Open Access Article
Open Access ArticleAntiproliferative activity of ruthenium(II) arene complexes with mono- and bidentate pyridine-based ligands†
Stefan
Richter
a,
Sushma
Singh
b,
Dijana
Draca
c,
Anup
Kate
b,
Anupa
Kumbhar
b,
Avinash S.
Kumbhar
*b,
Danijela
Maksimovic-Ivanic
c,
Sanja
Mijatovic
c,
Peter
Lönnecke
a and
Evamarie
Hey-Hawkins
*a
aUniversität Leipzig, Institut für Anorganische Chemie, Johannisallee 29, 04103 Leipzig, Germany. E-mail: hey@uni-leipzig.de
bDepartment of Chemistry, Savitribai Phule Pune University, Pune-411007, India
cInstitute for Biological Research “Sinisa Stankovic”, University of Belgrade, Bulevar despota Stefana 142, 11060 Belgrade, Serbia
First published on 24th May 2016
Abstract
A series of RuII arene complexes of mono- and bidentate N-donor ligands with carboxyl or ester groups and chlorido ancillary ligands were synthesised and structurally characterised. The complexes have a distorted tetrahedral piano-stool geometry. The binding interaction was studied with calf thymus DNA (CT-DNA) by absorption titration, viscosity measurement, thermal melting, circular dichroism, ethidium bromide displacement assay and DNA cleavage of plasmid DNA (pBR322), investigated by gel electrophoresis. The dichlorido complexes bind covalently to DNA in the dark, similar to cisplatin, while the monochlorido complexes bind covalently on irradiation, similar to cisplatin analogues. The compounds are selectively cytotoxic against several tumour cell lines and show specific nonlinear correlation between dose and activity. This phenomenon is closely related to their potential to act preferentially as inhibitors of cell division.
Introduction
Ruthenium complexes are emerging as promising alternatives to highly active but toxic platinum complexes as anticancer drugs,1 as corroborated by successful clinical trials of trans-[tetrachlorido(dimethyl sulfoxide)imidazole ruthenium(III)] (NAMI-A),2–9 indazolium trans-[tetrachloridobis(1H-indazole)ruthenium(III)] (KP1019)10–14 and sodium trans-[tetrachloridobis(1H-indazole)ruthenate(III)] (NKP1339).15,16 The mechanism involves the in vivo reduction of the RuIII prodrugs to the active RuII complexes in the hypoxic tumour cells at lower pH, which then bind to specific biomolecules to provide selective toxicity.17,18 The inherent properties of ruthenium, that is, variable oxidation states, stability in air, relative ease of preparation and purification, slow in vivo ligand exchange and lower toxicity, seem to be favourable compared to platinum. Several RuII polypyridyl complexes which were initially developed as structure- and site-specific reversible DNA binding agents more recently found applications as cellular imaging agents.19–21 The most widely studied organometallic ruthenium compounds are ruthenium arene and ruthenium cyclopentadienyl half-sandwich compounds with a piano-stool geometry, due to their structural diversity and varied binding modes to DNA. The first compound of this type was synthesised by Dale et al. by coordinating the known anticancer agent 1-β-hydroxyethyl-2-methyl-5-nitro-imidazole (metronidazole) to a ruthenium(II) benzene dichloride fragment resulting in better activity than metronidazole itself in an in vitro assay.22 This was followed by Sheldrick et al., who demonstrated that complexes of the type [RuII(η6-arene)(LL)X] (where LL = L-alanine and L-alanine methyl ester and X = halide) are coordinated by nitrogen atom N7 of guanine derivatives.23 Thereafter, in pioneering work Sadler et al.24–27 and Dyson et al.28–31 developed structure–activity relationships using [RuII(η6-arene)(en)Cl]+ (arene = biphenyl (Biph), tetrahydroanthracene (THA), dihydroanthracene (DHA), para-cymene (p-cym) or benzene; en = ethylenediamine) and the water-soluble [RuII(η6-arene)(pta)Cl2] complexes (pta = 1,3,5-triaza-7-phosphatricyclo[3.3.1.1]-decane), respectively, which are considered to be the prototypes of anticancer RuII arene complexes. Espino et al. have derived extensive structure–activity relationships for the anticancer properties of RuII arene complexes with 2,4-diamino-(2-pyridyl)-1,3,5-triazine,32,33 phenanthrolines,34 aminophosphines,35 benzimidazole35 and 2-aryldiazole.36 Recently, Wang et al. introduced Ru(arene)/BODIPY (BODIPY = boron dipyrromethene) hybrids, which on irradiation undergo fast release of BODIPY facilitating covalent binding of the resulting RuII complex fragment to DNA, resulting in high 1O2 quantum yields and DNA cleavage, and can thus act as a potential photoactivated anticancer agent (PACT).37 As shown by Mukherjee et al., the sterically encumbered imidazole-based Schiff base ligand N-[(1H-imidazol-2-yl)methylene]-2,6-diisopropylaniline slowed the hydrolysis, and the corresponding ruthenium complex [RuCl(p-cym){N-{(1H-imidazol-2-yl)methylene}-2,6-diisopropylaniline}] exhibited better activity under hypoxia, strong resistance to glutathione in vitro and thus strong anticancer activity.38 Dyson et al. recently demonstrated that the coordinating mode (N,N versus N,O) and the substituents on N-phenylpicolinamide39 or β-ketonamide40 ligands drastically affect the biological activity of the corresponding complexes. Complexes with N,N-coordinating ligands are hydrolysed quickly, show selective binding to guanine and therefore are cytotoxic, while complexes with N,O-coordinating ligands are not hydrolysed and therefore cannot be coordinated by guanine and are non-toxic.For this reason, complexes of mono- (1–3) and bidentate (4–6) N-donor ligands (A–E) with free carboxyl acid and ester groups (Scheme 1) were synthesised, and their binding interaction with DNA was evaluated by absorption titration, viscosity measurement, thermal melting, circular dichroism and ethidium bromide displacement assay. DNA cleavage was studied by gel electrophoresis, and cytotoxicity against human thyroid (8505C), melanoma (518A2), breast (MCF-7) and colon (SW-480) tumour cell lines was evaluated. Cytotoxic compounds with free carboxyl groups are potentially interesting for conjugation to tumour-targeting peptides for achieving better tumour selectivity.41,42
Experimental section
Materials
RuCl3·3H2O was purchased from Johnson Matthey. The precursor complex [{RuCl2(p-cym)}2] (p-cym = 1-iPr-4-Me-C6H4)43 and ligands B,44C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44 and D
44 and D![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 were prepared according to literature procedures. Yields, properties, NMR (1H, 13C) and IR data of B–D are given in the ESI.† The ligand 4-aminobenzoic acid (A, 4-ABA) was purchased from Sigma Aldrich. Calf thymus DNA (CT-DNA) and plasmid pBR322 DNA were purchased from SRL (India). Supercoiled pBR322 DNA (CsCl) purified was obtained from Bangalore Genei (Bangalore, India) and used as received. Ethidium bromide (EtBr) was purchased from S.D. Fine Chemicals, Mumbai, India. Deionised water was used for the preparation of the buffer solutions.
