Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dinuclear ruthenium(II) complexes containing one inert metal centre and one coordinatively-labile metal centre: syntheses and biological activities
Xin
Li
a,
Kirsten
Heimann
bc,
Fangfei
Li†
a,
Jeffrey M.
Warner
cd,
F.
Richard Keene
*cef and
J.
Grant Collins
*a
aSchool of Physical, Environmental and Mathematical Sciences, University of New South Wales, Australian Defence Force Academy, Canberra, ACT 2600, Australia. E-mail: g.collins@adfa.edu.au
bCollege of Marine & Environmental Sciences, James Cook University, Townsville, QLD 4811, Australia
cCentre for Biodiscovery and Molecular Development of Therapeutics, James Cook University, Townsville, QLD 4811, Australia
dCollege of Public Health, Medical and Veterinary Sciences, James Cook University, Townsville, QLD 4811, Australia
eCollege of Science, Technology & Engineering, James Cook University, Townsville, QLD 4811, Australia
fSchool of Physical Sciences, University of Adelaide, Adelaide, SA 5005, Australia. E-mail: Richard.keene@adelaide.edu.au
First published on 3rd February 2016
Abstract
A series of non-symmetric dinuclear polypyridylruthenium(II) complexes (Rubbn-Cl) that contain one inert metal centre and one coordinatively-labile metal centre, linked by the bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane ligand (“bbn” for n = 7, 12 and 16), have been synthesised and their potential as antimicrobial agents examined. The minimum inhibitory concentrations (MIC) of the ruthenium(II) complexes were determined against four strains of bacteria − Gram-positive Staphylococcus aureus (S. aureus) and methicillin-resistant S. aureus (MRSA), and Gram-negative Escherichia coli (E. coli) and Pseudomonas aeruginosa (P. aeruginosa). The Rubbn-Cl complexes displayed good antimicrobial activity, with Rubb12-Cl being the most active complex against both Gram-positive and Gram-negative strains. Interestingly, Rubb7-Cl was found to be eight- and sixteen-fold more active towards E. coli than against S. aureus and MRSA, respectively. The cytotoxicities of the Rubbn-Cl complexes against three eukaryotic cell lines – two kidney cell lines (BHK and HEK-293) and one liver cell line (HepG2) – were examined. The Rubbn-Cl complexes were found to be considerably less toxic towards eukaryotic cells than S. aureus, MRSA and E. coli, with Rubb12-Cl being thirty- to eighty-times more toxic to the bacteria than to BHK, HEK-293 or HepG2 cells. Unexpectedly, Rubb7-Cl was far more toxic to HepG2 cells (24 h-IC50 = 3.7 μM) and far less toxic to BHK cells (24 h-IC50 = 238 μM) than the Rubb12-Cl and Rubb16-Cl complexes. In order to understand the unexpected large differences in the cytotoxicities of the Rubbn-Cl complexes towards eukaryotic cells, a confocal microscopic study of their intracellular localisation was undertaken. The results suggest that the observed cytotoxicity might be related to the extent of DNA binding.
Introduction
The emergence of drug-resistant populations of microorganisms has become a serious worldwide health issue.1 There is clearly a need for new antimicrobials; however more importantly, new classes of antimicrobials are needed rather than drugs based upon analogues of known scaffolds.Traditionally, the design and development of new antimicrobial drugs has centred upon organic chemistry. However, due to the success of cisplatin as an anticancer drug and the established ability of transition metal complexes to bind DNA and RNA,2–6 there has been increasing interest in using metal complexes as antimicrobial agents.7–14 Among the transition metal complexes, ruthenium-based complexes have drawn increasing attention.9–14 Dwyer and co-workers were the first to report the biological activity of mononuclear tris(bidentate) inert polypyridyl metal complexes, in particular complexes with 1,10-phenanthroline