Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Silver nanoparticle–protein interactions in intact rainbow trout gill cells†
Yang
Yue
ab,
Renata
Behra
ac,
Laura
Sigg‡
ac,
Marc J.-F.
Suter
ac,
Smitha
Pillai
ac and
Kristin
Schirmer
*abc
aDepartment of Environmental Toxicology, Eawag, Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, CH 8600, Switzerland. E-mail: kristin.schirmer@eawag.ch
bÉcole Polytechnique Fédérale de Lausanne, School of Architecture, Civil and Environmental Engineering, Lausanne, CH 1015, Switzerland
cDepartment of Environmental Systems Science, ETH-Zürich, Zürich, CH 8092, Switzerland
First published on 17th August 2016
Abstract
Upon contact with biota, nanoparticles can bind to proteins, which coat the nanoparticles and form a nanoparticle-protein corona. Knowledge of corona proteins is therefore important for a mechanistic understanding of how nanoparticles interact with biomolecules in cells. Here we present the first study to reveal the identity of corona proteins from silver nanoparticle (AgNPs)-exposed living vertebrate cells. The cells are from a rainbow trout (Oncorhynchus mykiss) gill cell line, RTgill-W1, representing the interface between the aquatic environment and one of its model species. Subcellular fractionation allowed AgNP-protein corona complexes to be recovered from intact subcellular compartments and proteins lysed from the AgNPs to be detected by mass spectrometry. The identified proteins mark the trail of AgNPs processing in the cells like a forensic fingerprint: the cells take up the AgNPs via endocytic processes and store the particles in endosomal/lysosomal compartments. Moreover, stress response proteins were recovered in the AgNPs protein corona. In this way, we established a list of AgNPs susceptible proteins which can be investigated further in targeted nanoparticle–protein interaction. As a proof of principle, we demonstrate that Na+/K+-ATPase, identified from the corona and a known key protein in ion regulation in gill cells, is inhibited in its activity by AgNPs, confirming previously published in vivo experiments. The developed methodology is broadly applicable to other nanoparticles and cell types, representing a valuable tool for mechanistic nanoparticle–cell interaction studies, ranging from environmental and human risk assessment to biomedicine. In this way, our research also contributes to safer particle design.
Nano impactGiven the general importance of proteins in cell structure and function, identifying the proteins that interact with nanoparticles is instrumental to deciphering the mechanisms of nanoparticle–cell and eventually nanoparticle–organism interactions. However, there is a lack of broadly applicable methods to identify proteins that bind to nanoparticles in intact, living cells. For example, the gill is a confirmed entry port for nanoparticles into fish and therefore one of the initial sites of nanoparticle–protein interactions, but knowledge of proteins that bind to nanoparticles in gills is lacking. On this background, we developed a density gradient centrifugation method coupled with protein identification by mass spectrometry in order to discover the proteins that bind to silver nanoparticles in cultured fish gill cells. The identified proteins mark the trail of silver nanoparticle processing in the cells like a forensic fingerprint. The established list of susceptible proteins can be investigated further for targeted particle–protein interactions and can therefore help prioritize mechanistic investigations on nanoparticle–biomolecule interactions. Our method is broadly applicable to other types of animal or human cells as well as other metal-based nanoparticles. It contributes to improved environment and human health risk assessment as well as to safer particle design. |
Introduction
When taken up by cells via endocytic processes, nanoparticles accumulate in different compartments such as endosomes and lysosomes.1,2 Endocytic uptake routes and lysosome-related degradation processes play vital roles in cellular metabolism and homeostasis.3,4 Overloading these compartments with an exogenous stressor, such as nanoparticles, can lead to lysosomal dysfunction and other adverse effects in cells.5 Lysosome membrane permeabilization and destabilization are common causes of lysosome dysfunction. They can induce oxidative stress, lysosomal alkalization and osmotic swelling.5–7 Indeed, in an in vitro investigation of the impact of silver nanoparticles (AgNPs) on fish gill cells, lysosomal membrane integrity was shown to be more strongly affected than cellular metabolic activity and membrane integrity.8Due to the nanoparticles' very high surface-to-volume ratio, nanoparticles feature a high surface energy in comparison to bulk biomaterials.9,10 Hence, upon entry into cells, a variety of biomolecules may adsorb to the nanoparticle surface and reduce the nanoparticle surface energy by physical adsorption or chemical reactions.11–13 Based on their abundance and diversity, proteins are thought to play a dominant role in such types of interactions, coating the nanoparticle surface with a so-called protein corona.11,14 Thus, it is the NP-protein corona, not the original nanoparticle surface, that influences the interaction of nanoparticles with constituents in cells.15 In turn, corona-forming proteins may be depleted from the cellular machinery and become structurally and/or functionally impaired.16,17
Proteins that adsorb to nanoparticle surfaces are prone to alterations, which has been demonstrated using isolated proteins or protein mixtures extracted from cells. After adsorbing to a gold nanoparticle surface, the structure of serum albumin was different from that of the native form.18 The activity of tryptophanase (TNase) from an E. coli extract was significantly inhibited by AgNPs due to high affinity binding to the enzyme active site.19 Yet, knowledge of nanoparticle interactions with proteins in intact cells is scarce. Two recent studies made use of the unique magnetic property of magnetite nanoparticles to recover the protein corona by magnetic separation from intact cells and identified the recovered proteins by mass spectrometry.20,21 However, a universally applicable method to recover the metal nanoparticle-protein corona from cells does not yet exist. Accordingly, no previous study has attempted to determine the proteins in living cells that bind to AgNPs.
Here we explore the uptake, fate and interactions of AgNPs with proteins in intact cells of the rainbow trout (Oncorhynchus mykiss) gill cell line, RTgill-W1.22 These cells can survive in a low ionic strength exposure medium (d-L-15/ex, ESI† Table S1), which can stabilize citrate-coated AgNPs in suspension. In this way, the medium allows to more closely mimic the aquatic environment a gill cell would face.8 The AgNPs dispersed very well in d-L-15/ex medium with a Z-average size of 40–100 nm (PDI: 0.46–0.85) and a zeta potential of −20 mV.8 We have previously demonstrated that AgNPs elicit a particle-specific effect on RTgill-W1 cells. Specifically, lysosomal membrane integrity was significantly more sensitive than cell membrane integrity and cellular metabolic activity upon exposure to AgNPs. Moreover, scavenging silver ions stemming from AgNPs dissolution by a strong silver ion ligand, cysteine, only partially prevented the AgNPs impact on the lysosomes, further corroborating the particle-specific toxicity.8 These findings led us to hypothesize that AgNP-induced toxicity to RTgill-W1 cells may be elicited via lysosome related pathways and involve binding of lysosomal proteins to the AgNPs. Thus, we exploited the RTgill-W1 cell system to recover the AgNP-protein corona from intact cells. To do this, we first confirmed the presence of AgNPs in membrane-bound compartments and then performed subcellular fractionation of RTgill-W1 cells by means of density gradient centrifugation. The AgNP-protein corona was subsequently recovered from intact cellular compartments enriched in silver and the corona composition was analyzed. We explored silver nitrate (AgNO3) exposures in each step in parallel to account for processes attributable to silver ions potentially released from AgNPs.
