Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
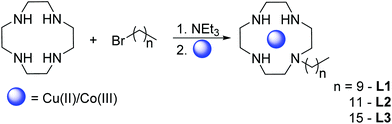
Significantly enhanced proteolytic activity of cyclen complexes by monoalkylation†
Chrischani
Perera-Bobusch
a,
Jan
Hormann
a,
Christoph
Weise
b,
Stefanie
Wedepohl
cd,
Jens
Dernedde
d and
Nora
Kulak
*a
aInstitut für Chemie und Biochemie, Freie Universität Berlin, Fabeckstr. 34/36, 14195 Berlin, Germany. E-mail: nora.kulak@fu-berlin.de
bInstitut für Chemie und Biochemie, Freie Universität Berlin, Thielallee 63, 14195 Berlin, Germany
cInstitut für Chemie und Biochemie, Freie Universität Berlin, Takustr. 3, 14195 Berlin, Germany
dCharité – Universitätsmedizin Berlin, Institut für Laboratoriumsmedizin, Klinische Chemie und Pathobiochemie, CVK, Augustenburger Platz 1, 13353 Berlin, Germany
First published on 1st June 2016
Abstract
A simple approach towards efficient artificial proteases based on the cyclen ligand is presented. We thus achieved an increase of the proteolytic activity of two orders of magnitude when compared to the unsubstituted cyclen complex. Amphiphilic Cu(II) and Co(III) complexes cut BSA and myoglobin as model substrates at μM concentrations. MALDI-ToF MS is used to identify the cleavage fragments.
The selective cleavage of proteins has numerous possible applications, e.g. in proteomics, for industrial applications or in medicinal chemistry for the degradation of pathogenic proteins.1–3 Misfolded proteins play a key role in amyloidogenic diseases like Alzheimer's disease, Parkinson's disease and type 2 diabetes mellitus.4 They aggregate to form soluble and insoluble oligomers, protofibrils and finally fibrils, which constitute the so-called plaques. With population aging and the subsequent rising numbers of patients suffering from those maladies, artificial proteases are of growing scientific and medicinal interest.3,5 Organic compounds and metal complexes have already been developed for this purpose.6–8
Metal complexes exhibit the advantage of performing different steps of the hydrolysis reaction on their own (e.g. polarization of the carbonyl group, nucleophilic attack at the amide group, stabilization of tetrahedral intermediates), in contrast the interaction of multiple functionalities would be necessary for organic residues.9,10 For the specific cleavage of proteins and peptides with metal complexes several examples have been reported in literature.11–13 Seminal work was published by Suh et al. who were able to drastically increase the cleavage activity of the known artificial protease Cu(II) cyclen (1,4,7,10-tetraazacyclododecane) by linkage to polystyrene and introduction of an aldehyde group in proximity to the macrocycle.14,15
Already more than 20 years ago, the mechanism for hydrolytic cleavage of unactivated amide bonds by Co(III) cyclen complexes16 as well as the cleavage of Cu(II) tacn (1,4,7-triazacyclononane) towards dipeptides and proteins was reported.13 Such hydrolytic reactions are based on the Lewis acidity of the metal center and the formation of a hydroxide nucleophile from coordinated aqua ligands like in [M(cyclen)(H2O)(OH)]+/2+. This mechanistic proposal was recently corroborated for several metal ions (Co(III), Cu(II), Ni(II), Cd(II), Zn(II), Pd(II)) by DFT calculations.10
Our goal was to increase the cleavage activity of Cu(II) and Co(III) cyclen complexes through micelle formation via self-assembly. A similar approach has already been shown to promote DNA cleavage with Zn(II) cyclen derivatives.17 Also, Zn(II) cyclen complexes with long alkyl chain substituents have been investigated before for the cleavage of activated esters like 4-nitrophenyl acetate in co-micellar solution with the surfactant Triton X-100.18 However, the application to proteins has not been reported before. This is surprising, since the cleavage of proteins and peptides is of potential medicinal interest and straightforward methods like gel electrophoresis and MALDI-ToF MS can be used for monitoring such cleavage reactions.
The self-assembly of the metal complexes of amphiphilic cyclen derivatives is supposed to increase the local density of catalytic units for protein cleavage. This approach can be compared with the linkage of hydrophilic compounds to polystyrene and therefore might increase the cleavage activity of cyclen complexes similarly.