45 were prepared according to literature procedures. Yields, properties, NMR (1H, 13C) and IR data of B–D are given in the ESI.† The ligand 4-aminobenzoic acid (A, 4-ABA) was purchased from Sigma Aldrich. Calf thymus DNA (CT-DNA) and plasmid pBR322 DNA were purchased from SRL (India). Supercoiled pBR322 DNA (CsCl) purified was obtained from Bangalore Genei (Bangalore, India) and used as received. Ethidium bromide (EtBr) was purchased from S.D. Fine Chemicals, Mumbai, India. Deionised water was used for the preparation of the buffer solutions.
Physical measurements
All complexation reactions were performed under N2 atmosphere by using standard Schlenk techniques. Solvents were dried with the solvent purification system SPS-800 SERIES (company MBRAUN). NMR spectra (1H, 13C) were recorded at 27 °C on a Bruker 300 or DRX 400 spectrometer. Chemical shifts are relative to internal standard (TMS). Numbering schemes are included in Scheme 1. IR spectra of the complexes were recorded as KBr disks on a Perkin Elmer FTIR Spektrum 2000 spectrometer. The elemental analyses were recorded on a Heraeus Vario Analyser and the melting points were determined in capillaries using a Gallenkamp instrument. Mass spectra were obtained as ESI MS with an FT ICR mass spectrometer (Bruker Daltonics).![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) (cm−1) 3384 (s), 2963 (w), 2545 (w), 2363 (w), 1667 (s), 1606 (s), 1528 (m), 1474 (m), 1435 (m), 1417 (m), 1316 (m), 1290 (s), 1175 (s), 1089 (w), 840 (w), 776 (m), 765 (m).
(cm−1) 3384 (s), 2963 (w), 2545 (w), 2363 (w), 1667 (s), 1606 (s), 1528 (m), 1474 (m), 1435 (m), 1417 (m), 1316 (m), 1290 (s), 1175 (s), 1089 (w), 840 (w), 776 (m), 765 (m).
Synthesis of complexes 1–6
13C{1H} NMR (75 MHz, DMSO-d6): δ 14.3 (s, CH3a), 17.8 (s, CH3b), 21.5 (s, CH3c), 39.9 (s, CHd), 60.0 (s, CH2e), 86.5 (s, CHf), 86.3 (s, CHg), 100.1 (s, Ch), 106.4 (s, Cj), 111.8 (s, CHk), 115.5 (s, CHl), 116.5 (s, CHm), 120.8 (s, Cn), 130.3 (s, Co), 137.6 (s, CHp), 146.2 (s, CHq), 147.2 (s, CHr), 155.1 (s, Cs), 165.6 (s, CO). IR: ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) (cm−1) 3444 (s), 3222 (s), 1712 (s), 1603 (s), 1568 (m), 1528 (m), 1464 (m), 1445 (m), 1344 (w), 1277 (s), 1177 (w), 1162 (w), 1108 (m), 1023 (w). ESI MS (pos. mode): m/z: 571.1 ([RuCl2(p-cym)(B) + Na]+).
(cm−1) 3444 (s), 3222 (s), 1712 (s), 1603 (s), 1568 (m), 1528 (m), 1464 (m), 1445 (m), 1344 (w), 1277 (s), 1177 (w), 1162 (w), 1108 (m), 1023 (w). ESI MS (pos. mode): m/z: 571.1 ([RuCl2(p-cym)(B) + Na]+).
Crystals of 2 were obtained from a saturated solution of 2 in methanol by slow evaporation of solvent at room temperature in air over 1 d.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) (cm−1) 3420 (s, br), 2962 (s), 2380 (w), 2370 (w), 1868 (w), 1844 (w), 1772 (w), 1716 (s), 1698 (s), 1683 (s), 1652 (s), 1635 (s), 1602 (s), 1522 (s), 1507 (s), 1457 (s), 1387 (m), 1338 (m), 1261 (s), 1175 (s), 1103 (s). ESI MS (pos. mode): m/z: 483.1 ([RuCl(p-cym)(C-2H)]+).
(cm−1) 3420 (s, br), 2962 (s), 2380 (w), 2370 (w), 1868 (w), 1844 (w), 1772 (w), 1716 (s), 1698 (s), 1683 (s), 1652 (s), 1635 (s), 1602 (s), 1522 (s), 1507 (s), 1457 (s), 1387 (m), 1338 (m), 1261 (s), 1175 (s), 1103 (s). ESI MS (pos. mode): m/z: 483.1 ([RuCl(p-cym)(C-2H)]+).
Crystals of 3 were obtained from a saturated solution of 3 in chloroform by slow evaporation of solvent at room temperature in air.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) (cm−1) 3418 (br, s), 3270 (s), 2960 (s), 1757 (s), 1736 (m), 1615 (m), 1432 (m), 1389 (m), 1360 (m), 1285 (m), 1237 (s), 1200 (s), 1095 (m), 1027 (m), 835 (s). ESI MS (pos. mode): m/z: 451.1 ([RuCl(p-cym)(DMe)]+).
(cm−1) 3418 (br, s), 3270 (s), 2960 (s), 1757 (s), 1736 (m), 1615 (m), 1432 (m), 1389 (m), 1360 (m), 1285 (m), 1237 (s), 1200 (s), 1095 (m), 1027 (m), 835 (s). ESI MS (pos. mode): m/z: 451.1 ([RuCl(p-cym)(DMe)]+).
Crystals of 4 were obtained from a solution of 4 in methanol by slow evaporation of solvent at room temperature in air.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) (cm−1): 3418 (m, br), 2322 (w), 2929 (w), 1723 (m), 1708 (m), 1601 (w), 1475 (w), 1417 (w), 1105 (w), 839 (s). ESI MS (pos. mode): m/z: 497.1 ([RuCl(p-cym)(E-2H)]+).
(cm−1): 3418 (m, br), 2322 (w), 2929 (w), 1723 (m), 1708 (m), 1601 (w), 1475 (w), 1417 (w), 1105 (w), 839 (s). ESI MS (pos. mode): m/z: 497.1 ([RuCl(p-cym)(E-2H)]+).