Materials and methods
RTgill-W1 culture
RTgill-W1 cells were routinely cultivated in L-15 medium (Invitrogen, Basel, Switzerland) supplemented with 5% fetal bovine serum (FBS, Gold, PAA Laboratories GmbH, Austria) and 1% penicillin/streptomycin (Sigma-Aldrich, Buchs, Switzerland) in 75 cm2 flasks. The L-15 medium containing these supplements is termed “complete L-15”. Cells are routinely cultured in the dark in a normal atmosphere at 19 °C.AgNP characteristics and exposure of cells
A stock solution of citrate-coated AgNPs (Z-average size: 19.4 nm; zeta potential: −30 mV) was purchased from NanoSys GmbH (Wolfhalden, Switzerland) as an aqueous suspension with a concentration of 1 g L−1 (9.27 mM referring to the total silver, pH 6.46). The stock AgNPs solution was stored in the dark and experimental solutions were prepared in d-L-15/ex exposure medium (for composition see ESI,† Table S1). This medium is a simple buffer containing only salts, sodium pyruvate and galactose, which supports short-term RTgill-W1 cell viability and is designed to mimic the aquatic environmental a gill cell would face.7 The AgNPs dispersed very well in d-L-15/ex medium, which was confirmed by UV-vis spectroscopy, dynamic light scattering and electron microscopy.8 Analysis of AgNPs in d-L-15/ex using Zetasizer showed that the Z-average size was 40–100 nm (PDI: 0.46–0.85) and the zeta potential was around −20 mV.8 A stock solution of AgNO3 (Sigma-Aldrich, Buchs SG, Switzerland) was prepared at a concentration of 10 mM in nanopure water (16–18 MΩ cm−1; Barnstead Nanopure Skan AG, Basel-Allschwil, Switzerland). All the AgNO3 was soluble at the concentrations used. Approximately 60% silver was still in free Ag+ form and 40% silver formed AgCln(n−1)− complexes, which was checked using Visual MINTEQ 3.1 (KTH, Sweden).8For exposure to AgNPs and AgNO3, cells were seeded in 24-well microtiter plates, 25 cm2 or 300 cm2 flasks, and cultured in complete L-15 medium. After being fully confluent, the cell monolayers were washed with d-L-15/ex medium, and AgNPs or AgNO3 suspended in d-L-15/ex was added. The exposure proceeded for 0.5–2 h at 19 °C, except for uptake studies at low temperature, which were carried out at 4 °C. Toxicity was assessed by means of three fluorescent indicator dyes: Alamar Blue (AB, Invitrogen, Basel, Switzerland) was used to measure the cellular metabolic activity; 5-carboxyfluorescein diacetate acetoxymethyl ester (CFDA-AM, Invitrogen, Basel, Switzerland) to measure the cell membrane integrity; and Neutral Red (NR, Sigma-Aldrich, Buchs, Switzerland) to measure the lysosomal membrane integrity.23–25
Dissolution of AgNPs in exposure medium
To measure the dissolution of AgNPs in exposure medium after incubation with cells for 2 h, dissolved silver was separated by centrifugal ultrafiltration with a nominal molecular weight cut-off of 3 kDa (Amicon ultra-4 centrifugal filter units, Millipore, Germany) and by ultra-centrifugation (145![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 3 h).8 The silver concentration was measured by inductively coupled plasma mass spectrometry (ICP-MS, Element 2, Thermo Finnigan, Bremen, Germany). The reliability of the measurements was determined using specific water references (M105A, IFA-Tull, Austria).
000g, 3 h).8 The silver concentration was measured by inductively coupled plasma mass spectrometry (ICP-MS, Element 2, Thermo Finnigan, Bremen, Germany). The reliability of the measurements was determined using specific water references (M105A, IFA-Tull, Austria).
Uptake of AgNPs by RTgill-W1 cells and cell-internal distribution
To prepare samples for electron microscopy, confluent RTgill-W1 cells were exposed to AgNPs in 24-well plates. Upon termination of exposure, cells were sequentially washed with PBS, 0.5 mM cysteine in PBS for 5 min, and Versene (Invitrogen/Gibco, Germany) to remove loosely bound AgNPs and dissolved silver located on the cell surface. Washed cells were fixed with glutaraldehyde and paraformaldehyde and post-fixed with osmium tetroxide (OsO4). After uranyl acetate block staining, samples were dehydrated with a gradient of ethanol and embedded in Epon. Ultrathin sections were cut using a Leica microtome and placed on carbon-coated copper grids. Images were taken for transmission electron microscopy (TEM, FEI Morgagni 268, 100 kV). A scanning transmission electron microscope (STEM, Hitachi HD-2700) was used to perform the energy dispersive X-ray (EDX) spectroscopy analyses.To quantify the cell associated silver, RTgill-W1 cells were cultured in 25 cm2 flasks until confluency and then exposed to AgNPs or AgNO3 in d-L-15/ex medium. After exposure, the medium with AgNPs or AgNO3 was removed and cells were washed as described above. Cells were then trypsinized. Detached cells were re-suspended in complete L-15 medium. Cell suspensions were centrifuged at 1000g for 3 min to pellet the cells. Cell pellets were re-suspended in 550 μL of PBS and the cell density was determined using an electronic cell counter (CASY1 TCC, Schärfe System, Germany). A volume of 500 μL of cell supernatant was digested with 4.5 mL of 65% HNO3 in a high-performance microwave digestion unit (MLS-1200 MEGA, Oberwil, Switzerland) at a maximal temperature of 195 °C for 20 min. The digests were diluted 50 times and measured by ICP-MS as described above.
