Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Inactivation of urease by 1,4-benzoquinone: chemistry at the protein surface†
L.
Mazzei
a,
M.
Cianci
 b,
F.
Musiani
a and
S.
Ciurli
*a
b,
F.
Musiani
a and
S.
Ciurli
*a
aLaboratory of bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Viale G. Fanin 40, I-40127, Bologna, Italy. E-mail: stefano.ciurli@unibo.it
bEuropean Molecular Biology Laboratory, DESY, Notkestraße 85, 22607 Hamburg, Germany
First published on 2nd March 2016
Abstract
The high activity of urease, a Ni(II) enzyme, has several adverse effects on human health and agriculture, and its modulation needs the use of inhibitors. 1,4-Benzoquinone (BQ) irreversibly inactivates Sporosarcina pasteurii urease (SPU), with first order kinetics for both the inhibitor and the enzyme. This reaction is stoichiometrically quenched in the presence of sulphite. The 2.07 Å crystal structure of SPU bound to BQ shows the presence of a 1,4-hydroquinone moiety covalently bound to the thiol group of αCys322, a key residue found on the mobile flap regulating the substrate access to the active site. The 1.75 Å crystal structure obtained when sulphite is added to a solution of SPU previously incubated with BQ shows the presence of a 2,5-dihydroxy-benzenesulphonate moiety bound to the αCys322 thiol group. These data reveal how the active site cysteine reacts with a prototypical BQ moiety, found in a large number of natural substances potentially suitable to control the urease activity.
The rapid hydrolysis of urea catalysed by the Ni(II)-containing enzyme urease1 in bacteria, fungi, algae and plants causes an abrupt increase of pH and, consequently, negative side effects on human health2 and the environment.3 Ureases show a quaternary structure made of a functionally minimal trimer, differently assembled in higher order structures depending on the biological source.1 Each unit of the trimer hosts one enzyme active site, constituted by two Ni(II) ions (Ni1 and Ni2) bridged by a carbamylated lysine residue (αLys220* in SPU) and by a hydroxide ion (Fig. 1A). The Ni(II) ions are further coordinated by two histidine residues, and are distinguished by the presence of a carboxylate O atom from an aspartate residue bound to Ni2. The coordination geometry of each Ni(II) ion is completed by a water molecule, leading to a pseudo-square pyramidal penta-coordinated Ni1 and a pseudo-octahedral hexa-coordinated Ni2. A third water molecule completes, together with the bridging hydroxide, a cluster of solvent-derived O atoms that defines a tetrahedral cavity that is assumed to stabilize the transition state of the catalysed reaction.4 The active site is found at the end of a ca. 15 Å-deep pocket characterized by the presence of a helix-turn-helix flap located at its entrance (Fig. 1B and 1-ESI†). This flap was observed to adopt different conformations, and has been suggested to modulate both the access of substrate to the enzyme active site and the dimensions of the catalytic cavity.4a,5
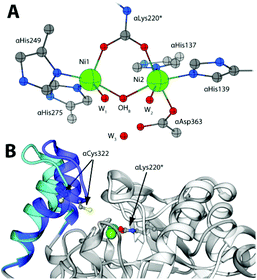 | ||
| Fig. 1 (A) Coordination environment of the Ni(II) ions in the active site of SPU (PDB code 4CEU). (B) Superimposition of the open (white and light blue ribbons, PDB code 4CEU) and closed (grey and blue ribbons, PDB code 3UBP) conformations of the flexible flap in SPU, highlighting the side chain of αLys220* and αCys322. | ||
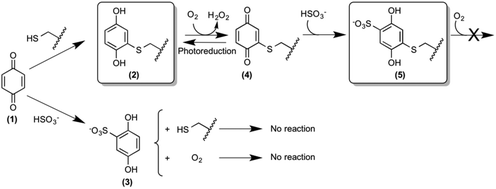
The design and synthesis of novel and potent urease inhibitors necessary to modulate its catalytic activity to counterbalance its negative effects require the knowledge of the structural details of the inhibition mechanism. Several urease structures reveal how the Ni(II) ions in the active site could be targeted by competitive inhibitors.1b,6 On the other hand, the most efficient urease inhibitors known so far contain the 1,4-benzoquinone moiety (BQ, 1, Scheme 1).7 Quinones are known to react with thiols, leading to the formation of the corresponding thiol-substituted benzene-1,4-diol.8 The sequence of the urease mobile flap contains the highly conserved residue αCys322 (residue numbering as in SPU), which is crucial for the activity of urease,9 and whose position with respect to the active site is also modulated by the flap movement (Fig. 1B). Covalent modifications of this residue by the formation of a disulphide bond in the presence of 2-mercaptoethanol, demonstrated by crystallography,10 lead to enzyme inhibition.11 These observations support the hypothesis that the conserved cysteine in the flap is the target for quinone-based inhibitors, as shown in Scheme 1, by the formation of 2.7d Here, we describe a kinetic characterization of the inhibition of SPU by 1,4-benzoquinone, and report, for the first time, the high-resolution crystal structure of the urease–BQ covalent complex, shedding light on the long-awaited molecular details of this interaction.