We synthesized monosubstituted ligands with alkyl chains of 10, 12 and 16 carbon atoms to investigate the influence of the chain length on the cleavage activity. Among the many protocols available for monofunctionalization of cyclen19 (see ref. 4 and 5 therein for an overview), we have decided for a facile one-step procedure: Cyclen was applied in a fourfold excess with the corresponding alkyl bromide and triethylamine as base (cf. ESI, S-1.1†).20,21
We were able to isolate crystalline Cu(II) and Co(III) complexes of L3 (Scheme 1). However, X-ray crystallography did not provide molecular structures with satisfactory resolution due to the floppiness of the long alkyl chain. For complexation Cu(ClO4)2·6H2O was employed, whereas for the Co(III) complex Na3[Co(CO3)3]·3H2O was used as a precursor (cf. ESI, S-1.2†).22
For cleavage activity studies towards proteins the complexes were generated in situ with Cu(NO3)2 and CoCl2, respectively, with a 5% excess of the ligand if not stated otherwise (cf. ESI, S-1.2†). Thus we were able to ensure that cleavage is induced by the complex and not by free metal ions. Oxidation of the Co(II) to the Co(III) metal center of cyclen complexes was accomplished by exposing the solution to air.
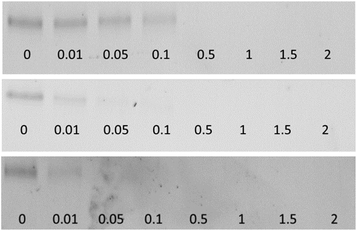
The proteolytic activity was examined via SDS-PAGE (cf. ESI, S-2.1†). BSA and myoglobin were applied as model proteins. Protein cleavage was observable in the fading of the protein bands upon addition of the complexes compared to the untreated reference protein.
All six complexes, namely the Cu(II) and Co(III) cyclen complexes of all three chain lengths, exhibit a remarkable cleavage activity (Fig. 1, S-1†). The three Cu(II) complexes reveal a cleavage activity towards BSA more than two orders of magnitude higher compared to the parent compound Cu(II) cyclen. For Cu(II) cyclen there is barely any activity observable at a concentration of 15 mM (Fig. S-2†), whereas for Cu(II) L3 the substrate protein is already fully cleaved at a concentration of 0.05 mM. We could observe an increase of the proteolytic activity with increased length of the alkyl chain (Fig. 1). This is obvious for L1 and L2 in the presence of both metals, however the difference of L2 to L3 is hardly noticeable. For Co(III) L1 the protein is fully cleaved at a concentration of 0.5 mM, whereas for the corresponding L2 and L3 complexes full cleavage is already achieved at a complex concentration of 0.05 mM. The cleavage activity of the Co(III) complexes is slightly higher compared to the Cu(II) complexes (cf. ESI, Fig. S-1†). With respect to potential in vivo applications, Co(III) complexes would be preferable because of their exchange inertness.23
Since hydrolytic cleavage of biomolecules using Zn(II) is ubiquitous in nature (e.g. in the endonuclease S1 and the endopeptidase thermolysin) and is often reported in the field of bioinorganic model chemistry (for DNA and RNA cleavage,24 however, interestingly only rarely seen for protein cleavage25), we have also tested Zn(II) L3 (cf. ESI, Fig. S-9†). This complex has shown similar activity like the Cu(II) and Co(III) analogs corroborating the above mentioned hydrolytic cleavage mechanism.10 In addition, catalytic activity of the Cu(II) and Co(III) complexes in the absence of any reducing agent (like ascorbate, thiols or H2O2), which is decisive for the activation of redoxactive metal ions and subsequent generation of reactive oxygen species (ROS), gives further indication to a hydrolytic mechanism.26
A comparison of the complexes formed in situ with the isolated complexes of L3 showed the same cleavage activity. Therefore we could ensure that the cleavage originates indeed from the complexes (Fig. S-3†). Another concern regarding the identity of the active cleavage agent, refers to the involvement of Tris buffer molecules, which are also potential ligands for Cu(II) and Co(III). Thus, Tris and MOPS, the latter being a non-coordinating buffer, were exemplarily compared for the cleavage reaction catalyzed by Cu(II) L3 (Fig. S-10†). The cleavage was slightly better in Tris buffer. When the stability constants for Cu(II) cyclen (23.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 27) and Cu(II) Tris (around 6
27) and Cu(II) Tris (around 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28) and the concentration ranges used (0.1–1 mM vs. 50 mM) are considered, however, one can exclude formation of any Cu(II) Tris species.