Subcellular fractionation
The isolation of intact cell compartments was based on a lysosome enrichment kit (Thermo Fisher Scientific, No. 89839, USA). All fractionation steps and subsequent NP-protein corona isolation work was performed at 4 °C or on ice and all isolation buffers were added with protease inhibitor (Halt™ Protease Inhibitor Single-Use Cocktail EDTA-Free, Thermo Fisher Scientific, No. 78425, USA) in order to minimize protein degradation.About 3 × 108 RTgill-W1 cells were used for this work. After washing with 0.5 mM cysteine solution in PBS, cells treated with silver or untreated cells were harvested with trypsin digestion and centrifuged to pellet the cells as described above. Cell pellets were suspended in lysis buffer and lysed by two subsequent sonication pulses of 10 s at 90 W (LABSONIC® M, Sartorius AG, Germany). Trypan blue staining was used to check the percentage of broken cells and ensure that at least 80% of cells were lysed. The cell lysate was centrifuged at 500g for 10 min to spin down unbroken cells and big fragments such as some remaining nuclei. The centrifuged lysate was loaded in a density gradient buffer with 15% to 30% Optiprep and ultra-centrifuged at 145![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 2 h. After centrifugation, several bands formed in the gradient. These bands were separated into 12 fractions according to density (F1–F12, low to high density). Each fraction was mixed with 2–3 volumes of PBS to decrease the fraction density and centrifuged at 18
000g for 2 h. After centrifugation, several bands formed in the gradient. These bands were separated into 12 fractions according to density (F1–F12, low to high density). Each fraction was mixed with 2–3 volumes of PBS to decrease the fraction density and centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 minutes. Pellets were surface washed with 200 μL of gradient dilution buffer and centrifuged at 18
000g for 30 minutes. Pellets were surface washed with 200 μL of gradient dilution buffer and centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 minutes. Each pellet was tested for protein concentration, acid phosphatase activity, silver content and LysoTracker staining.
000g for 30 minutes. Each pellet was tested for protein concentration, acid phosphatase activity, silver content and LysoTracker staining.
The protein quantity was determined using the Bradford assay (Coomassie Plus™ (Bradford) Assay Kit, Thermo Fisher Scientific, No. 23236, USA). Acid phosphatase activity was checked using an acid phosphatase assay kit (Sigma, CS0740, USA). Silver content was measured with ICP-MS as described above. LysoTracker (LysoTracker® Red DND-99, L7528, Invitrogen, USA) staining was used to visualize the intact acidic compartment, such as late endosomes and lysosomes. After the compartment intactness was confirmed, each pellet was frozen at −80 °C for subsequent work.
AgNP-protein corona isolation
The protein corona was isolated by two different approaches. The first approach was to isolate endosome–lysosome fractions and cell membrane–mitochondria–nucleus fractions from intact cells after exposed to AgNPs. The second approach was to fractionate cells as described above and expose cell compartment extracts from endosome–lysosome fractions and cell membrane–mitochondria–nucleus fractions to AgNPs.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 45 minutes. After removing the supernatant with the unbound proteins, 50 μL of TBS was added to each pellet and centrifuged at 10
000g for 45 minutes. After removing the supernatant with the unbound proteins, 50 μL of TBS was added to each pellet and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 minutes for washing. The resulting supernatant was again removed; the pellet now contained the AgNP-protein corona. The same procedure was followed after AgNO3 exposure as a control.
000g for 30 minutes for washing. The resulting supernatant was again removed; the pellet now contained the AgNP-protein corona. The same procedure was followed after AgNO3 exposure as a control.
Protein identification
To detach proteins from the AgNP-protein corona, 35–50 μL of TBS with 1% SDS and 50 mM DTT were added to each sample and incubated at 95 °C for 15 min. Samples were centrifuged at room temperature for 15 min at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g to pellet AgNPs. Supernatants containing isolated proteins were collected and quantified using Nanodrop and a Direct Detect® Infrared Spectrometer. All samples were stored at −20 °C for further analysis.
000g to pellet AgNPs. Supernatants containing isolated proteins were collected and quantified using Nanodrop and a Direct Detect® Infrared Spectrometer. All samples were stored at −20 °C for further analysis.
In order to identify the corona proteins by mass spectrometry, recovered protein samples were run in SDS-PAGE for a short time to remove CHAPS. Protein bands were excised and digested in-gel with trypsin and analyzed by electrospray liquid chromatography mass spectrometry (LC-MS/MS) using a label-free quantitative mass spectrometry approach. On average, approximately 3000 peptide counts were recorded for each sample. The MS raw data were analyzed by Scaffold 4 (version 4.3.4) and searched for rainbow trout protein recordings in NCBI. It has to be noted that the NCBI database does not yet contain full annotation of the rainbow trout genome. Therefore, as long as the analysis has to rely on the NCBI database alone, certain proteins not recorded there are lost in the above protein identification step.
In the protein identification process, the protein thresholds were set as minimum 99.0% probability and minimum 2 identified peptides. Together, 1223 proteins were identified in all samples. The relative abundance of each identified protein in each sample was normalized to the total peptide count (3000) by the following equation:
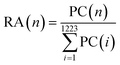 | (1) |
DAVID ontology analysis
After data evaluation of the MS spectra, >2-fold enriched proteins in AgNPs samples were selected for further analysis. Currently, genome information for rainbow trout is still comparatively limited. For this reason and also to obtain as much functional annotation as possible, all selected rainbow trout proteins were blasted against the NCBI human database in order to obtain the human protein GI number and a better protein functional annotation. All identified human proteins were submitted to a DAVID (Database for Annotation, Visualization and Integrated Discovery) protein ontology analysis (http://david.abcc.ncifcrf.gov/) and functional annotation clustering. Cluster enrichment and p-values were calculated for each item.Inhibition of AgNPs on Na+/K+-ATPase activity
Na+/K+-ATPase from porcine cerebral cortex was purchased from Sigma-Aldrich (No. A7510). The protein concentration of Na+/K+-ATPase was determined using the Bradford assay. The Na+/K+-ATPase activity was measured in a buffer containing 20 mM Tris-HCl, 0.60 mM EDTA, 5 mM MgCl2, 3 mM KCl and 133 mM NaCl (pH 7.8). Na+/K+-ATPase (0.5 U mL−1, 19.5 μg mL−1) and AgNPs (0.195–3.9 μg mL−1) or Ag+ (in AgNO3 form, 0.0975–0.39 μg mL−1) were added in buffer. Samples were pre-incubated at 37 °C for 15 min. The reaction started with the addition of ATP (2.7 mM, No. A9062, Sigma-Aldrich) and lasted for 30 min. The reaction was stopped by adding 20% trichloroacetic acid.26 Each sample was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 3 min to clarify the solution and pellet AgNPs. The resulting inorganic phosphate in suspension was determined by adding Taussky–Shorr reagent and reading the absorption at 660 nm.27 The relative activity of Na+/K+-ATPase was calculated relative to the sample without AgNP or AgNO3 addition.