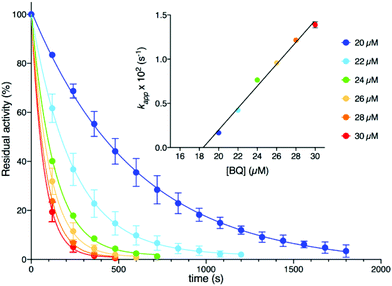
Kinetic measurements (see the ESI†) were performed at room temperature by using a spectrophotometric assay that monitors the decrease of urease activity as a function of time in the presence of increasing amounts of BQ (Fig. 2). The data were optimally fitted to a single exponential decay, and the resulting time constant kapp (s−1) showed a linear dependence on the concentration of BQ (inset of Fig. 2). This behaviour is consistent with an irreversible process that has two reactants (SPU and BQ) and is first-order with respect to both of them, leading to an inactive form of SPU, hypothetically represented by compound 2 in Scheme 1:
| v = k [BQ] [SPUactive] = kapp [SPUactive]; kapp = k [BQ] |
The value for the second-order kinetic constant resulting from the linear fit is k = 1.24 ± 0.06 × 103 M−1 s−1. The intercept of the linear fit in the inset of Fig. 2 crosses the abscissa at a value of 18.4 ± 1.5 μM; this value indicates the minimum concentration of BQ above which an inhibition effect can be detected. This observation was interpreted by considering the presence of ca. 20 μM sodium sulphite in the enzymatic assay solution, added as a stabilizer of urease6a but also known to react stoichiometrically with BQ to give 2,5-dihydroxy-benzenesulphonate (3, Scheme 1).12 The evident lack of reactivity between 3 and SPU, compared to the reactivity of 1, could be ascribed to the increase of the reduction potential caused by the electron withdrawing effects of the sulphonate moiety, which prevents the oxidation of 3 by dissolved molecular oxygen to the quinone level necessary for the reaction with the cysteine thiol (Scheme 1).12,13
In order to elucidate the structural basis of urease inactivation by BQ, SPU was incubated with 1 for two hours in the absence of sulphite in solution before crystallization. The resulting 2.07 Å crystal structure of SPU inhibited by BQ (see Table 1 and the ESI† for full details of crystallization, X-ray data collection, elaboration and analysis) confirmed the well-established architecture of the enzyme, as well as the rigidity of the protein scaffold, with a root mean square deviation (RMSD) for the Cα residue atoms of only 0.12 Å with respect to the structure of native urease (PDB code 4CEU).