ligands.9,10 [Ru(phen)3]2+ was found to be inactive; however, the introduction of methyl substituents on the phen ligands dramatically increased the activity against all bacteria.9,10 More recently, it has been demonstrated that polypyridylruthenium(II) complexes which bind DNA by intercalation have significant bactericidal activity, particularly against Gram-positive strains.11,12 While DNA binding is generally thought to be responsible for the antimicrobial activity of polypyridylruthenium(II) complexes, Lam et al. suggested that the antimicrobial activity of a bis(bipyridine)ruthenium(II) complex containing a N-phenyl-substituted diazafluorene ligand might be due to DNA damage caused by the formation of reactive oxygen species.15 In addition, a range of labile ruthenium(II) and ruthenium(III) complexes have shown antimicrobial activity.16,17
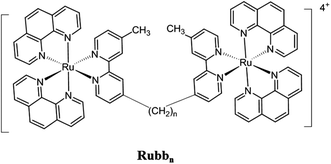
We have recently demonstrated that dinuclear polypyridylruthenium(II) complexes containing a flexible bis[4(4′-methyl-(-2,2′-bipyridyl)]-1,n-alkane (bbn) bridging ligand (see Fig. 1) have good antimicrobial activity.18–21 These ruthenium complexes were highly active against a range of pathogenic bacteria, particularly Gram-positive strains,18 and maintained the activity against drug-resistant bacteria, including strains that are of considerable current concern, e.g. methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE). Furthermore, preliminary toxicity experiments indicated the dinuclear Rubbn complexes were significantly less toxic to eukaryotic cells.22,23 Based upon the good antimicrobial activity and cell selectivity of the Rubbn complexes, corresponding tri- and tetra-nuclear inert ruthenium complexes were subsequently synthesised.24 These complexes generally showed better activities than the dinuclear analogues and were more active against Gram-positive species.24
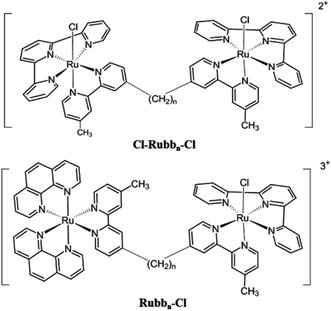
In another approach, we have also examined the effects of incorporating labile chlorido groups into dinuclear ruthenium(II) complexes linked by the bbn ligand, [{Ru(tpy)Cl}2{μ-bbn}]2+ (Cl-Rubbn-Cl; where tpy = 2,2′:6′,2′′-terpyridine),25 see Fig. 2. The symmetrical Cl-Rubbn-Cl complexes showed good activity against both Gram-positive bacteria and Gram-negative species. However, incorporation of the chlorido groups did significantly affect the relative activities of the ruthenium complexes, compared to the corresponding inert Rubbn complexes. Whereas the order of activities for the inert complexes was Rubb16 ≥ Rubb12 > Rubb7, it was found that Cl-Rubb12-Cl was the most active of the Cl-Rubbn-Cl complexes and Cl-Rubb16-Cl was slightly less active than the Cl-Rubb7-Cl complex. Taken together, the combined results highlight the balance between cationic charge and lipophilicity; however, it is not yet clear where the optimal charge/lipophilicity balance lies. In order to help clarify this issue, we aimed to synthesise and examine the antimicrobial activities of dinuclear ruthenium complexes that contained one inert metal centre and one metal centre that incorporated a chlorido ligand (Rubbn-Cl complexes, see Fig. 2).
In this study, the synthesis and the antimicrobial properties of the non-symmetrical Rubbn-Cl complexes (for n = 7, 12 and 16) against Gram-positive S. aureus and MRSA, and Gram-negative Escherichia coli and Pseudomonas aeruginosa were examined. As the clinical potential of any new drug is determined by both the antimicrobial activity and the associated toxicity towards eukaryotic cells, the cytotoxicities of the Rubbn-Cl complexes against three eukaryotic cell lines were also examined. In order to understand the unexpected large differences in the cytotoxicities of the Rubbn-Cl complexes towards eukaryotic cells, a confocal microscopic study of their intracellular localisation was also undertaken.