X-ray data collection and structure refinement
The data of complexes 2 to 6 were collected at 130 K on a Gemini diffractometer (Rigaku Corp.) by using Mo-Kα radiation (λ = 71.073 pm) and ω-scan rotation. Data reduction was performed with CrysAlis Pro47 including the program SCALE3 ABSPACK for empirical absorption correction. All structures were solved by direct methods,48,49 and the refinement of all non-hydrogen atoms was performed with SHELXL-2013 or SHELXL-2014.49 Excluding strongly overlapping disordered parts of the structures, all non-hydrogen atoms were refined with anisotropic thermal parameters. A difference-density Fourier map was used to locate all hydrogen atoms of compound 6 and OH and NH hydrogen atoms of 3 and 4. All other H atoms were calculated on idealised positions by using the riding model. Structure figures were generated with DIAMOND-3.50 CCDC 1421200 (2), 1421201 (3), 1421202 (4), 1421203 (5), and 1421204 (6) contain the supplementary crystallographic data for this paper.DNA binding studies
Absorption spectra were measured with a Shimadzu 1800 spectrophotometer. The concentration of calf thymus DNA was determined by UV absorbance at 260 nm by taking the molar absorption coefficient as 6600 M−1 cm−1. Solutions of CT-DNA in phosphate buffer gave a ratio of UV absorbance at 260 and 280 nm, A260/A280, of 1.8–1.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, indicating that the DNA was sufficiently free of protein.46 Absorption titration experiments were performed by maintaining a constant metal-complex concentration (100 μM) and varying nucleotide concentration (0–200 μM) in phosphate buffer (pH = 7.2). After addition of DNA to the metal complex, absorption readings were noted. The emission spectra were measured with a Jasco spectrofluorometer; the excitation slit widths employed were 5 nm each. The change in emission intensity of metal complex at fixed metal concentration (100 μM) with increasing concentration of DNA was measured on excitation at λmax (260–380 nm).
1, indicating that the DNA was sufficiently free of protein.46 Absorption titration experiments were performed by maintaining a constant metal-complex concentration (100 μM) and varying nucleotide concentration (0–200 μM) in phosphate buffer (pH = 7.2). After addition of DNA to the metal complex, absorption readings were noted. The emission spectra were measured with a Jasco spectrofluorometer; the excitation slit widths employed were 5 nm each. The change in emission intensity of metal complex at fixed metal concentration (100 μM) with increasing concentration of DNA was measured on excitation at λmax (260–380 nm).
The mode of binding was also studied by a competitive binding assay with ethidium bromide (EtBr)-bound DNA in phosphate buffer (pH = 7.2). The emission was observed at 580 nm on excitation at 530 nm. Ethidium bromide in phosphate buffer shows quenched emission intensity, but in the presence of DNA, EtBr shows enhanced emission intensity.
The viscosity measurements were carried out with a Schott-Geräte ViscoSystem AVS 370 maintained at 28.0 ± 1 °C. Flow time of solutions in phosphate buffer (pH = 7.2) was recorded in triplicate for each sample with a digital stop watch, and an average flow time was calculated. Data were presented as (η/η0)1/3versus binding ratio, where η is the viscosity of DNA in the presence of the complex and η0 is the viscosity of DNA alone.
DNA melting experiments were carried out by monitoring the absorption at 260 nm of CT-DNA (100 μM) with a Jasco V-630 spectrophotometer equipped with a Peltier temperature-controlling programmer ETC-717 (0.1 °C) in phosphate buffer at various temperatures in the absence and presence of the complexes (20 μM). UV melting profiles were obtained by scanning A260 absorbance monitored at a heating rate of 1 °C min−1 for solutions of CT-DNA (100 μM) in the temperature range 20–90 °C. The melting temperature Tm which is defined as the temperature at which half of the total base pairs are unbound, was determined from the midpoint of the melting curves.
The CD spectra of CT-DNA (20 μM) were monitored in absence and presence of metal complex in phosphate buffer by using a Jasco J-1500 spectrophotometer.
DNA cleavage studies
The electrophoretic mobility experiments were carried out by agarose gel electrophoresis51–53 on a 10 μl total sample volume solution containing pBR322 DNA (200 ng) and the respective ruthenium(II) complex. Stock solutions (10 μM and 20 μM) of 1–4 and 6 were prepared in 2% DMSO and water. Supercoiled pBR322 DNA was treated with the complexes (2–200 μM) and the mixtures were incubated in the dark for 30 min at 37 °C. The samples were analysed by 1% agarose gel electrophoresis [TBE buffer (TBE = TRIS/borate/EDTA, TRIS = tris(hydroxymethyl)aminomethane), pH = 7.8] for 3 h at 60 V. The gel was stained with a 0.5 μg ml−1 ethidium bromide, visualised by UV light and photographed for analysis with an Alpha Innotech Gel documentation system (Alphamager 2200).Cytotoxicity studies
Foetal calf serum (FCS), RPMI-1640 medium, phosphate-buffered saline (PBS), dimethylformamide (DMF) and propidium iodide (PI) were purchased from Sigma (St. Louis, MO). Annexin V-FITC (AnnV) was purchased from Biotium (Hayward, CA) and Apostat from R&D (R&D Systems, Minneapolis, MN USA). Human 518A2, 8505C, MCF-7 and SW-480 were cultivated in HEPES-buffered RPMI-1640 medium supplemented with 10% FCS, 2 mM L-glutamine, 0.01% sodium pyruvate and antibiotics (culture medium) at 37 °C in a humidified atmosphere with 5% CO2. After standard trypsinisation, cells were seeded at (1–2) × 103 cells per well in 96-well plates for viability determination and 2 × 105 cells per well in 6-well plates for flow-cytometric analysis. All compounds were dissolved in DMF immediately before treatment and solutions used were prepared in 10% FCS-RPMI-1640. Controls were exposed to equivalent amounts of DMF in culture medium.Sulforhodamine B test (SRB test)
For evaluation of the effect of complexes 1–4, 6 and ligands A–E on the viability of tumour cells, an SRB test was used.54 Tumour cells were treated for 96 h with a wide range of concentrations of these compounds. At the end of cultivation, cells were fixed with 10% of trichloroacetic acid (TCA) for 2 h at 4 °C. After fixation, cells were washed with distilled water and additionally stained for 30 min at room temperature with 0.4% SRB solution. Then the cells were washed with 1% acetic acid and dried overnight. The dye was dissolved in 10 mM TRIS buffer, and after 20 min of incubation at room temperature, the absorbance was measured at 540 nm with the reference wavelength at 670 nm. Results were expressed as percentage of untreated cells (control) and presented as mean ± standard deviation (SD).AnnexinV-FITC/PI and caspase detection
Cells were incubated with a 2× IC50 dose of a selected complex (2) for 72 h. At the end of cultivation, cells were stained with AnnV-FITC/PI or Apostat according to the manufacturer's instructions. Cells were analysed with CyFlow® Space Partec by using the Partec FloMax® software.55Cell staining with carboxyfluorescein succinimidyl ester (CFSE)
Before exposure to complex 2, cells were stained with 1 μM of CFSE for 10 min at 37 °C.56 Thereafter, cells were washed and additionally cultivated for 72 h with an IC50 dose of 2. Finally, cells were examined with CyFlow® Space Partec using Partec FloMax® software.55Statistical analysis
Results are presented as mean ± standard deviation (SD) of triplicate observations obtained from three individually repeated experiments. The significance of the differences between control and treated culture was evaluated by two-tailed Student's t-test. A p value of less than 0.05 was considered significant.Results and discussion
Synthesis
The ruthenium complex [{RuCl2(p-cym)}2] is an excellent starting material for synthesis of novel RuII complexes with mono- or bidentate amine or phosphine ligands.57,58 Complex 1 was prepared according to a literature procedure and the identity and purity were verified by 1H and 13C NMR spectroscopy;46 complexes 2–5 were synthesised accordingly by treating [{RuCl2(p-cym)}2] with the corresponding amine ligand B–E under inert atmosphere (Scheme 1). Complexes 2 and 3 form orange solids within several minutes even at room temperature, underlining the reactivity of the ruthenium complex [{RuCl2(p-cym)}2] toward Lewis bases such as amines. In air, complex 5 reacts with oxygen with elimination of hydrogen and formation of water to give complex 6, which exhibits an imine group. Gomez et al. have described similar ruthenium complexes [Ru(p-cym)(pyridine-NH-arene)] and their oxidation to similar products.59 The products were characterised by 1H and 13C{1H} NMR spectroscopy, IR spectroscopy and ESI MS, and the single-crystal X-ray structures of 2–6 were obtained. The spectroscopic data are as expected.57,58Stock solutions of the compounds in DMSO were stored at 4 °C to ensure the stabilities of the investigated solutions. Furthermore, the stock solutions were checked by time-resolved UV/VIS measurements before use.
