environ. Sci. Technol., 2013, 47, 11004–11012 CrossRef CAS PubMed.
- O. C. Romero-Maraccini, J. L. Shisler and T. H. Nguyen, Appl. Environ. Microbiol., 2015, 81, 4090–4097 CrossRef CAS PubMed.
- N. W. Seidler, GAPDH: Biological Properties and Diversity, Advances in Experimental Medicine and Biology, vol. 985, Springer, New York, USA, 2013 Search PubMed.
- R. Bhowmick, U. C. Halder, S. Chattopadhyay, M. K. Nayak and M. Chawla-Sarkar, J. Virol., 2013, 87, 6840–6850 CrossRef CAS PubMed.
- L. K. McConnell, R. C. Sims and B. B. Barnett, Appl. Environ. Microbiol., 1984, 48, 818–825 CAS.
- G. G. Robeck, N. A. Clarke, K. A. Dostal and H. O. Hartung, J. - Am. Water Works Assoc., 1962, 1275–1292 CAS.
- L. Gutierrez and T. H. Nguyen, Environ. Sci. Technol., 2012, 46, 8705–8713 CrossRef CAS PubMed.
- A. Jada, R. A. Akbour and J. Douch, Chemosphere, 2006, 64, 1287–1295 CrossRef CAS PubMed.
- J. C. Crittenden, R. R. Trussell, D. W. Hand, K. J. Howe and G. Tchobanoglous, MWH's Water Treatment: Principles and Design, John Wiley & Sons, 2012 Search PubMed.
- A. J. Sisson, P. J. Wampler, R. R. Rediske and A. R. Molla, J. Water, Sanit. Hyg. Dev., 2013, 3, 51–60 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ew00297d |
| This journal is © The Royal Society of Chemistry 2016 |