Experimental
Physical measurements and materials
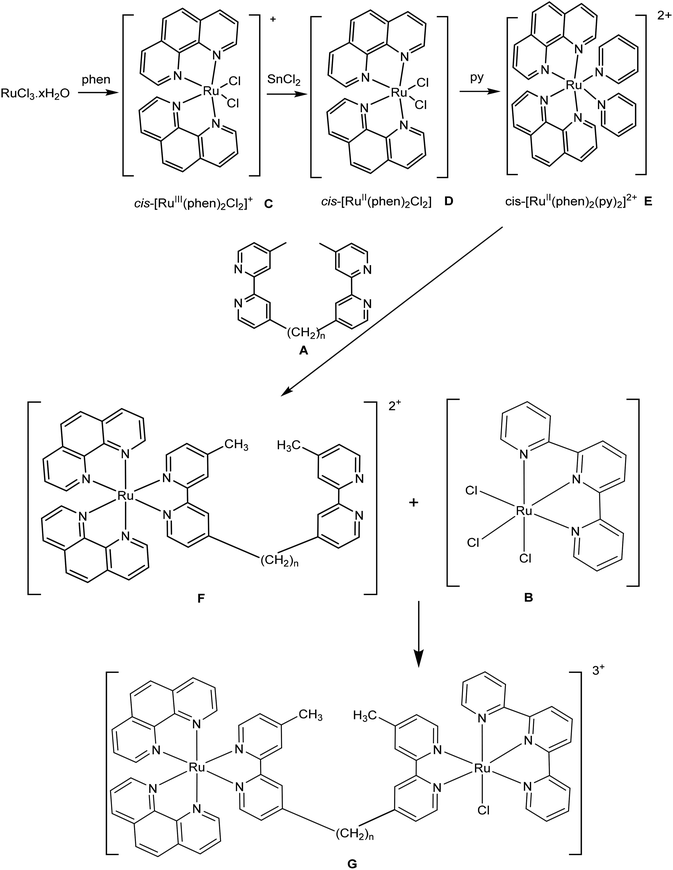
1H NMR spectra were recorded on a Varian Unityplus 400 MHz spectrometer with chemical shifts reported as δ values relative to the signal of tetramethylsilane. The NMR solvents D2O (99.9%), CD2Cl2 (99.8%), CDCl3 (99.8%), CD3CN (99.8%) and DMSO-d6 (99.9%) were obtained from Cambridge Isotope Laboratories. Microanalyses were performed by the Microanalytical Unit, Research School of Chemistry, Australian National University. High-resolution mass spectral measurements were made using a Waters LCT mass spectrometer (Research School of Chemistry, Australian National University). 4,4′-Dimethyl-2,2′-bipyridine (Me2bpy), lithium di-isopropylamide (LDA), 1,5-dibromopentane, 1,10-dibromodecane, 1,14-dibromotetradecane, 2;2′,6′;2′′-terpyridine (tpy), and 1,10-phenanthroline (phen) were purchased from Sigma-Aldrich, and RuCl3·xH2O from American Elements. For the nucleotides used, 5′-GMP (di-sodium salt) was obtained from Sigma-Aldrich while 5′-AMP, 5′-CMP and 5′-UMP (sodium salts) were from Apollo chemicals, Burlington, US. All chemicals were used as supplied.The mononuclear ruthenium complex [Ru(tpy)(Me2bpy)Cl]Cl was synthesised as previously described.26 The synthesis of the bridging ligands bbn (n = 7, 12 and 16; A) were performed in a similar manner to that reported in the literature.27 The precursors [Ru(tpy)Cl3] (B) [Ru(phen)2Cl2]Cl (C), [Ru(phen)2Cl2] (D), [Ru(phen)2(py)2]Cl2 (E; py = pyridine) and rac-[Ru(phen)2(bbn)](PF6)2 (F) were prepared according to previously reported methods.27–29
Synthesis of [Ru(phen)2(μ-bbn)Ru(tpy)Cl]Cl3 (Rubbn-Cl, n = 7, 12 and 16, G)
For [Ru(phen)2(μ-bb7)Ru(tpy)Cl]Cl3, solid [Ru(tpy)Cl3] (10 mg, 0.032 mmol) and [Ru(phen)2(bb7)](PF6)2 (3.7 mg, 0.032 mmol) were refluxed in ethanol/water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10 mL) for 3 h. After cooling, excess NH4PF6 was added to precipitate a dark-brown material which was filtered and washed with ethanol. The crude product was then loaded onto a Sephadex LH20 size-exclusion column and eluted with acetone. The [Ru(phen)2(μ-bb7)Ru(tpy)Cl](PF6)3 fraction was obtained as the major dark-brown band which was isolated and evaporated to dryness. The PF6− salt was converted to the chloride (metathesis) by dissolving the solid in the minimum amount of acetone followed by the dropwise addition of a saturated solution of tetraethylammonium chloride in acetone with stirring for 30 min. The resulting fluffy precipitate was centrifuged, decanted, washed several times with cold acetone and dried under reduced pressure to afford [Ru(phen)2(μ-bb7)Ru(tpy)Cl]Cl3 (yield 70–80%). Separation of the possible geometric isomers (the chlorido ligand in the coordinatively-labile metal centre can potentially be either cis or trans to the pyridine ring of the bbn ligand bearing the methyl group – the trans disposition is shown in Fig. 2) was not attempted.