28) and the concentration ranges used (0.1–1 mM vs. 50 mM) are considered, however, one can exclude formation of any Cu(II) Tris species.
To make sure that the complexes do not cleave BSA exclusively, myoglobin was applied as a second substrate. For the Co(III) complex of L3 the cleavage activity towards myoglobin was comparable to BSA but for the respective Cu(II) complex complete cleavage was only observable at the lowest complex concentrations (0.01 mM) applied (Fig. S-4†). With increasing protease concentration the proteolytic activity surprisingly decreased. This indicates that in case of the heme-containing protein myoglobin high Cu(II) concentrations lead to some kind of side reaction inhibiting the actual cleavage process. Investigations elucidating this effect with heme and non-heme proteins are underway.
At physiological pH and temperature cleavage towards BSA was still observable but markedly decreased (Fig. S-5†) in comparison to the conditions otherwise applied (pH 9, 50 °C). Elevation of pH and temperature have been proven necessary for other artificial proteases as well.15,29,30
A time-dependent study with BSA revealed that for Co(III) L3 only 16 h of incubation were necessary for complete cleavage at 0.05 mM, whereas 39 h were necessary for Cu(II) L3 (Fig. S-6†). After 4 h of incubation just a very faint protein band was detectable on the gel for both complexes but the total cleavage of the protein took 4 and 10 times longer, respectively. This proteolytic activity is comparable to that of other artificial proteases of even more complex structures.14,29
Analyses by SDS-PAGE performed with metal salts corresponding to the in situ generated complexes substantiated that the substrate cleavage originates from the synthesized complexes and could not be ascribed to free metal ions (Fig. S-7†). CoCl2 showed no cleavage activity towards BSA at all, whereas Cu(NO3)2 induced only very weak cleavage. Those results clearly indicate that the observed cleavage originates from the cyclen complexes.
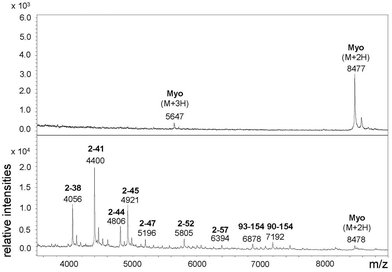
For the above described SDS-PAGE experiments a Tris-glycine buffer system was used, which is suitable for the detection of proteins >30 kDa. In order to estimate the size of the myoglobin fragments, however, the Tris-tricine buffer system was applied since this system is more suitable for low-molecular mass proteins and peptides (1–30 kDa).31 The size of the myoglobin fragments was thus estimated to be in a range of 4–7 kDa (Fig. S-8†).31 To further characterize these fragments MALDI-ToF MS was performed. The spectrum showed fragments between 4.0 and 7.2 kDa (Fig. 2). Fragments with a similar size have already been reported, in particular the fragment 93–154, the N terminus of which is close to the heme moiety (proximal His94), has been previously identified by Suh et al.15
In order to evaluate the cytotoxicity of the complexes, the MTT assay was applied for the Cu(II) and Co(III) complexes of L2 in comparison to Cu(II) cyclen and Co(III) cyclen. In Fig. S-11,† the dose–response relationship of these complexes incubated for 48 h on A549 lung cancer cells is shown. The calculated IC50 values for Cu(II) and Co(III) cyclen complexes were both about 1.7 mM, 9.5 μM for Cu(II) L2 and 24.5 μM for Co(III) L2. The exact mechanism of this enhanced cytotoxicity is not known yet, but an interaction of the alkyl chains with the cell membranes due to their composition of amphiphilic lipid molecules is highly probable.32 Nevertheless, the IC50 values are comparable to some approved drugs: at the same conditions in A549 cells e.g. 64–70 μM for the anticancer drug cisplatin and 2–3 μM for imatinib, a drug originally developed as an anticancer drug but found to inhibit amyloid-β formation.33,34 Curcumin (16 μM) and resveratrol (26 μM) have been shown to deplete Aβ plaques.35–37