| Data collection | 5FSE (BQ) @ 2.07 Å | 5FSD (BQ + HSO3−) @ 1.75 Å |
|---|---|---|
| a Highest resolution bin in parentheses. b R sym = ∑hkl ∑j|Ij − <|>|/∑hkl ∑jIj, where I is the intensity of a reflection, and <|> is the mean intensity of all symmetry related reflections j. c R p.i.m. = ∑hkl{[1/(N − 1)]∑j|Ij − <|>|}1/2/∑hkl∑jIj, where I is the intensity of a reflection, and <|> is the mean intensity of all symmetry related reflections j, and N is the multiplicity.15 d Taken from REFMAC;16Rfree is calculated using 5% of the total reflections that were randomly selected and excluded from refinement. e DPI = Rfactor·Dmax·compl−1/3·[Natoms/(Nrefl − Nparams)]1/2, where Natoms is the number of the atoms included in the refinement, Nrefl is the number of the reflections included in the refinement, Dmax is the maximum resolution of reflections included in the refinement, compl is the completeness of the observed data, and for isotropic refinement, Nparams ≈ 4Natoms.17 f Taken from BAVERAGE.18 g Taken from PROCHECK.19 | ||
| Wavelength (Å) | 0.954 | 1.214 |
| Crystal-to-detector distance (mm) | 378.1 | 256.9 |
| Oscillation angle (°) | 0.10 | 0.10 |
| Number of images | 400 | 1200 |
| Space group | P6322 | P6322 |
| Unit cell (a, b, c, Å) | 131.83, 131.83, 188.58 | 131.31, 131.31, 188.92 |
| Resolution rangea (Å) | 188.58–2.07 (2.13–2.07) | 97.43–1.75 (1.78–1.75) |
| Total number of reflectionsa | 257![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 019 (18 019 (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 993) 993) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 243 243![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 307 (51 307 (51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 825) 825) |
| Unique reflectionsa | 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 491 (4412) 491 (4412) |
96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 874 (4696) 874 (4696) |
| Redundancya | 4.4 (4.3) | 12.8 (11.0) |
| Completenessa (%) | 98.8 (97.6) | 100.0 (100.0) |
R
sym![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a,b (%) a,b (%) |
13.5 (90.8) | 10.1 (145.4) |
R
pim![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a,c (%) a,c (%) |
8.8 (55.1) | 4.2 (66.5) |
| Mean I half-set correlation CC(1/2) | 0.620 | 0.723 |
| Mean I/σ(I)a | 9.0 (1.6) | 20.0 (1.6) |
| Refinement statistics | ||
| Number of urease subunits in the asymmetric unit | 3 | 3 |
R
factor![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) d (%) d (%) |
14.4 | 13.4 |
R
free![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) d (%) d (%) |
19.1 | 16.9 |
| Cruickshank's DPI for coordinate errore based on Rfactor (Å) | 0.15 | 0.08 |
| Wilson plot B-factor (Å2) | 22.6 | 21.5 |
| Average all atom B-factorf (Å2) | 30.77 | 29.16 |
| Cys322 B-factorf (Å2) (Cβ, Sγ) | 63.2, 76.5 | 52.4, 61.7 |
| Cys555 B-factorf (Å2) (Cβ, Sγ) | 37.9, 42.2 | 39.4, 42.1 |
| Average ligand – Cys322 B-factorf (Å2) | 66.5 | 55.6 |
| Average ligand – Cys555 B-factorf (Å2) | 41.1 | 43.1 |
| RMS (bonds)d | 0.016 | 0.019 |
| RMS (angles)d | 1.803 | 1.973 |
| Total number of atomsc | 6800 | 7012 |
| Total number of water molecules | 465 | 642 |
| Solvent content (%) | 55.50 | 55.23 |
| Matthews coefficient (Å3/Da) | 2.76 | 2.75 |
| Ramachandran plot | ||
| Most favoured region (%) | 90.1 | 91.6 |
| Additionally allowed region (%) | 9.0 | 7.5 |
| Generously allowed region (%) | 0.6 | 0.8 |
| Disallowed region (%) | 0.3 | 0.2 |
No significant changes were observed for the Ni(II) coordination environment (Table 1-ESI†). The mobile flap covering the active site cavity is found in the open conformation, as in all SPU structures determined so far with the notable exception of the complex with diamidophosphate (DAP), an analogue of the intermediate or the transition state of the reaction.4a The unbiased omit electron density map calculated with Fourier coefficients Fo–Fc and phases refined using the model of the citrate-inhibited SPU (PDB code 4AC7, 1.50 Å resolution14), shows an additional electron density in the vicinity of the flap and proximal to the αCys322 residue (Fig. 2A-ESI†). This electron density could be successfully modelled with full