000g for 3 min to clarify the solution and pellet AgNPs. The resulting inorganic phosphate in suspension was determined by adding Taussky–Shorr reagent and reading the absorption at 660 nm.27 The relative activity of Na+/K+-ATPase was calculated relative to the sample without AgNP or AgNO3 addition.
Results and discussion
RTgill-W1 cells take up silver in a time- and concentration-dependent manner
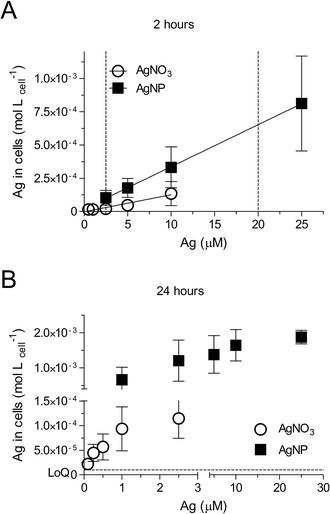
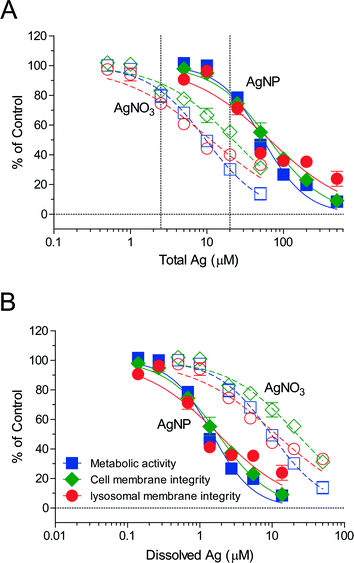
The total silver uptake into cells was quantified after 2 and 24 h of exposure to AgNO3 (0.1–10 μM, 11–1079 μg L−1) or AgNPs (citrate coated, 1–25 μM, 108–2697 μg L−1, referring to the concentration of total silver), followed by a cysteine wash to remove any loosely adsorbed silver prior to ICP-MS analysis (ESI† Fig. S1). Cell internal silver levels differed greatly between comparable total silver exposures of AgNO3 and AgNPs (Fig. 1). For example, a 2.5 μM total silver exposure led to a 4.8-fold higher level after 2 h and to 10.2-fold higher levels after 24 h of exposure to AgNPs compared to AgNO3. These values suggest that the RTgill-W1 cells take up AgNPs and silver ions via different routes. As shown in our previous work, Ag+ and AgCl0(aq) are the major dissolved silver species present in d-L-15/ex.8 Other research studies indicated that cells take up Ag+via Cu+ transporters in fish gill cells28 and algae29 while the AgCl0(aq) complex enters fish gill cells by passive diffusion.30 In contrast, AgNPs uptake was demonstrated in human mesenchymal stem cells31 and human neuroblastoma cells (SK-N-SH)32 to be via endocytic pathways. The cell associated silver in RTgill-W1 cells was comparable with the cell associated silver in other cell types, such as mouse erythroleukemia cells33 and HepG2 cells,34 upon exposure to similar concentrations of AgNPs.Uptake of silver by RTgill-W1 cells linearly correlated with exposure concentrations for the 2 h time point and reached near steady state after 24 h (Fig. 1). Aiming to capture the early dynamics of AgNPs entering cells prior to reaching steady state, we established concentration–cell viability response relationships for 2 h of exposure (Fig. 2, ESI† Table S2). Based on these, we identified concentrations of AgNO3 and AgNPs that are sub-toxic but detectable. For equal levels of cytotoxicity, these were 2.5 μM (270 μg L−1) AgNO3 and 20 μM (2157 μg L−1) AgNPs (Fig. 2A, dashed vertical lines). Interestingly, in contrast to previously established concentration–response relationships after 24 h of exposure to the same AgNPs,8 a specific impact on the lysosomes is not apparent at the early exposure time. Conceivably, the time of uptake and internal processing is a determinant for the lysosome-specific toxicity to proceed.
Based on the dissolved silver content in the exposure medium, d-L-15/ex (ESI† Table S3), re-calculation of the 2 h exposure concentration–response curves as a function of dissolved silver showed that AgNPs elicited a higher toxicity than AgNO3 (Fig. 2B), indicating that the AgNPs elicited a particle-specific effect on the cells. In the context of our study, this finding provided additional support that we were capturing the early dynamics of AgNPs uptake and intracellular processing, including the presence of intact lysosomes.
AgNPs are visible in endocytic compartments in RTgill-W1 cells
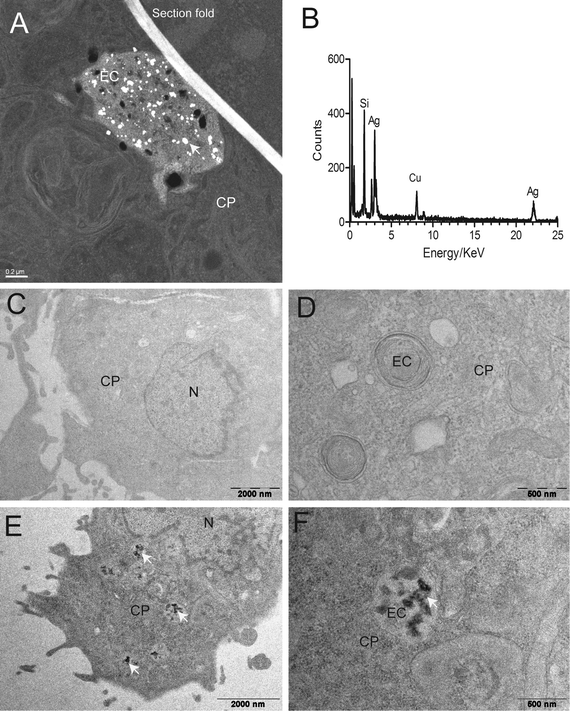
Electron microscopy, coupled with energy-dispersive X-ray analysis, suggested the internalization of AgNPs into cells and their presence in endocytic compartments (Fig. 3A and B). Furthermore, no or very little uptake was seen at 4 °C exposure temperature (Fig. 3C and D) compared to 19 °C (Fig. 3E and F). This indicates that the cellular uptake of AgNPs is temperature- and therefore energy-dependent, as is the case for endocytic processes.35 Previously, uptake of tungsten carbide nanoparticles was demonstrated to proceed in RTgill-W1 cells whether or not particles were presented in little or strongly agglomerated form.36 The fact that RTgill-W1 cells incorporate particles when exposed in a simple medium, such as d-L-15/ex, points toward endocytosis by gill cells as a likely uptake mechanism of AgNPs in fish. An in vivo study demonstrated silver accumulation in the fish gill upon water borne exposure of fish to AgNPs.37 Indeed, the d-L-15/ex better reflects the environment to which gill cells of freshwater fish would be exposed compared to common, complex cell culture media.8AgNPs exposure leads to a distinctly different intracellular silver distribution pattern compared to AgNO3
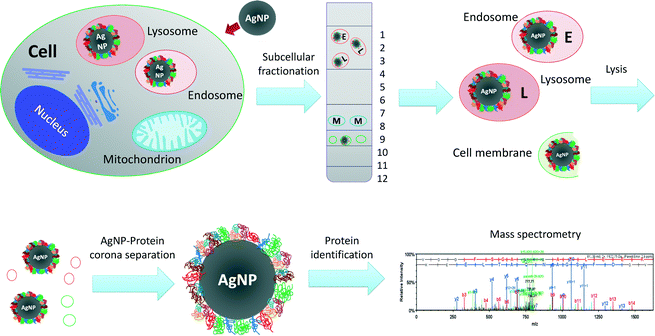
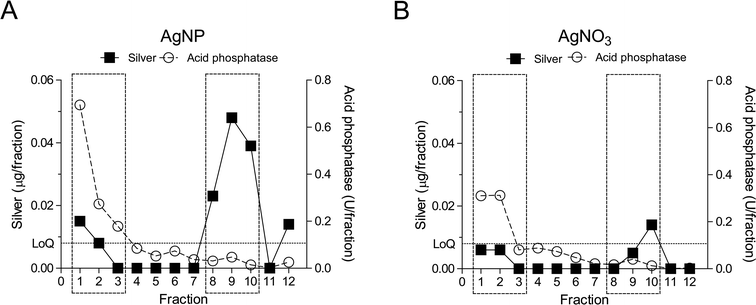