The structure–activity relationship of the amphiphilic metal complexes was investigated with the established “pyrene 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio” method (Fig. S-12 and Table S-2†). This technique utilizes the change of the ratio of the first and third vibronic peak (I1/I3) of fluorescing pyrene while changing its environment from hydrophilic to hydrophobic due to micelle formation.38,39 Above the critical micellar concentration (cmc) pyrene is supposed to be located in the hydrophobic core of the micelles causing an increase of I3. The peak I3 could not be detected for the complexes presented here and a drastic decrease of the fluorescence intensity was observed below the cmc. This can be explained with fluorescence quenching of pyrene which has been described before for Cu(II) and Co(II/III) surfactant systems.40,41 However, regarding the ratio of the intensity at 375 and 385 nm, values reported in the literature for I1 and I3, a sigmoidal curve could be generated as expected for this method (Fig. S-12†).42 The values for the cmc of the Cu(II) complexes of L1–L3 and Co(III) L3 calculated from these measurements can be taken from Table S-2.† The results indicate that particle assembly requires complex concentrations above the amount essential for complete protein cleavage. Therefore micelle formation can be ruled out as the origin of the improved cleavage activity but an alternative method for the determination of the cmc should be utilized to verify the values presented here. One might expect discrepancies of the two experiments due to temperature differences (50 °C (323 K) for the cleavage experiments and only 25 °C (298 K) for the cmc determination). For cationic amphiphiles it was found, however, that the cmc is almost constant over a wide range of temperature, e.g. hexamethyltrimethylammonium bromide (cmc around 0.02–0.04 mM in a range of 280 to 360 K).43
3 ratio” method (Fig. S-12 and Table S-2†). This technique utilizes the change of the ratio of the first and third vibronic peak (I1/I3) of fluorescing pyrene while changing its environment from hydrophilic to hydrophobic due to micelle formation.38,39 Above the critical micellar concentration (cmc) pyrene is supposed to be located in the hydrophobic core of the micelles causing an increase of I3. The peak I3 could not be detected for the complexes presented here and a drastic decrease of the fluorescence intensity was observed below the cmc. This can be explained with fluorescence quenching of pyrene which has been described before for Cu(II) and Co(II/III) surfactant systems.40,41 However, regarding the ratio of the intensity at 375 and 385 nm, values reported in the literature for I1 and I3, a sigmoidal curve could be generated as expected for this method (Fig. S-12†).42 The values for the cmc of the Cu(II) complexes of L1–L3 and Co(III) L3 calculated from these measurements can be taken from Table S-2.† The results indicate that particle assembly requires complex concentrations above the amount essential for complete protein cleavage. Therefore micelle formation can be ruled out as the origin of the improved cleavage activity but an alternative method for the determination of the cmc should be utilized to verify the values presented here. One might expect discrepancies of the two experiments due to temperature differences (50 °C (323 K) for the cleavage experiments and only 25 °C (298 K) for the cmc determination). For cationic amphiphiles it was found, however, that the cmc is almost constant over a wide range of temperature, e.g. hexamethyltrimethylammonium bromide (cmc around 0.02–0.04 mM in a range of 280 to 360 K).43
A possible explanation for the improved proteolytic activity might rather be the attraction of the alkyl chains to the hydrophobic domains of the target proteins, which would facilitate the attack of the catalytic unit. The dependence of the proteolytic activity on the alkyl chain length – the longer the chain, i.e. the more hydrophobic the molecule, the higher the cleavage activity – is also a hint in this direction. Indeed, BSA exhibits a hydrophobic region, subdomain IIA, and the heme moiety of myglobin is known to be located in a hydrophobic pocket.44,45 Computational studies have revealed oxygenated myoglobin as a transporter and sequestering agent for fatty acids in muscles with the hydrophobic heme pocket being the binding site. The hydrophobic tail of the fatty acids is located within the pocket lined with hydrophobic amino acids whereas the hydrophilic head group interacts with two Lys amino acids.46 Along the lines of these findings the cuts of the amphiphilic cyclen complexes in myoglobin close to the heme moiety can be conclusively explained. Molecular modelling studies are underway to prove this hypothesis.