occupancy using the aromatic dihydroxylated moiety of the inhibitor, as shown by the 2Fo–Fc map (Fig. 3A). The final refined structure features a covalent bond between αCys322 Sγ and an aromatic C atom of the inhibitor, with a C–S distance of 1.64 Å (Fig. 3B). The same covalent modification is also observed for αCys555, a residue located on the protein surface far from the active site, with a similar C–S distance of 1.67 Å (Fig. 3-ESI†). The remaining αCys520, protected from the solvent, is not affected by this modification. This result finally provides full structural support to the mechanism by which the solvent-exposed side chain thiols of cysteine residues in ureases are the reactive groups involved in the formation of covalent adducts with BQ.7d
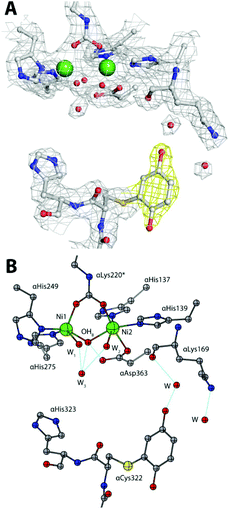 | ||
| Fig. 3 An atomic model of the active site of BQ-inactivated SPU (PDB code 5FSE) in the absence of sulphite. In panel (A), the nickel-coordination environment is shown superimposed on the final 2Fo–Fc electron density map contoured at 1σ; the map of the inhibitor is shown in yellow. In panel (B), the corresponding structural model is shown. H-bonds are shown as thin blue lines. | ||
In the case of αCys322, the O atom in the meta position with respect to the thiol substituent points towards the entrance of the active site channel (Fig. 3B), forming a hydrogen bond with a water molecule (at 2.85 Å) that is also stabilized by H-bonds with αLeu365 O (at 2.66 Å) and αLys169 O (at 2.76 Å).
The 1.75 Å resolution structure of SPU obtained from the crystals generated by adding 50 mM sodium sulphite to the crystallization drop obtained as before discloses further details of the reactivity of the enzyme inactivated by BQ. In this case, while the overall protein structure is maintained (as revealed by a value of 0.10 Å for the RMSD with respect to the structure obtained in the absence of sulphite, and no changes around the Ni(II) ions, see Table 1-ESI†), an additional electron density was identified in the unbiased omit Fo–Fc map calculated using the refined model of the 2.07 Å structure described above, in the proximity of the inhibitor aromatic ring bound to αCys322 (Fig. 2B-ESI†). This additional electron density was successfully modelled as a sulphonate group covalently bound to the aromatic ring in the para position with respect to the cysteine thiol, with 70% occupancy (Fig. 4A). One of the O atoms of the sulphonate group forms a H-bond with αLys169 Nζ (at 2.85 Å, Fig. 4B). The same covalent modification is also observed for αCys555, albeit with a lower occupancy (Fig. 4-ESI†). A comparative analysis of the solvent-excluded surfaces of the enzyme active site channel, in the absence and presence of the covalent modifications of αCys322 with and without sulphite in the crystallization milieu (Fig. 5-ESI†) shows that the channel is not physically closed in the presence of the bound inhibitor, independently of the sulphonate modification, suggesting that the inactivation of the enzyme is not caused by the blockage of substrate transport through the path leading into the active site, but it is rather due to the prevention of the flap to close, as necessary for the catalysis to occur.5 A site-directed mutation of the conserved cysteine with tyrosine in K. aerogenes urease, supposedly having a similar effect on the active site entrance, causes the abolition of catalytic activity.9
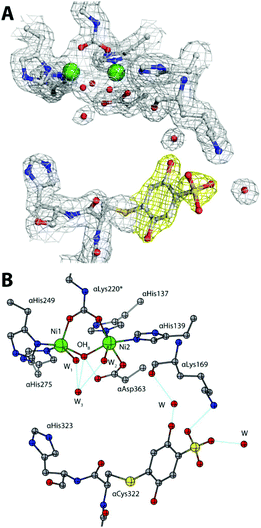 | ||
| Fig. 4 An atomic model of the active site of BQ-inactivated SPU (PDB code 5FSD) in the presence of sulphite. (A) The nickel-coordination environment superimposed on the final 2Fo–Fc electron density 1σ map; (B) the corresponding structural model. The figure settings are the same as in Fig. 3. | ||