1, 10 mL) for 3 h. After cooling, excess NH4PF6 was added to precipitate a dark-brown material which was filtered and washed with ethanol. The crude product was then loaded onto a Sephadex LH20 size-exclusion column and eluted with acetone. The [Ru(phen)2(μ-bb7)Ru(tpy)Cl](PF6)3 fraction was obtained as the major dark-brown band which was isolated and evaporated to dryness. The PF6− salt was converted to the chloride (metathesis) by dissolving the solid in the minimum amount of acetone followed by the dropwise addition of a saturated solution of tetraethylammonium chloride in acetone with stirring for 30 min. The resulting fluffy precipitate was centrifuged, decanted, washed several times with cold acetone and dried under reduced pressure to afford [Ru(phen)2(μ-bb7)Ru(tpy)Cl]Cl3 (yield 70–80%). Separation of the possible geometric isomers (the chlorido ligand in the coordinatively-labile metal centre can potentially be either cis or trans to the pyridine ring of the bbn ligand bearing the methyl group – the trans disposition is shown in Fig. 2) was not attempted.
[Ru(phen)2(μ-bb12)Ru(tpy)Cl]Cl3 and [Ru(phen)2(μ-bb16)Ru(tpy)Cl]Cl3 were synthesised using an analogous procedure as that for [Ru(phen)2(μ-bb7)Ru(tpy)Cl]Cl3 but with the appropriate bbn bridging ligand.
Δ-[Ru(phen)2(μ-bbn)Ru(tpy)Cl]Cl3 complexes were synthesised following the same procedure as that for the racemic mixtures, using Δ-[Ru(phen)2(bbn)](PF6)2 (n = 7, 12 and 16) as precursors. CD spectra: Δ-Rubb7-Cl {λ/nm (Δε/cm−1 M−1) H2O}: 290 (−374.3), 281 (−294.2), 272 (−405.1), 261 (469.3), 233 (27.3), 221 (77.5). Δ-Rubb12-Cl {λ/nm (Δε/cm−1 M−1) H2O}: 290 (−356.7), 281 (−244.3), 271 (−352.3), 261 (412.8), 228 (10.5), 213 (107.4). Δ-Rubb16-Cl {λ/nm (Δε/cm−1 M−1) H2O}: 288 (−364.4), 279 (−285.9), 271 (−358.7), 260 (393.6), 227 (39.2), 221 (80.5).
Aquation of Rubbn-Cl complexes
The ruthenium complexes [Ru(tpy)(Me2bpy)Cl]Cl and Δ-Rubb7-Cl were dissolved in D2O (650 μL) to give 1.0 mM solutions. 1H NMR spectra were then recorded as a function of time at 25 °C.Reaction of ruthenium complexes with nucleotides
Reactions with nucleotides were carried out after the aquation of the chlorido-containing ruthenium complexes had reached equilibrium. The nucleotides 5′-GMP, 5′-AMP, 5′-CMP, and 5′-UMP were dissolved in D2O and separately added to the ruthenium complexes (1 mM) dissolved in D2O at the desired [Ru complex]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [nucleotide] ratio. 1H NMR spectra were recorded as a function of time after mixing the sample thoroughly. NOESY experiments were conducted using the method of States et al.,30 with 1024 data points in t2 for 256 t1 values, a pulse repetition delay set to 1.5 s and mixing times from 100 to 500 ms. Correlation spectroscopy experiments (DQFCOSY) were recorded using the same t1, t2 and pulse repetition values.
[nucleotide] ratio. 1H NMR spectra were recorded as a function of time after mixing the sample thoroughly. NOESY experiments were conducted using the method of States et al.,30 with 1024 data points in t2 for 256 t1 values, a pulse repetition delay set to 1.5 s and mixing times from 100 to 500 ms. Correlation spectroscopy experiments (DQFCOSY) were recorded using the same t1, t2 and pulse repetition values.
Molecular modelling was performed using HyperChem.31 Energy minimisation by Polak–Ribiere conjugate-gradient refinement was carried out with the metal complex treated as a rigid group. The ruthenium complex was manually docked to the GMP to reflect observed intermolecular NOEs.