Conclusions
In only two steps alkylated Cu(II) and Co(III) cyclen complexes were obtained, which were shown to act as efficient artificial proteases. Compared to other artificial proteases this simple system can keep up with far more complex structures regarding their proteolytic activity.14,29 With SDS-PAGE cleavage of BSA and myoglobin as model proteins could be shown by the intensity loss of the protein bands, whereas MALDI-ToF MS allowed to identify cleavage sites for myoglobin. The cytotoxicity of the novel complexes was comparable to other drugs discussed for the treatment of e.g. Alzheimer's disease, a long-term goal for the presented complexes due to their conceivable interactions with disease-related Aβ peptides. Hydrophobic interactions with the proteins to be cleaved probably account for the high cleavage activity, rather than micelle formation of the amphiphilic complexes.Acknowledgements
Financial support by the FU Focus Area NanoScale is gratefully acknowledged. We would like to acknowledge the assistance of the Bio-MS unit of the Core Facility BioSupraMol supported by the Deutsche Forschungsgemeinschaft. We thank Dr Kai Licha from IC Discovery GmbH for generous gifts of cyclen, Prof. Dr Michael Gradzielski from TU Berlin for help with cmc determination and the Kulak group for proofreading of the manuscript.Notes and references
- F. Miskevich, A. Davis, P. Leeprapaiwong, V. Giganti, N. M. Kostić and L. A. Angel, J. Inorg. Biochem., 2011, 105, 675 CrossRef CAS PubMed.
- M. B. Rao, A. M. Tanksale, M. S. Ghatge and V. V. Deshpande, Microbiol. Mol. Biol. Rev., 1998, 62, 597 CAS.
- C. M. Dobson, Science, 2004, 304, 1259 CrossRef CAS PubMed.
- C. M. Dobson, Nature, 2003, 426, 884 CrossRef CAS PubMed.
- D. M. Holtzman, J. C. Morris and A. M. Goate, Sci. Transl. Med., 2011, 3, 77sr1 Search PubMed.
- H. Kim, H. Paik, M.-S. Kim, Y.-S. Chung and J. Suh, Bioorg. Med. Chem. Lett., 2002, 12, 2557 CrossRef CAS PubMed.
- A. Suzuki, M. Hasegawa, M. Ishii, S. Matsumura and K. Toshima, Bioorg. Med. Chem. Lett., 2005, 15, 4624 CrossRef CAS PubMed.
- K. B. Grant and M. Kassai, Curr. Org. Chem., 2006, 10, 1035 CrossRef CAS.
- J. Suh, Acc. Chem. Res., 1992, 25, 273 CrossRef CAS.
- T. Zhang, X. Zhu and R. Prabhakar, Organometallics, 2014, 33, 1925 CrossRef CAS.
- P. A. Sutton and D. A. Buckingham, Acc. Chem. Res., 1987, 20, 357 CrossRef CAS.
- L. Zhu and N. M. Kostic, J. Am. Chem. Soc., 1993, 115, 4566 CrossRef CAS.
- E. L. Hegg and J. N. Burstyn, J. Am. Chem. Soc., 1995, 117, 7015 CrossRef CAS.
- C. E. Yoo, P. S. Chae, J. E. Kim, E. J. Jeong and J. Suh, J. Am. Chem. Soc., 2003, 125, 14580 CrossRef CAS PubMed.
- S. H. Yoo, B. J. Lee, H. Kim and J. Suh, J. Am. Chem. Soc., 2005, 127, 9593 CrossRef CAS PubMed.
- B. K. Takasaki, J. H. Kim, E. Rubin and J. Chin, J. Am. Chem. Soc., 1993, 115, 1157 CrossRef CAS.
- B. Gruber, E. Kataev, J. Aschenbrenner, S. Stadlbauer and B. König, J. Am. Chem. Soc., 2011, 133, 20704 CrossRef CAS PubMed.
- E. Kimura, H. Hashimoto and T. Koike, J. Am. Chem. Soc., 1996, 118, 10963 CrossRef CAS.
- V. Boldrini, G. B. Giovenzana, R. Pagliarin, G. Palmisano and M. Sisti, Tetrahedron Lett., 2000, 41, 6527 CrossRef CAS.
- J. Massue, S. E. Plush, C. S. Bonnet, D. A. Moore and T. Gunnlaugsson, Tetrahedron Lett., 2007, 48, 8052 CrossRef CAS.
- W. C. Baker, M. J. Choi, D. C. Hill, J. L. Thompson and P. A. Petillo, J. Org. Chem., 1999, 64, 2683 CrossRef CAS PubMed.
- H. F. Bauer and W. C. Drinkard, J. Am. Chem. Soc., 1960, 82, 5031 CrossRef CAS.