The oxidation state (BQ or 1,4-hydroquinone, HQ) of the inhibitor bound to αCys322 and αCys555 was also a matter of investigation. In order to discern between these two possibilities, both moieties were used as initial models for crystallographic refinement, yielding a negligible difference in data statistics. Quantum-mechanical calculations were thus carried out on the two possible redox moieties (see the ESI†): a comparison of the key C–O distances for the covalently modified urease cysteines obtained by crystallography with those derived from these calculations (see Tables 3, 4-ESI and Fig. 6, 7-ESI†), did not allow us to unequivocally discern between the oxidized or reduced form in the crystal. However, a comparison of the C1–C2–Sγ–Cβ dihedral angle (see Scheme 1-ESI†) suggests the presence of the reduced form, characterized by a value close to 90° both in the crystal structure and in the theoretical model (see Tables 5, 6-ESI and Fig. 6, 7-ESI†). This particular conformation is known to be due to the formation of an intra-molecular hydrogen-bond between a S atom p-orbital and the O–H group of the HQ.20 The final refinement for both structures was thus carried out using the reduced form of BQ. The reaction of the BQ-inactivated enzyme with sulphite in solution, leading to the crystallographically characterized compound 5 (Scheme 1), requires however the presence of the oxidized form 4, which must be obtained from the initial product 2 by reaction with dissolved molecular oxygen (Scheme 1). The presence of the reduced form 2 in the solid state could thus be due to photo-reduction. In the case of the sulphonate-substituted compound 5, on the other hand, the presence of the reduced form is consistent with its known stabilization by the electron-withdrawing properties of the sulphonate group, as discussed for compound 3.
In this paper we have described and discussed the reactivity of αCys322, a key residue for the catalytic activity of urease, towards 1,4-benzoquinone, and established the molecular structural details of the resulting irreversibly inactivated enzyme. This information is essential for the development of structure-based design and structure–activity relationship studies of novel urease inhibitors based on this moiety. Moreover, considering the conservation of the cysteine on the mobile flap of ureases from other sources, such as the pathogenic bacterium Helicobacter pylori, the results of our study have wider general applications. Finally, the large class of quinonoid compounds, containing 1 as their basic core, is widely distributed in nature and play pivotal roles as electron transfer agents in primary metabolic processes such as photosynthesis, respiration and oxidative phosphorylation.21 A large number of chemical derivatives of differently substituted quinones are also involved in pharmacological applications for their activity as antibiotics, antitumor, antimalarial, antineoplastic, anticoagulant and herbicidal agents. Therefore, the structural information obtained in our study could have far reaching consequences in the comprehension of the reactivity of this class of compounds with thiol groups in proteins, enzymes and enzyme cofactors.22
This research was funded by CIRMMP (Consorzio Interuniversitario di Risonanze Magnetiche di Metallo-Proteine) and by Specialty Fertilizer Products (Leawood, KS, USA). We thank Dr Vara for his assistance in S. pasteurii cell growth. LM is a Ph.D. fellow from the University of Bologna. FM is a research assistant supported by a FARB Program of the University of Bologna. X-ray diffraction data were collected under the beam time award number MX-333 from the European Molecular Biology Laboratory (EMBL, Petra III, c/o DESY, Hamburg, Germany).
Notes and references
- (a) B. Zambelli, F. Musiani, S. Benini and S. Ciurli, Acc. Chem. Res., 2011, 44, 520–530 CrossRef CAS PubMed; (b) M. J. Maroney and S. Ciurli, Chem. Rev., 2014, 114, 4206–4228 CrossRef CAS PubMed.
- J. C. Rutherford, PLoS Pathog., 2014, 10, e1004062 Search PubMed.
- S. Kiss and M. Simihaian, Improving efficiency of urea fertilizers by inhibition of soil urease activity, Kluwer Academic Publishers, Dordrecht, The Netherlands, 2002 Search PubMed.
- (a) S. Benini, W. R. Rypniewski, K. S. Wilson, S. Miletti, S. Ciurli and S. Mangani, Structure, 1999, 7, 205–216 CrossRef CAS PubMed; (b) S. Ciurli, S. Benini, W. R. Rypniewski, K. S. Wilson, S. Miletti and S. Mangani, Coord. Chem. Rev., 1999, 190–192, 331–355 CrossRef CAS.
- F. Musiani, E. Arnofi, R. Casadio and S. Ciurli, J. Biol. Inorg. Chem., 2001, 6, 300–314 CrossRef CAS PubMed.