Molecular structures of complexes 2–6
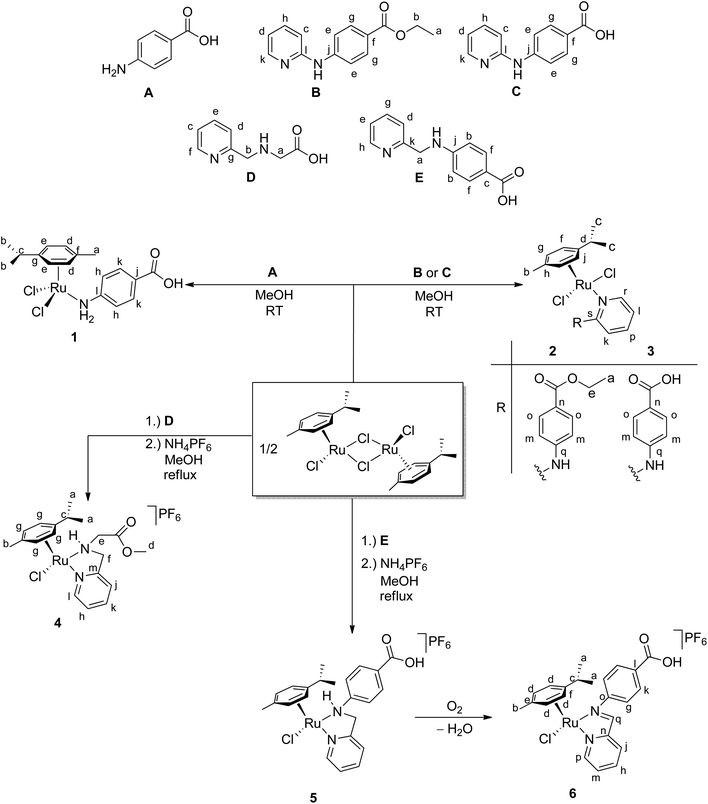
Single crystals suitable for X-ray structure analysis were obtained for complexes 2–6 (Table SI-1, ESI†). Complexes 2 and 3 with a monodentate N-donor ligand crystallised from methanol (compound 2) as light red needles in the monoclinic space group P21/c (2, Fig. 1a) or from chloroform (compound 3) as red needles in the triclinic space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) with two molecules of chloroform per formula unit (3·2CHCl3, Fig. 1b). Selected bond lengths and angles of 2 and 3 are listed in Table 1. In both complexes the ruthenium atom is coordinated by para-cymene, the monodentate amine ligand B or C and two chlorido ligands in a typical distorted piano-stool geometry. The environment of the RuII atom in 2 and 3 (Table 1) is similar to those previously found in related complexes,57e.g., [RuCl2(p-cym)(2-aminopyridine)].60 Furthermore, a hydrogen bond [Cl2⋯N2 3.090(4) (for 2) and 3.076(3) Å (for 3)] between the chlorido ligand and the NH proton is observed.60 In 3, centrosymmetric dimers are formed by moderately strong hydrogen bonding between the carboxyl groups (O1⋯O2′ 2.599(4) Å).61
with two molecules of chloroform per formula unit (3·2CHCl3, Fig. 1b). Selected bond lengths and angles of 2 and 3 are listed in Table 1. In both complexes the ruthenium atom is coordinated by para-cymene, the monodentate amine ligand B or C and two chlorido ligands in a typical distorted piano-stool geometry. The environment of the RuII atom in 2 and 3 (Table 1) is similar to those previously found in related complexes,57e.g., [RuCl2(p-cym)(2-aminopyridine)].60 Furthermore, a hydrogen bond [Cl2⋯N2 3.090(4) (for 2) and 3.076(3) Å (for 3)] between the chlorido ligand and the NH proton is observed.60 In 3, centrosymmetric dimers are formed by moderately strong hydrogen bonding between the carboxyl groups (O1⋯O2′ 2.599(4) Å).61
| 2 | 3 | |
|---|---|---|
| Ru–N1 | 2.172(3) | 2.168(4) |
| Ru–N2 | 3.231(3) | 3.234(4) |
| Ru–Cl1 | 2.4343(8) | 2.407(1) |
| Ru–Cl2 | 2.4059(9) | 2.440(1) |
| Ru–C1 | 2.198(5) | 2.195(3) |
| Ru–C2 | 2.169(5) | 2.183(3) |
| Ru–C3 | 2.178(5) | 2.181(3) |
| Ru–C4 | 2.197(5) | 2.197(4) |
| Ru–C5 | 2.176(5) | 2.167(3) |
| Ru–C6 | 2.175(5) | 2.154(3) |
| Ru–Cen(p-cym) | 1.6626(4) | 1.6606(3) |
| Cl2⋯N2 | 3.090(4) | 3.075(3) |
| O1⋯O2′ | — | 2.599(4) |
| Cl1–Ru–Cl2 | 85.46(4) | 85.46(4) |
| Cl1–Ru–N1 | 88.7(1) | 86.41(8) |
| Cl2–Ru–N1 | 89.2(1) | 90.02(8) |
| N1–Ru–Cen(p-cym) | 125.6(1) | 125.54(8) |
| Cl2–Ru–Cen(p-cym) | 127.23(3) | 127.81(3) |
| Cl1–Ru–Cen(p-cym) | 127.65(3) | 128.27(3) |
| N1–C15–N2 | 115.6(4) | 115.6(3) |
| Ru–N1–C11 | 117.6(3) | 123.4(3) |
| Ru–N1–C15 | 117.1(2) | 123.9(2) |
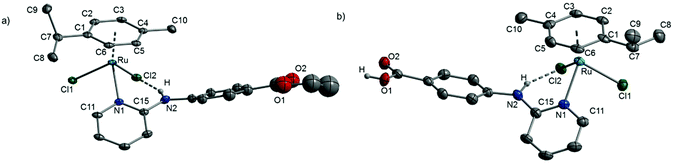
Complexes 4, 5 and 6 with a bidentate N-donor ligand crystallised from methanol (Table SI-1, ESI†). [RuCl(p-cym){D(Me)}]PF6 (4) crystallised as dark-red needles in the orthorhombic space group Pna21. Esterification had occurred in refluxing methanol (Fig. 2a, Table 2). An intramolecular hydrogen bond (Cl1⋯H 2.0(1) Å) between the chlorido ligand and the NH proton is observed.60 Unexpectedly, orange crystals obtained on recrystallising 5 from methanol in inert atmosphere at −4 °C after 48 h contained two different cations: two pseudo-symmetry-related cations of the target compound [RuCl(p-cym)(E)]+ (5+) (Fig. 2b) with hydrogen bonds between the carboxyl groups (O1⋯O4 2.575(4), O2⋯O3 2.592(5) Å) (Table 2), and the dinuclear complex [Ru2Cl3(p-cym)2]+ (I+). [Ru2Cl3(p-cym)2]+ (I+) is located on a crystallographic inversion centre with disordered chlorine atoms. The cation was described previously.43,62 The compound crystallises in the monoclinic space group P21/c as [RuCl(p-cym)(E)]2[Ru2Cl3(p-cym)2][PF6]3·1MeOH (5+I+[PF6]3·1MeOH) with four formula units in the unit cell. The dimers of the cation [RuCl(p-cym)(E)]+ (5+) are located on a non-crystallographic centre of inversion. For this compound, pseudo-translation symmetry of a/2 is detectable with the exception of the non-disordered PF6− anion [P(1), F(1) to F(6)] and the methanol molecule [C(67) and O(5)]. An originally determined smaller a axis led to unacceptable structure parameters and a wrong stoichiometry because of an overlap of the hidden methanol molecule with a PF6− anion. By considering relatively weak reflections, the correct a axis could be determined.