To further study the intracellular distribution of AgNPs, subcellular fractionation of RTgill-W1 cells exposed to either AgNPs or AgNO3 for 2 h was performed using density gradient centrifugation (Fig. 4, ESI† Fig. S2). Twelve fractions were isolated and each was analyzed for silver content and for acid phosphatase activity as a typical biomarker of endocytic compartments.38The intracellular distribution of silver in AgNP- vs. AgNO3-exposed cells was clearly distinct. In support of an endocytic pathway for AgNPs, silver accumulated in the low-density fractions, which represent the endosome–lysosome compartments (Fig. 5A, fractions 1–3). The presence of intact lysosomes was also confirmed by LysoTracker® staining (ESI† Fig. S3A–C). In contrast, silver content in the AgNO3-exposed cells in those same fractions was below the level of quantification (Fig. 5B). Significant silver accumulation occurred as well in fractions 8–10 of AgNP-exposed cells (Fig. 5A). Very little to no accumulation was detected in these fractions in AgNO3-exposed cells. Based on their density, these fractions represent nuclei, mitochondria and peroxisomes. As for the presence of high silver levels in these fractions after AgNPs exposure, we do not attribute it to an accumulation of AgNPs in mitochondria and nuclei because AgNPs were never seen in these organelles in TEM images. Another study using fish gill cells has also pointed out that nanoparticles were never observed in nuclei, especially after short exposure times.36 Localization of nanoparticles in mitochondria has been reported only once where human lung cells were exposed to gold nanorods (A549).39 One explanation for the silver content in these fractions might be silver ions liberated from the AgNPs; detected silver levels measured after AgNO3 exposure in fraction 10 are in support of this. However, the silver content in fractions 8–10 upon AgNP exposure was much higher than on exposure to AgNO3. We therefore argue that the fractions may also contain some endocytosis related organelles which were overloaded with AgNPs and cell membrane debris containing AgNPs. Then, due to large amounts of AgNPs, these fragments would have a significantly higher density than the endosome–lysosome fraction and therefore elute in the heavy fractions 8–10. The total recovery of silver in the organelle debris from AgNO3 exposures was only 13.2%. This indicates that most of the silver ions must have been present in the cytosol. Indeed, when quantifying subcellular silver distribution after AgNP and AgNO3 exposures of worms, a high silver content was found in cell organelles from AgNP exposures while AgNO3 exposures led to silver accumulation in the cell cytosol (metallothionein like protein fractions).40
383 different proteins were specifically enriched on AgNPs isolated from intact RTgill-W1 cells
Based on the combined parameters of acid phosphatase activity and silver content, fractions 1–3 (the endosome–lysosome fractions) and 8–10 (the cell membrane–mitochondria–nucleus fractions) were harvested, pooled and subjected to AgNP-protein corona recovery. Samples from AgNO3 exposures served as controls. Relevant fractions were lysed by freezing and thawing in TBS with 1% CHAPS, a non-denaturing solvent, in order to minimize changes to the protein corona composition during preparation. The AgNP-protein corona was pelleted from the lysates by centrifugation. Proteins were detached by denaturants and digested with trypsin. Proteins in the AgNP-protein corona were identified by a label-free quantitative mass spectrometry approach using nano-LC-MS/MS (Fig. 4; ESI† Fig. S2A).A total of 1223 proteins were identified in both AgNP and AgNO3 exposures. Among those, 383 proteins were selected for further analysis: 267 proteins were specifically present in AgNP-exposed cells and 116 proteins were enriched in AgNP-exposed cells with an enrichment factor of at least two,21 relative to AgNO3 exposures (SI spectral counts of proteins file). An additional experiment was performed to offer further proof of the identity of proteins binding to AgNPs. Here, AgNPs were added to extracts of fractions separated as described above but from untreated cells. After incubation of these extracts with AgNPs or AgNO3, the same protocol was used for AgNP-protein corona separation and protein identification (ESI† Fig. S2B). 236 proteins were specifically enriched in the AgNP-protein corona using the same criteria as above (SI spectral counts of proteins file). Among the proteins detected in the isolations starting from intact exposed cells, 82 proteins were confirmed in this AgNP-extraction experiment. The low percentage of overlap between these two experiments indicates that AgNP–protein interactions in intact cells differ from those possible in a simpler exposure study using extracted cell organelles. This difference is probably due to AgNPs following the endocytosis pathway when interacting with intact fish cells. AgNPs could come into contact with more proteins from the cellular uptake and internal transport pathways than in the cell compartment extracts experiment. Moreover, when intact cells are exposed to AgNP suspensions, both AgNPs and dissolved silver can elicit proteomic responses, while the cell compartment extracts could not mount such proteomic responses.