- S. J. Lippard and J. M. Berg, Principles of Bioinorganic Chemistry, University Science Books, Mill Valley, 1994 Search PubMed.
- F. Mancin and P. Tecilla, New J. Chem., 2007, 31, 800 RSC.
- M. Yashiro, Y. Kawakami, J.-I. Taya, S. Arai and Y. Fujii, Chem. Commun., 2009, 1544 RSC.
- E. R. Stadtman, Free Radical Biol. Med., 1990, 9, 315 CrossRef CAS PubMed.
- R. D. Hancock, S. M. Dobson, A. Evers, P. W. Wade, M. P. Ngwenya, J. C. A. Boeyens and K. P. Wainwright, J. Am. Chem. Soc., 1988, 110, 2788 CrossRef CAS.
- J. Nagaj, K. Stokowa-Sołtys, E. Kurowka, T. Frączyk, M. Jeżowska-Bojczuk and W. Bal, Inorg. Chem., 2013, 52, 13927 CrossRef CAS PubMed.
- M. G. Kim, S. H. Yoo, W. S. Chei, T. Y. Lee, H. M. Kim and J. Suh, J. Biol. Inorg. Chem., 2010, 15, 1023 CrossRef CAS PubMed.
- H. M. Kim, B. Jang, Y. E. Cheon, M. P. Suh and J. Suh, J. Biol. Inorg. Chem., 2009, 14, 151 CrossRef CAS PubMed.
- H. Schägger, Nat. Protoc., 2006, 1, 16 CrossRef PubMed.
- H. R. Mellor, F. M. Platt, R. A. Dwek and T. D. Butters, Biochem. J., 2003, 374, 30 CrossRef PubMed.
- P. Zhang, W. Y. Gao, S. Turner and B. S. Ducatman, Mol. Cancer, 2003, 2, 1 CrossRef PubMed.
- G. He, W. Luo, P. Li, C. Remmers, W. J. Netzer, J. Hendrick, K. Bettayeb, M. Flajolet, F. Gorelick, L. P. Wennogle and P. Greengard, Nature, 2010, 467, 95 CrossRef CAS PubMed.
- J. Zhang, Y. Du, C. Wu, X. Ren, X. Ti, J. Shi, F. Zhao and H. Yin, Oncol. Rep., 2010, 24, 1217 CAS.
- J.-Y. Wu, K.-W. Tsai, J.-I. Shee, Y.-Z. Li, C.-H. Chen, J.-I. Chuang and Y.-W. Liu, Acta Pharmacol. Sin., 2010, 31, 81 CrossRef CAS PubMed.
- K. Rajasekhar, M. Chakrabarti and T. Govindaraju, Chem. Commun., 2015, 51, 13434 RSC.
- K. Kalyanasundaram and J. K. Thomas, J. Am. Chem. Soc., 1977, 99, 2039 CrossRef CAS.
- T. Chakraborty, I. Chakraborty and S. Ghosh, Arabian J. Chem., 2011, 4, 265 CrossRef CAS.
- R. Konuk, J. Cornelisse and S. P. McGlynn, J. Phys. Chem., 1989, 93, 7405 CrossRef CAS.
- R. S. Kumar, P. Paul, A. Riyasdeen, G. Wagniéres, H. van den Bergh, M. A. Akbarsha and S. Arunachalam, Colloids Surf., B, 2011, 86, 35 CrossRef CAS PubMed.
- A. I. Mitsionis and T. C. Vaimakis, Chem. Phys. Lett., 2012, 547, 110 CrossRef CAS.
- H.-U. Kim and K.-H. Lim, Colloids Surf., A, 2004, 235, 121 CrossRef CAS.
- B. K. Paul, A. Samanta and N. Guchhait, J. Phys. Chem. B, 2010, 114, 6183 CrossRef CAS PubMed.
- T. Uchida, K. Ishimori and I. Morishima, J. Biol. Chem., 1997, 272, 30108 CrossRef CAS PubMed.
- S. V. Chintapalli, G. Bhardwaj, R. Patel, N. Shah, R. L. Patterson, D. B. van Rossum, A. Anishkin and S. H. Adams, PLoS One, 2015, 10, e0128496 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, protein cleavage experiments, cytotoxicity studies, cmc determination. See DOI: 10.1039/c6dt00681g |
| This journal is © The Royal Society of Chemistry 2016 |