| 4 | 5 | 6 | |
|---|---|---|---|
| Ru–N1 | 2.094(3) | 2.102(3) [2.088(3)] | 2.082(1) |
| Ru–N2 | 2.12(2) | 2.156(3) [2.154(3)] | 2.089(2) |
| Ru–Cl | 2.51(1) | 2.404(1) [2.407(1)] | 2.3939(5) |
| Ru–C1 | 2.224(4) | 2.235(4) [2.224(4)] | 2.215(2) |
| Ru–C2 | 2.199(4) | 2.192(4) [2.203(4)] | 2.196(2) |
| Ru–C3 | 2.200(4) | 2.191(4) [2.184(4)] | 2.210(2) |
| Ru–C4 | 2.204(4) | 2.199(4) [2.194(4)] | 2.202(2) |
| Ru–C5 | 2.171(4) | 2.181(4) [2.177(4)] | 2.179(2) |
| Ru–C6 | 2.193(4) | 2.191(4) [2.186(4)] | 2.193(2) |
| Ru–Cen(p-cym) | 1.6831(3) | 1.6856(4) [1.677(4)] | 1.6841(3) |
| C16–N2 | 1.52(2) | 1.478(5) [1.496(5)] | 1.290(2) |
| H⋯Cl | 2.0(1) | — | — |
| O1⋯O4 | — | 2.575(4) [2.592(5)] | — |
| O1⋯O3 | — | — | 2.618(3) |
| N1–Ru–N2 | 79.9(4) | 75.3(1) [75.7(1)] | 76.92(6) |
| N1–Ru–Cl | 86.1(2) | 85.6(1) [85.1(1)] | 85.68(4) |
| N2–Ru–Cl | 79.9(6) | 82.29(9) [83.76(9)] | 85.48(4) |
| Cl–Ru–Cen(p-cym) | 120.3(2) | 128.11(3) [128.72(3)] | 127.53(2) |
| N1–Ru–Cen(p-cym) | 132.82(8) | 131.82(9) [131.74(1)] | 131.18(4) |
| N2–Ru–Cen(p-cym) | 138.5(5) | 133.31(9) [133.73(9)] | 132.17(4) |
Complex 6 crystallised from methanol in air as red needles in the monoclinic space group P21/n with one molecule of methanol per formula unit. In contrast to 5+, no dimers are formed via carboxyl groups; here, the carboxyl group interacts with a methanol molecule via hydrogen bonding (O1⋯O3 2.618(2) Å) (Fig. 2c, Table 2).
Complexes 4+, 5+ and 6+ (Fig. 2) exhibit the typical distorted tetrahedral three-legged piano-stool geometry with η6-coordinated p-cymene, the chelating amine D(Me) (4+), E (5+) or E-2H (6+) and one chlorido ligand. Bond lengths and angles at RuII in 4+–6+ are very similar. Complexes 4+ and 5+ have two chiral centres (Ru and N2). In the examined crystals, only the rac isomers are present (RRu,RN or SRu,SN). Complex 6+ has only one chiral centre (Ru; RRu or SRu). In the air-sensitive dimeric cationic complex 5+, the five-membered rings exhibit C–N bond lengths of 1.478(5) Å (C16–N2) and 1.496(5) Å (C39–N4), which are in the typical range of a C–N single bond, whereas the C16-N2 bond length in 6+ of 1.290(2) Å is indicative of a double bond. Chow et al. reported a comparable RuII complex with a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N double bond of 1.293(6) Å.63
N double bond of 1.293(6) Å.63
Biological investigations
Electronic absorption spectroscopy is one of the most useful techniques for studying DNA binding by metal complexes. Intercalation of a complex in DNA results in hypochromism and a redshift,64 while for non-intercalative binding (covalent, electrostatic and hydrogen bonding) no hypochromism or redshift is observed. The absorption spectra of complexes 1–4 and 6 in the absence and presence of CT-DNA show negligible change (Fig. SI-1†) suggesting non-intercalative binding.
Competitive binding studies with ethidium bromide (EtBr)-bound DNA were performed to further elucidate the binding nature of complexes 1–4 and 6. Binding of a second molecule to DNA by stacking interactions between adjacent DNA base pairs displaces the ethidium bromide and enhances the emission intensity. However, with complexes 1–4 and 6, no enhancement in the emission intensity is observed, indicating non-intercalative binding (Fig. SI-2†).
CT-DNA melts at 58.00 ± 1 °C (phosphate buffer, pH = 7.2) in the absence of complex. The melting temperature of DNA increased by up to 3 °C on interaction with complexes 1–4 and 6, which also indicates non-intercalative binding (Table SI-2†).
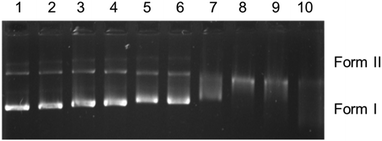
Hydrodynamic measurements are sensitive to changes in length of DNA and considered to be the most critical and least ambiguous tests for evaluating binding modes in solution. The relative specific viscosities of DNA in the absence and presence of complexes 1–4 and 6 plotted against [complex]/[DNA] are shown in (Fig. 3). The relative viscosities of CT-DNA bound complexes increased negligibly, in contrast to the known intercalator ethidium bromide, and this suggests covalent binding of complexes 1–4 and 6 with CT-DNA.