It is also important to note that it is inherently difficult to isolate pure endo–lysosome fractions. Endosomes and lysosomes are heterogeneous in size and density and therefore the fractions of endosomes, lysosomes, mitochondria and peroxisomes overlap in density gradient centrifugation. Furthermore, lysosomal fractions very likely contain non-degraded or partially degraded macromolecules from other cell compartments.41 Therefore, validation of identified proteins is an important direction for future work.
Identified corona proteins do not have a distinct abundance of cysteine
Given the high affinity of the amino acid cysteine to bind ionic silver, we hypothesized that the identity of corona proteins correlates with their cysteine abundance. Thus, the amino acid composition was compared between unbound proteins (154 proteins) and bound proteins (236 proteins) from the AgNP-extraction experiment (SI cysteine abundance analysis). No statistically relevant trend was observed for linking protein adsorption and cysteine abundance by t-test. This is consistent with a previous study where Eigenheer et al. correlated cysteine abundance in the AgNP-protein corona, isolated from the incubation of a yeast protein extract with AgNPs, with AgNP coatings and particle sizes. They found that cysteine abundance neither was higher in AgNP corona proteins nor corresponded to AgNP coating or size.42Corona proteins allow reconstruction of major paths of AgNP–cell interactions
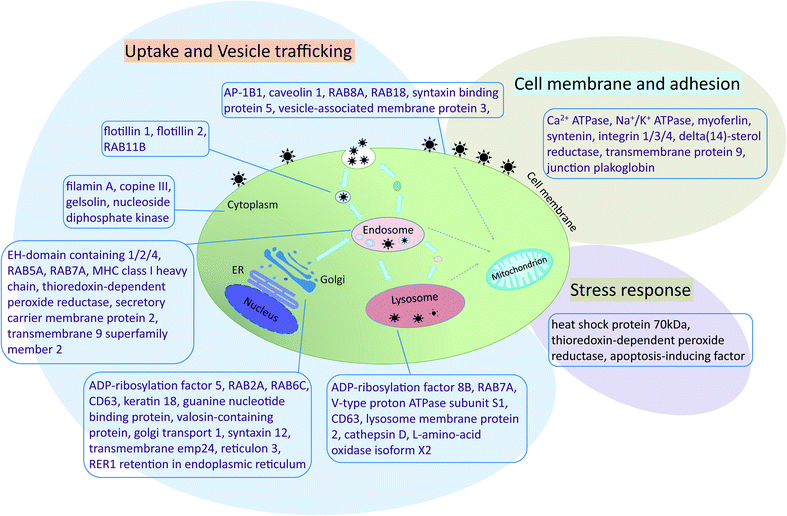
To identify protein functions and potential pathways of the 383 proteins isolated from the AgNPs corona, protein ontology was analyzed using DAVID.43 Proteins not included in protein ontology analysis were checked in UniProt. Proteins identified from the AgNP corona belong to the cell membrane, cytoplasm, endoplasmic reticulum (ER), endosome, Golgi, lysosome, mitochondrion and nucleus (ESI† Tables S4–S8). Two biological pathways were identified that relate to AgNP uptake: endocytosis pathways (GO:0006897 – endocytosis, ESI† Table S6) and vesicle-mediated transport pathways (GO:0016192 – vesicle-mediated transport, ESI† Table S7). Also, a number of stress response proteins were identified. Based on these results, the interaction of AgNPs with RTgill-W1 cells was reconstructed along three main routes of interaction: cell membrane and adhesion, uptake and vesicle trafficking, and stress response (Fig. 6).AP-1B1 is a necessary factor in clathrin-mediated endocytosis by facilitating both the recruitment of clathrin to membranes and the recognition of sorting signals of clathrin-coated vesicles.47 Two other proteins participating in clathrin-coated vesicle formation, namely syntaxin binding protein 5 and RAB5A, were also identified in the AgNP-protein corona. Greulich and colleagues reported that several specific clathrin-mediated endocytosis inhibitors reduced the AgNP uptake by human mesenchymal stem cells.31 However, also caveolin 1, a biomarker of caveolae-mediated endocytosis, was identified in the protein corona. This finding points to a possible clathrin-independent uptake in this study in addition to a clathrin-dependent pathway. Flotillin 1 and flotillin 2 form flotillin vesicles and mediate a clathrin-independent, caveolae-like endocytotic pathway.48,49 Knockdown of these proteins decreased magnetic nanoparticle uptake in HeLa cells.21
Once inside the cells, vesicles carrying AgNPs are apparently transported to different compartments, e.g. early endosome and multi-vesicular bodies. For example, in addition to clathrin-coated vesicle formation, RAB5A is also required for the fusion of plasma membranes and early endosome intracellular membrane trafficking. RAB7A is an important marker of late endosomes and lysosomes and plays a key role in the regulation of endo–lysosomal trafficking.50 Live cell imaging by fluorescence microscopy revealed that polystyrene particles travelled in HeLa Kyoto cells and retinal pigment epithelial cells from early endosomes (marked with fluorescence-labeled RAB5) to late endosomes and lysosomes (marked with fluorescence-labeled RAB7).2,49 RAB5A and RAB7A were also identified in the corona of magnetic nanoparticles separated from HeLa cells.21
AgNPs are transported and stored in late endosomes and lysosomes. Several previous studies showed that nanoparticles co-localize with different endosome or lysosome protein markers.2,21,33 As a confirmation of these findings, we identified a number of proteins associated with the lysosome: lysosome membrane protein 2, cathepsin D, and L-amino-acid oxidase isoform X2 (ESI† Table S8). The vacuolar ATP synthase 16 kDa proteolipid subunit was another important protein identified in the AgNP-protein corona. This synthase is a hydrogen ion transport protein with proton-transporting ATPase activity in lysosomes. The protein is a subunit of the membrane integral V0 complex of vacuolar ATPase (v-ATPase). Coupled with ATP hydrolysis, v-ATPase is responsible for acidifying lysosomes and late endosomes. Indeed, gold nanoparticles were shown to cause lysosome alkalinization through dissociation of v-ATPase in normal rat kidney cells.51 A similar mechanism may therefore have led to the lysosome-specific AgNPs effect previously observed upon one day of exposure of the RTgill-W1 cells to AgNPs.8 Moreover, considering the similarity of proteins identified from coronas of different types of metal-based nanoparticles, it appears that the binding of the endosome- and lysosome-related proteins relates to the physical nature of the particles rather than to their elemental composition.