- (a) L. Mazzei, M. Cianci, S. Benini, L. Bertini, F. Musiani and S. Ciurli, J. Inorg. Biochem., 2016, 154, 42–49 CrossRef CAS PubMed; (b) S. Benini, M. Cianci, L. Mazzei and S. Ciurli, J. Biol. Inorg. Chem., 2014, 19, 1243–1261 CrossRef CAS PubMed.
- (a) J. M. Bremner and L. A. Douglas, Soil Biol. Biochem., 1971, 3, 297–307 CrossRef CAS; (b) R. L. Mulvaney and J. M. Bremner, Soil Biol. Biochem., 1978, 10, 297–302 CrossRef CAS; (c) W. Zaborska, M. Kot and K. Superata, J. Enzyme Inhib. Med. Chem., 2002, 17, 247–253 CrossRef CAS PubMed; (d) W. Zaborska, B. Krajewska, M. Kot and W. Karcz, Bioorg. Chem., 2007, 35, 233–242 CrossRef CAS PubMed; (e) M. Kot and W. Zaborska, J. Enzyme Inhib. Med. Chem., 2006, 21, 537–542 CrossRef CAS PubMed; (f) B. Krajewska, J. Mol. Catal. B: Enzym., 2009, 59, 9–21 CrossRef CAS.
- J. M. Snell and A. Weissberger, J. Am. Chem. Soc., 1939, 61, 450–453 CrossRef CAS.
- P. R. Martin and R. P. Hausinger, J. Biol. Chem., 1992, 267, 20024–20027 CAS.
- S. Benini, W. R. Rypniewski, K. S. Wilson, S. Ciurli and S. Mangani, J. Biol. Inorg. Chem., 1998, 3, 268–273 CrossRef CAS.
- M. J. Todd and R. P. Hausinger, J. Biol. Chem., 1989, 264, 15835–15842 CAS.
- T. H. James, J. M. Snell and A. Weissberger, J. Am. Chem. Soc., 1938, 60, 2084–2093 CrossRef CAS.
- (a) T. H. James and A. Weissberger, J. Am. Chem. Soc., 1938, 60, 98–104 CrossRef CAS; (b) T. H. James and A. Weissberger, J. Am. Chem. Soc., 1939, 61, 442–450 CrossRef CAS.
- S. Benini, P. Kosikowska, M. Cianci, L. Mazzei, A. G. Vara, L. Berlicki and S. Ciurli, J. Biol. Inorg. Chem., 2013, 18, 391–399 CrossRef CAS PubMed.
- M. S. Weiss, J. Appl. Crystallogr., 2001, 34, 130–135 CrossRef CAS.
- (a) G. N. Murshudov, A. A. Vagin and E. J. Dodson, Acta Crystallogr., 1997, 53, 240–255 CAS; (b) G. N. Murshudov, A. A. Vagin, A. Lebedev, K. S. Wilson and E. J. Dodson, Acta Crystallogr., 1999, 55, 247–255 CAS.
- D. W. J. Cruickshank, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1999, 55, 583–601 CrossRef CAS.
- M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. W. Leslie, A. McCoy, S. J. McNicholas, G. N. Murshudov, N. S. Pannu, E. A. Potterton, H. R. Powell, R. J. Read, A. Vagin and K. S. Wilson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 235–242 CrossRef CAS PubMed.
- R. A. Laskowski, M. W. MacArthur, D. S. Moss and J. M. Thornton, J. Appl. Crystallogr., 1993, 26, 283–291 CrossRef CAS.
- P. Politzer, J. S. Murray and P. Lane, Int. J. Quantum Chem., 2007, 107, 3046–3052 CrossRef CAS.
- I. Abraham, R. Joshi, P. Pardasani and R. T. Pardasani, J. Braz. Chem. Soc., 2011, 22, 385–421 CrossRef CAS.
- W.-W. Li, J. Heinze and W. Haehnel, J. Am. Chem. Soc., 2005, 127, 6140–6141 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, structural parameters and unbiased electron density maps of BQ inhibited SPU, details of the SPU surface, and the results of the quantum mechanical calculations. See DOI: 10.1039/c6dt00652c |
| This journal is © The Royal Society of Chemistry 2016 |