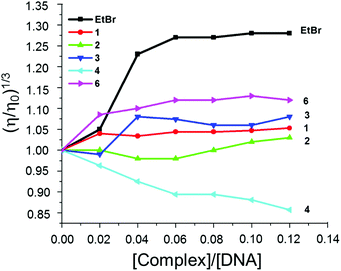 | ||
Fig. 3 Effect of increasing amount of EtBr (■), 1 ( ), 2 ( ), 2 ( ), 3 ( ), 3 ( ), 4 ( ), 4 ( ), 6 ( ), 6 ( ) on the relative viscosity of CT-DNA at 28 ± 1 °C, [DNA] = 300 μM. ) on the relative viscosity of CT-DNA at 28 ± 1 °C, [DNA] = 300 μM. | ||
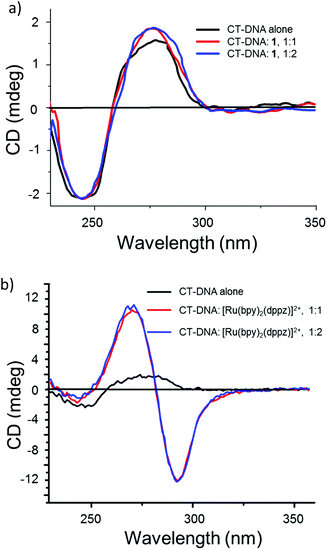
Circular dichroism (CD) spectroscopy was used to monitor the conformational change of CT-DNA on addition of complexes 1–4 and 6. There was no change in the CD spectrum of CT-DNA (20 μM) on addition of complexes 1–4 and 6, while a drastic change in the CD spectrum occurred when a known intercalator, namely, [Ru(bpy)2(dppz)]2+,65 was added (Fig. 4 and SI-3, ESI†) confirming the non-intercalative and, probably, covalent binding of these complexes, which was investigated further by electrophoretic mobility studies.
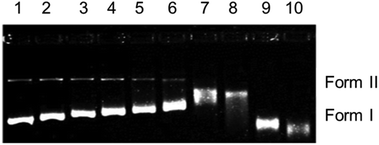
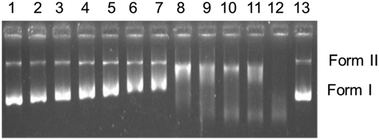
The cationic complexes 4 and 6 are structurally different from 1–3, as they have a bidentate pyridine derivative and one chlorido ligand. Complexes 4 and 6 did not show any DNA cleavage in the dark (Fig. SI-4†). Contrary to this, after exposure to UVA light (365 nm) for 1 h, only 6 shows a regular decrease in the mobility of form I (Fig. SI-5†) with increasing concentration. These results are similar to those obtained for 1–3 in the dark. It is assumed that photoexcitation facilitates hydrolysis of the monochlorido species to a monoaquo species facilitating adduct formation with DNA leading to local unwinding and retarded mobility at lower concentration (Fig. SI-5,† lane 8, 50 μM).
In summary, the electrophoretic mobility studies confirm covalent binding of 1–3 in the dark, while complexes 4 and 6 bind covalently on irradiation.
| 8505C | MCF-7 | SW-480 | 518A2 | |
|---|---|---|---|---|
| 1 | >100 | >100 | >100 | >100 |
| 2 | 69.1 ± 2.5 | 36.3 ± 2.6 | 94.1 ± 8.3 | 97.7 ± 3.2 |
| 3 | 90.2 ± 13.9 | 42.5 ± 0 | >100 | >100 |
| 4 | >100 | >100 | >100 | >100 |
| 6 | >100 | >100 | >100 | >100 |
| A | >100 | >100 | >100 | >100 |
| B | 79.6 ± 4.5 | 20.2 ± 3.5 | 91.1 ± 4.9 | >100 |
| C | >100 | 55.8 ± 0.3 | >100 | >100 |
| D | >100 | >100 | >100 | >100 |
| E | 58.2 ± 11.6 | >100 | 44.1 ± 5.1 | 40.5 ± 4.5 |
| Cisplatin | 4.8 ± 0.1 | 2.2 ± 0.2 | 3 ± 0.4 | 2 ± 0.4 |
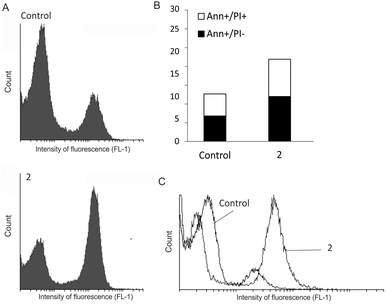
A relationship between decrease of cell viability and dose as well as an evident plateau effect indicated that complex 2 affected cellular proliferation. Cell staining with CFSE was used to explore the influence of 2 on cell division. According to the obtained data (Fig. 8A), complex 2 resulted in strongly suppressed proliferation, manifested by a larger quantity of undivided cells in comparison to the control culture. As induction of apoptotic cell death is the typical profile of cytostatic drug action, the presence of apoptotic cell death in cultures exposed to 2 was estimated by Ann/PI double staining. Flow-cytometric analysis of cells after 72 h of treatment revealed moderate accumulation of Ann+/PI− cells, recognised as early apoptotic as well as double positive, necrotic cells (Fig. 8B). This process was synchronised with enhanced caspase activation (Fig. 8C). Taken together, the anticancer capacity of newly synthesised complex 2 is basically related to inhibition of cell proliferation and subsequent caspase-dependent apoptosis. Similar RuII complexes of the general formula [Ru(p-cym)Cl2(L)] (L = amine ligand) induced cell death via inhibition of DNA synthesis.67 The proposed mechanism of RuII arene complexes, primarily based on inhibition of cellular proliferation with moderate caspase-dependent apoptosis, can explain higher IC50 values than determined for cisplatin, which is a cytocidal agent.68
Conclusions
Several RuII arene complexes with mono- and bidentate N-donor ligands were synthesised and characterised by analytical and structural methods. The single-crystal X-ray structure analyses showed complexes 2–6 to have distorted piano-stool geometry. Several spectral, thermal and hydrodynamic measurements of the interaction of complexes 1–4 and 6 with calf thymus DNA indicated covalent binding to DNA. Complexes 1–3 bind covalently to DNA in the dark, similar to cisplatin, while the cationic complexes 4 and 6 covalently bind to DNA on irradiation, similar to cisplatin analogues. Complexes 2 and 3 are cytotoxic against various cell lines, with highest efficacy for MCF-7 cells. In parallel, ligands B and E revealed cytotoxicity against almost all tested cell lines. This antitumour activity was shown to be preferentially realised through inhibition of cell division accompanied by caspase-dependent apoptosis.Acknowledgements
Financial support from the Free State of Saxony (project number 100099597) and the Graduate School “Leipzig School of Natural Sciences – Building with Molecules and Nano-objects” (BuildMoNa) is gratefully acknowledged (E. H.-H., S. R.). A. S. K. and E. H.-H. thank the DST and DAAD for financial support of a joint Indo-German research project (INT/FRG/DAAD/P-225/2013). A. S. K. and A. A. K. thank DST (FIST, PURSE) and UGC (CAS) for funding of the Department of Chemistry, SPPU. D. M.-I., D. D. and S. M. would like to acknowledge financial support from the DAAD (PPP project) and the Ministry of Education, Science and Technological Development of the Republic of Serbia (project No. 173013).Notes and references
- S. Medici, M. Peana, V. M. Nurchi, J. I. Lachowicz, G. Crisponi and M. A. Zoroddu, Coord. Chem. Rev., 2015, 284, 329 CrossRef CAS.