AgNPs inhibit Na+/K+-ATPase activity – proof of concept on the effect of AgNPs on corona proteins
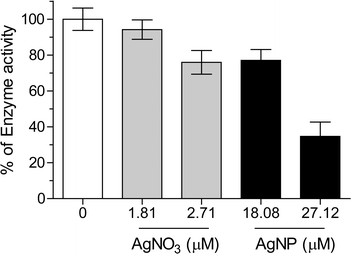
The protocol presented here is the first to recover nanoparticle corona proteins from live cells without relying on magnetism for isolating the nanoparticle-corona complex. The thereby derived protein list provides a valuable resource to study nanoparticle–protein interactions in a targeted way. To demonstrate the utility of this list, we selected Na+/K+-ATPase and explored how AgNPs exposure impacts on the enzymatic activity of the isolated protein. Exposures to AgNO3 were again performed as controls.Indeed, co-exposure of Na+/K+-ATPase with either AgNPs or AgNO3 significantly inhibited Na+/K+-ATPase activity (Fig. 7). A concentration of 2.71 μM AgNO3 led to the same level of inhibition as 18.08 μM AgNPs. Considering that the percentage of dissolved silver in AgNPs suspensions is around 2.37% (ESI† Table S3), the dissolved silver only contributed 16% of inhibition. This finding indicates that the inhibition of enzyme activity is attributable primarily to a particle-specific rather than a dissolved silver ion effect. Schultz reported that citrate-coated AgNP, i.e. the same type of particle as that used in this work, led to significant inhibition of Na+/K+ ATPase activity in juvenile rainbow trout gill in vivo. They found that fish exposed to AgNPs showed a nano-specific effect on sodium influx in the fish.54
Conclusions
This is the first study to reveal the identity of corona proteins from AgNP-exposed living cells. Subcellular fractionation allowed AgNP-protein corona complexes to be recovered from intact subcellular compartments and proteins lysed from the AgNPs to be analyzed. In this way, 383 AgNP corona proteins were identified. This analysis constitutes a snapshot of corona proteins in living cells. Nanoparticle–protein interactions are dynamic55 and we are capturing an early moment of the interaction of the AgNPs with the gill cells. The density gradient centrifugation method coupled with protein identification by mass spectrometry is of great value because it is applicable to any type of metal-based nanoparticle. The list of proteins obtained in this manner can guide subsequent targeted research on protein–nanoparticle interactions, as demonstrated here, for the impact of AgNPs on Na+/K+-ATPase activity. Our study focused on fish gill cells as important gatekeepers for nanoparticle uptake into fish, thereby providing a mechanistic understanding in the context of environmental risk assessment. Yet, other cell types, such as mammalian cells in a medical or human health risk assessment context, can be subjected to the same basic method. In this way, we anticipate that this research not only contributes to a mechanism-based risk assessment but also to safer particle design.Acknowledgements
We thank C. Buser, A. Sologubenko, and M. Günthert (Scientific Center for Optical and Electron Microscopy, ETH Zürich, Switzerland) for EM analysis and D. Kistler (Eawag, Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, Switzerland) for ICP-MS measurements. We thank R. Hamelin and D. Chiappe from the Proteomics Core Facility (PCF, EPFL) for their support in protein identification. This research was supported by the Swiss National Science Foundation in the framework of the Swiss National Research Program 64 (NRP 64) “Opportunities and Risks of Nanomaterials” (Project number: 406440-131240).References
- T.-G. Iversen, T. Skotland and K. Sandvig, Nano Today, 2011, 6, 176–185 CrossRef CAS.
- P. Sandin, L. W. Fitzpatrick, J. C. Simpson and K. A. Dawson, ACS Nano, 2012, 6, 1513–1521 CrossRef CAS PubMed.
- G. Kroemer and M. Jaattela, Nat. Rev. Cancer, 2005, 5, 886–897 CrossRef CAS PubMed.
- L. Rodriguez-Lorenzo, K. Fytianos, F. Blank, C. von Garnier, B. Rothen-Rutishauser and A. Petri-Fink, Small, 2014, 10, 1341–1350 CrossRef CAS PubMed.
- S. Stern, P. Adiseshaiah and R. Crist, Part. Fibre Toxicol., 2012, 9, 20 CrossRef CAS PubMed.
- A. H. Futerman and G. van Meer, Nat. Rev. Mol. Cell Biol., 2004, 5, 554–565 CrossRef CAS PubMed.
- T. Xia, M. Kovochich, M. Liong, J. I. Zink and A. E. Nel, ACS Nano, 2007, 2, 85–96 CrossRef PubMed.
- Y. Yue, R. Behra, L. Sigg, P. Fernández Freire, S. Pillai and K. Schirmer, Nanotoxicology, 2015, 9, 54–63 CrossRef CAS PubMed.
- M. Auffan, J. Rose, J.-Y. Bottero, G. V. Lowry, J.-P. Jolivet and M. R. Wiesner, Nat. Nanotechnol., 2009, 4, 634–641 CrossRef CAS PubMed.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- W. Liu, J. Rose, S. Plantevin, M. Auffan, J.-Y. Bottero and C. Vidaud, Nanoscale, 2013, 5, 1658–1668 RSC.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- C. Ge, J. Tian, Y. Zhao, C. Chen, R. Zhou and Z. Chai, Arch. Toxicol., 2015, 89, 519–539 CrossRef CAS PubMed.
- E. Sanfins, C. Augustsson, B. Dahlbäck, S. Linse and T. Cedervall, Nano Lett., 2014, 14, 4736–4744 CrossRef CAS PubMed.
- J. Wang, U. B. Jensen, G. V. Jensen, S. Shipovskov, V. S. Balakrishnan, D. Otzen, J. S. Pedersen, F. Besenbacher and D. S. Sutherland, Nano Lett., 2011, 11, 4985–4991 CrossRef CAS PubMed.
- N. S. Wigginton, A. D. Titta, F. Piccapietra, J. Dobias, V. J. Nesatyy, M. J. F. Suter and R. Bernier-Latmani, Environ. Sci. Technol., 2010, 44, 2163–2168 CrossRef CAS PubMed.
- F. Bertoli, G.-L. Davies, M. P. Monopoli, M. Moloney, Y. K. Gun'ko, A. Salvati and K. A. Dawson, Small, 2014, 10, 3307–3315 CrossRef CAS PubMed.
- D. Hofmann, S. Tenzer, M. B. Bannwarth, C. Messerschmidt, S.-F. Glaser, H. Schild, K. Landfester and V. Mailänder, ACS Nano, 2014, 8, 10077–10088 CrossRef CAS PubMed.
- N. C. Bols, A. Barlian, M. Chirinotrejo, S. J. Caldwell, P. Goegan and L. E. J. Lee, J. Fish. Dis., 1994, 17, 601–611 CrossRef.