- R. Trondl, P. Heffeter, C. R. Kowol, M. A. Jakupec, W. Berger and B. K. Keppler, Chem. Sci., 2014, 5, 2925 RSC.
- E. Alessio, G. Mestroni, A. Bergamo and G. Sava, Curr. Top. Med. Chem., 2004, 4, 1525 CrossRef CAS PubMed.
- A. Bergamo and G. Sava, Dalton Trans., 2007, 1267 RSC.
- G. Sava, A. Bergamo, S. Zorzet, B. Gava, C. Casarsa, M. Cocchietto, A. Furlani, V. Scarcia, B. Serli, E. Iengo, E. Alessio and G. Mestroni, Eur. J. Cancer, 2002, 38, 427 CrossRef CAS PubMed.
- M. A. Jakupec, M. Galanski, V. B. Arion, C. G. Hartinger and B. K. Keppler, Dalton Trans., 2008, 183 RSC.
- G. Sava and A. Bergamo, Int. J. Oncol., 2000, 17, 353 CAS.
- I. Bratsos, T. Gianferrara, E. Alessio, C. G. Hartinger, M. A. Jakupec and B. K. Keppler, in Bioinorganic Medicinal Chemistry, Wiley Online Library, 2011, 151 Search PubMed.
- A. Vacca, M. Bruno, A. Boccarelli, M. Coluccia, D. Ribatti, A. Bergamo, S. Garbisa, L. Sartor and G. Sava, Br. J. Cancer, 2002, 86, 993 CrossRef CAS PubMed.
- B. K. Keppler, M. Henn, U. M. Juhl, M. R. Berger, R. Niebl and F. E. Wagner, Prog. Clin. Biochem. Med., 1989, 10, 41 CAS.
- G. Sava, E. Alessio, A. Bergamo and G. Mestroni, Topics in Biological Inorganic Chemistry, ed. M. J. Clarke and P. J. Sadler, Springer-Verlag, Berlin, 1999, vol. 1, p. 143 Search PubMed.
- A. Bergamo, S. Zorzet, B. Gava, A. Sorc, E. Alessio, E. Iengo and G. Sava, Anti-Cancer Drugs, 2000, 11, 665 CrossRef CAS PubMed.
- J. M. Rademaker-Lakhai, D. Van Den Bongard, D. Pluim, J. H. Beijnen and J. H. M. Schellens, Clin. Cancer Res., 2004, 10, 3717 CrossRef CAS PubMed.
- M. Groessl, C. G. Hartinger, K. Połeć-Pawlak, M. Jarosz, P. J. Dyson and B. K. Keppler, Chem. Biodiversity, 2008, 5, 1609 CAS.
- P.-S. Kuhn, V. Pichler, A. Roller, M. Hejl, M. A. Jakupec, W. Kandioller and B. K. Keppler, Dalton Trans., 2015, 44, 659 RSC.
- D. S. Thompson, G. J. Weiss, S. Fields Jones, H. A. Burris, R. K. Ramanathan, J. R. Infante, A. Ogden and D. D. von Hoff, J. Clin. Oncol., 2012, 30, 2678 CrossRef PubMed.
- P. Schluga, C. G. Hartinger, A. Egger, E. Reisner, M. Galanski, M. A. Jakupec and B. K. Keppler, Dalton Trans., 2006, 1796 RSC.
- S. K. Singh and D. S. Pandey, RSC Adv., 2014, 4, 1819 RSC.
- M. R. Gill and J. A. Thomas, Chem. Soc. Rev., 2012, 41, 3179 RSC.
- B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565 RSC.
- V. Fernandez-Moreira, F. L. Thorp-Greenwood and M. P. Coogan, Chem. Commun., 2010, 46, 186 RSC.
- L. D. Dale, J. H. Tocher, T. M. Dyson, D. I. Edwards and D. A. Tocher, Anti-Cancer Drug Des., 1992, 7, 3 CAS.
- W. S. Sheldrick and S. Heeb, Inorg. Chim. Acta, 1990, 168, 93 CrossRef CAS.
- R. E. Morris, R. E. Aird, P. S. Murdoch, H. Chen, J. Cummings, N. D. Hughes, S. Pearsons, A. Parkin, G. Boyd, D. I. Jodrell and P. J. Sadler, J. Med. Chem., 2001, 44, 3616 CrossRef CAS PubMed.
- N. P. E. Barry and P. J. Sadler, Chem. Commun., 2013, 49, 5106 RSC.
- A. F. A. Peacock and P. J. Sadler, Chem. – Asian J., 2008, 3, 1890 CrossRef CAS PubMed.
- A. L. Noffke, A. Habtemariam, A. M. Pizarro and P. J. Sadler, Chem. Commun., 2012, 48, 5219 RSC.
- C. G. Hartinger, M. Groessl, S. M. Meier, A. Casini and P. J. Dyson, Chem. Soc. Rev., 2013, 42, 6186 RSC.
- C. G. Hartinger, N. Metzler-Nolte and P. J. Dyson, Organometallics, 2012, 31, 5677 CrossRef CAS.
- P. J. Dyson and G. Sava, Dalton Trans., 2006, 1929 RSC.
- K. J. Kilpin, S. M. Cammack, C. M. Clavel and P. J. Dyson, Dalton Trans., 2013, 42, 2008 RSC.
- N. Busto, J. Valladolid, C. Aliende, F. A. Jalon, B. Manzano, A. M. Rodríguez, J. F. Gaspar, C. Martins, C. T. Biver, G. Espino, J. M. Leal and B. García, Chem. – Asian J., 2012, 12, 788 CrossRef PubMed.
- N. Busto, J. Valladolid, M. Martínez-Alonso, H. J. Lozano, F. A. Jalon, B. R. Manzano, A. M. Rodríguez, M. C. Carrion, T. Biver, J. M. Leal, G. Espino and B. García, Inorg. Chem., 2013, 52, 9962 CrossRef CAS PubMed.
- J. Valladolid, C. Hortiguela, N. Busto, G. Espino, A. M. Rodríguez, J. M. Leal, F. A. Jalon, B. R. Manzano, A. Carbayo and B. García, Dalton Trans., 2014, 43, 2629 RSC.
- C. Aliende, M. Perez-Manrique, F. A. Jalon, B. R. Manzano, A. M. Rodrıguez, J. V. Cuevas, G. Espino, M. A. Martınez, A. Massaguer, M. Gonzalez-Bartulos, R. de Llorens and V. J. Moreno, J. Inorg. Biochem., 2012, 117, 171 CrossRef CAS PubMed.
- M. Martínez-Alonso, N. Busto, F. A. Jalon, B. R. Manzano, J. M. Leal, A. M. Rodríguez, B. García and G. Espino, Inorg. Chem., 2014, 53, 11274 CrossRef PubMed.
- T. Wang, Y. Hou, Y. Chen, K. Li, X. Cheng, Q. Zhou and X. Wang, Dalton Trans., 2015, 44, 12726 RSC.
- K. Purkait, S. Karmakar, S. Bhattacharyya, S. Chatterjee, S. K. Dey and A. Mukherjee, Dalton Trans., 2015, 44, 5969 RSC.
- R. Pettinari, C. Pettinari, F. Marchetti, C. M. Clavel, R. Scopelliti and P. J. Dyson, Organometallics, 2013, 32, 309 CrossRef CAS.
- R. Pettinari, F. Marchetti, C. Pettinari, A. Petrini, R. Scopelliti, C. M. Clavel and P. J. Dyson, Inorg. Chem., 2014, 53, 13105 CrossRef CAS PubMed.
- F. Barragán, P. López-Senín, L. Salassa, S. Betanzos-Lara, A. Habtemariam, V. Moreno, P. J. Sadler and V. Marchán, J. Am. Chem. Soc., 2011, 133, 14098 CrossRef PubMed.
- R. Frank, V. M. Ahrens, S. Boehnke, A. G. Beck-Sickinger and E. Hey-Hawkins, ChemBioChem, 2016, 17, 308 CrossRef CAS PubMed.
- M. A. Bennett and A. K. Smith, J. Chem. Soc., Dalton Trans., 1974, 233 RSC.
- K. Hino, H. Nakamura, Y. Nagai, H. Uno and H. Nishimura, J. Med. Chem., 1983, 26, 222 CrossRef CAS PubMed.
- X. Wang and J. J. Vittal, Inorg. Chem., 2003, 42, 5135 CrossRef CAS PubMed.
- A. Bacchi, G. Cantoni, M. Granelli, S. Mazza, P. Pelagatti and G. Rispoli, Cryst. Growth Des., 2011, 11, 5039 CAS.
- CrysAlis Pro: Data collection and data reduction software package, Rigaku Corp Search PubMed.
- SIR92: A. Altomare, G. Cascarano, C. Giacovazzo, A. Guagliardi, M. C. Burla, G. Polidori and M. Camalli, J. Appl. Crystallogr., 1994, 27, 435 Search PubMed.
- SHELX: G. M. Sheldrick, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2015, 71, 3 CrossRef PubMed.
- K. Brandenburg, Diamond, Version 3.2i, Crystal Impact, GbR, Bonn, Germany Search PubMed.
- S. S. Bhat, A. S. Kumbhar, A. A. Kumbhar and A. Khan, Chem. – Eur. J., 2012, 18, 16383 CrossRef CAS PubMed.
- S. Bhat, A. S. Kumbhar, A. A. Kumbhar, A. Khan, P. Lönnecke and E. Hey-Hawkins, Chem. Commun., 2011, 47, 11068 RSC.
- H. S. Sahoo, D. Tripathy, S. Chakrabortty, S. S. Bhat, A. Kumbhar and D. K. Chand, Inorg. Chim. Acta, 2013, 400, 42 CrossRef CAS.
- G. Ludwig, I. Ranđelović, D. Maksimović-Ivanić, S. Mijatović, M. Z. Bulatović, D. Miljković, M. Korb, H. Lang, D. Steinborn and G. N. Kaluđerović, ChemMedChem, 2014, 9, 1586 CrossRef CAS PubMed.
- D. Maksimović-Ivanić, S. Mijatović, I. Mirkov, S. Stošić-Grujičić, D. Miljković, T. J. Sabo, V. Trajković and G. N. Kaluđerović, Metallomics, 2012, 4, 1155 RSC.
- D. Maksimovic-Ivanic, S. Mijatovic, D. Miljkovic, L. Harhaji- Trajkovic, G. Timotijevic, M. Mojic, D. Dabideen, K. F. Cheng, J. A. McCubrey, K. Mangano, Y. Al-Abed, M. Libra, G. Garotta, S. Stosic- Grujicic and F. Nicoletti, Mol. Cancer Ther., 2009, 8, 1169 CrossRef CAS PubMed.
- T. Eichhorn, E. Hey-Hawkins, D. Maksimović-Ivanić, M. Mojić, J. Schmidt, S. Mijatović, H. Schmidt and G. N. Kaluđerović, Appl. Organomet. Chem., 2015, 29, 20 CrossRef CAS.
- G. N. Kaluđerović, T. Krajnović, M. Momcilovic, S. Stosic-Grujicic, S. Mijatović, D. Maksimović-Ivanić and E. Hey-Hawkins, J. Inorg. Biochem., 2015, 153, 315 CrossRef PubMed.
- J. Gomez, G. Garcia-Herbosa, J. V. Cuevas, A. Arnaiz, A. Carbayo, A. Munoz, L. Falvello and P. E. Fanwick, Inorg. Chem., 2006, 45, 2483 CrossRef CAS PubMed.
- R. Aronson, M. R. J. Elsegood, J. W. Steed and D. A. Tocher, Polyhedron, 1991, 10, 1727 CrossRef CAS.
- G. A. Jeffrey, An Introduction to Hydrogen Bonding, Oxford University Press, Oxford, 1997 Search PubMed.
- D. A. Tocher and M. D. Walkinshaw, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1982, 38, 3083 CrossRef CAS (with BPh4 as anion); T. Sumiyoshi, T. B. Gunnoe, J. L. Petersen and P. D. Boyle, Inorg. Chim. Acta, 2008, 361, 3254 CrossRef (with B{3,5-(CF3)2C6H3}4 as anion).
- M. J. Chow, C. Licona, D. Y. Q. Wong, G. Pastorin, C. Gaiddon and W. H. Ang, J. Med. Chem., 2014, 57, 6043 CrossRef CAS PubMed.
- B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565 RSC.
- H. Song, J. T. Kaiser and J. K. Barton, Nat. Chem., 2012, 4, 615 CrossRef CAS PubMed.
- B. Rosenberg, Cisplatin – Chemistry and Biochemistry of a Leading Anticancer Drug, ed. B. Lippert, Verlag Helvetica Chimica Acta, Zürich, 2nd edn, 1999 Search PubMed.
- W. M. Motswainyana and P. A. Ajibade, Adv. Chem., 2015, 859730 Search PubMed.
- S. Gómez-Ruiz, D. Maksimović-Ivanić, S. Mijatović and G. N. Kaluđerović, Bioinorg. Chem. Appl., 2012, 140284 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1421200–1421204. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c6dt01782g |
| This journal is © The Royal Society of Chemistry 2016 |