- K. Schirmer, A. G. J. Chan, B. M. Greenberg, D. G. Dixon and N. C. Bols, Toxicol. In Vitro, 1997, 11, 107–119 CrossRef CAS PubMed.
- K. Schirmer, D. G. Dixon, B. M. Greenberg and N. C. Bols, Toxicology, 1998, 127, 129–141 CrossRef CAS PubMed.
- K. Tanneberger, M. Knöbel, F. J. M. Busser, T. L. Sinnige, J. L. M. Hermens and K. Schirmer, Environ. Sci. Technol., 2013, 47, 1110–1119 CrossRef CAS PubMed.
- R. Kinne, J.-E. Schmitz and E. Kinne-Saffran, Pfluegers Arch., 1971, 329, 191–206 CrossRef CAS PubMed.
- H. H. Taussky and E. Shorr, J. Biol. Chem., 1953, 202, 675–685 CAS.
- N. R. Bury, M. Grosell, A. K. Grover and C. M. Wood, Toxicol. Appl. Pharmacol., 1999, 159, 1–8 CrossRef CAS PubMed.
- S. Pillai, R. Behra, H. Nestler, M. J. F. Suter, L. Sigg and K. Schirmer, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3490–3495 CrossRef CAS PubMed.
- N. R. Bury and C. Hogstrand, Environ. Sci. Technol., 2002, 36, 2884–2888 CrossRef CAS PubMed.
- C. Greulich, J. Diendorf, T. Simon, G. Eggeler, M. Epple and M. Köller, Acta Biomater., 2011, 7, 347–354 CrossRef CAS PubMed.
- L. Q. Chen, S. J. Xiao, P. P. Hu, L. Peng, J. Ma, L. F. Luo, Y. F. Li and C. Z. Huang, Anal. Biochem., 2012, 84, 3099–3110 CAS.
- Z. Wang, S. Liu, J. Ma, G. Qu, X. Wang, S. Yu, J. He, J. Liu, T. Xia and G.-B. Jiang, ACS Nano, 2013, 7, 4171–4186 CrossRef CAS PubMed.
- W. Liu, Y. Wu, C. Wang, H. C. Li, T. Wang, C. Y. Liao, L. Cui, Q. F. Zhou, B. Yan and G. B. Jiang, Nanotoxicology, 2010, 4, 319–330 CrossRef CAS PubMed.
- M. M. Fretz, N. A. Penning, S. Al-Taei, S. Futaki, T. Takeuchi, I. Nakase, G. Storm and A. T. Jones, Biochem. J., 2007, 403, 335–342 CrossRef CAS PubMed.
- D. Kühnel, W. Busch, T. Meißner, A. Springer, A. Potthoff, V. Richter, M. Gelinsky, S. Scholz and K. Schirmer, Aquat. Toxicol., 2009, 93, 91–99 CrossRef PubMed.
- M.-H. Jang, W.-K. Kim, S.-K. Lee, T. B. Henry and J.-W. Park, Environ. Sci. Technol., 2014, 48, 11568–11574 CrossRef CAS PubMed.
- G. Griffiths, R. Matteoni, R. Back and B. Hoflack, J. Cell Sci., 1990, 95, 441–461 CAS.
- L. Wang, Y. Liu, W. Li, X. Jiang, Y. Ji, X. Wu, L. Xu, Y. Qiu, K. Zhao, T. Wei, Y. Li, Y. Zhao and C. Chen, Nano Lett., 2010, 11, 772–780 CrossRef PubMed.
- J. García-Alonso, F. R. Khan, S. K. Misra, M. Turmaine, B. D. Smith, P. S. Rainbow, S. N. Luoma and E. Valsami-Jones, Environ. Sci. Technol., 2011, 45, 4630–4636 CrossRef PubMed.
- B. A. Schröder, C. Wrocklage, A. Hasilik and P. Saftig, Proteomics, 2010, 10, 4053–4076 CrossRef PubMed.
- R. Eigenheer, E. R. Castellanos, M. Y. Nakamoto, K. T. Gerner, A. M. Lampe and K. E. Wheeler, Environ. Sci.: Nano, 2014, 238–247 RSC.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Nat. Protoc., 2008, 4, 44–57 CrossRef PubMed.
- P. N. Bernatchez, A. Sharma, P. Kodaman and W. C. Sessa, Am. J. Physiol., 2009, 297, C484–C492 CrossRef CAS PubMed.
- P. Cowin, H.-P. Kapprell, W. W. Franke, J. Tamkun and R. O. Hynes, Cell, 1986, 46, 1063–1073 CrossRef CAS PubMed.
- B. B. Manshian, C. Pfeiffer, B. Pelaz, T. Heimerl, M. Gallego, M. Möller, P. del Pino, U. Himmelreich, W. J. Parak and S. J. Soenen, ACS Nano, 2015, 9, 10431–10444 CrossRef CAS PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- M. Meister and R. Tikkanen, Membranes, 2014, 4, 356–371 CrossRef PubMed.
- D. Vercauteren, H. Deschout, K. Remaut, J. F. J. Engbersen, A. T. Jones, J. Demeester, S. C. De Smedt and K. Braeckmans, ACS Nano, 2011, 5, 7874–7884 CrossRef CAS PubMed.
- B. D. Grant and J. G. Donaldson, Nat. Rev. Mol. Cell Biol., 2009, 10, 597–608 CrossRef CAS PubMed.
- X. Ma, Y. Wu, S. Jin, Y. Tian, X. Zhang, Y. Zhao, L. Yu and X.-J. Liang, ACS Nano, 2011, 5, 8629–8639 CrossRef CAS PubMed.
- E. Farmen, H. N. Mikkelsen, Ø. Evensen, J. Einset, L. S. Heier, B. O. Rosseland, B. Salbu, K. E. Tollefsen and D. H. Oughton, Aquat. Toxicol., 2012, 108, 78–84 CrossRef CAS PubMed.
- I. Passagne, M. Morille, M. Rousset, I. Pujalté and B. L'Azou, Toxicology, 2012, 299, 112–124 CrossRef CAS PubMed.
- A. G. Schultz, K. J. Ong, T. MacCormack, G. Ma, J. G. C. Veinot and G. G. Goss, Environ. Sci. Technol., 2012, 46, 10295–10301 CrossRef CAS PubMed.
- P. D. Pino, B. Pelaz, Q. Zhang, P. Maffre, G. U. Nienhaus and W. J. Parak, Mater. Horiz., 2014, 1, 301–313 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6en00119j |
| ‡ Current address: Wattstr. 13a, 8307 Effretikon, Switzerland. |
| This journal is © The Royal Society of Chemistry 2016 |