Bacterial strains
Note: the bacterial strains used in this study are classified as risk group 2 according to the Australian/New Zealand Standard (AS/NZS 2243.4:2010) and accordingly were manipulated in a PC2 class laboratory. Gram-positive isolates {a methicillin-susceptible S. aureus strain (ATCC 25923) and a clinical, multidrug-resistant, MRSA strain (JCU culture collection)}, and two Gram-negative isolates {E. coli (ATCC 25922) and P. aeruginosa (ATCC 27853)}, were used for antimicrobial studies.Determination of minimum inhibitory concentration (MIC)
MIC values were determined in duplicate by standard micro-dilution methodology in CAMHB,32 using gentamicin as the positive control, as previously described.18,19Determination of minimum bactericidal concentration (MBC)
The MBC tests were performed in duplicate according to a standard microbiological techniques protocol,33 as previously described.18,19Cell culture
Two kidney cell lines {BHK (baby hamster kidney) and HEK-293 (embryonic kidney)}, and one liver cell line {HepG2 (liver carcinoma)} were used in this study. All cell lines were generously supplied by the Australian Army Malaria Institute (AMI, Enoggera, QLD, Australia), and originated from the American Type Culture Collection (ATCC, Manassas, USA). Cells were cultured as previously described.22 Cells used in the study were in the logarithmic growth phase and were grown to 70% confluence, and then trypsinised with 0.25% trypsin–0.02% EDTA (Sigma-Aldrich) for detachment and used in the assays described below.Half-maximal inhibitory concentration (IC50) cytotoxicity assay
Half-maximal inhibitory concentrations (IC50) of the ruthenium complexes were assessed using the Alamar Blue cytotoxicity assay (Invitrogen, Carlsbad, USA) as previously described.22,34Cellular localisation of the ruthenium complexes
Trypsinised HepG2 or BHK cells were seeded in Lumox® multiwell plates (Sarstedt, Numbrecht, Germany), and incubated with 5 to 100 μM ruthenium complexes at 37 °C with 5% CO2 for 20 h. Following incubation, cells were stained with 100 nM Mitotracker® Green FM (Invitrogen, Carlsbad, USA) for mitochondrial staining, 100 nM DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) for nuclear staining and/or 50 nM SYTO 9 (Invitrogen) for nucleolus staining. Staining was carried out in RPMI-1640 medium under standard cultivation conditions as per the manufacture's instructions. Following staining, cells were gently rinsed with phosphate buffer solution (PBS; pH = 7.1) prior to confocal laser scanning microscopy.The cellular localisation of the ruthenium complexes was determined using a Zeiss laser scanning confocal microscope (LSM 700, Carl Zeiss, Göttingen, Germany). Samples were viewed under a 63× oil immersion lens. The ruthenium complexes (λex = 450 nm, λem = 610 nm), Mitotracker Green FM (λex = 490 nm, λem = 516 nm) and SYTO 9 (λex = 486 nm, λem = 501 nm) were excited using a blue argon laser (λex = 488 nm), and emissions were collected over the range 570–650 nm for the metal complexes, 470–550 nm for Mitotracker and 495–510 nm for SYTO 9. For DAPI excitation, a diode laser (λex = 405 nm) was used and the emission detected at 430–500 nm. Image data acquisition and processing was performed using Zen software 2009 (Carl Zeiss).
Results
Synthesis of Rubbn-Cl (n = 7, 12 and 16) complexes
The syntheses of the symmetric oligonuclear polypyridine ruthenium complexes Rubbn, Tri-Rubbn, Tetra-Rubbn and Cl-Rubbn-Cl have been previously reported and their biological properties have been thoroughly studied.18–25 To obtain a better structure–activity relationship, a new class of non-symmetric dinuclear ruthenium complexes Rubbn-Cl was designed and synthesised. The Rubbn-Cl complexes were prepared from [Ru(phen)2(py)2]2+ (E), [Ru(tpy)Cl3] (B) and the bbn ligand (A) following the synthetic route shown in Scheme 1, with the final dinuclear product Rubbn-Cl (G) obtained through reaction of [Ru(phen)2bbn]2+ (F) with [Ru(tpy)Cl3]. [Ru(phen)2(py)2]2+ and [Ru(phen)2bbn]2+ were purified by cation-exchange chromatography on an SP Sephadex C-25 column, whereas the final Rubbn-Cl complexes were purified by size-exclusion on a Sephadex LH20 column. The Rubbn-Cl complexes were characterised by 1H and 13C NMR, microanalysis and ESI-MS.Aquation of chlorido-containing ruthenium complexes
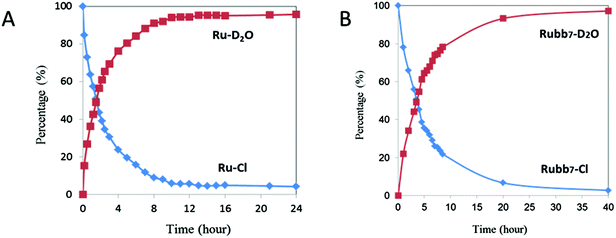
As the Rubbn-Cl complexes are activated towards covalent bond formation with intracellular targets by aquation, a study of the aquation of Rubb7-Cl and the parent complex [Ru(tpy)(Me2bpy)Cl]+ was carried out. The rate of aquation of [Ru(tpy)(Me2bpy)Cl]+ has been previously determined by 1H NMR spectroscopy,26 where the concomitant disappearance of the resonance from the H6 of the Me2bpy ligand from the chlorido complex and the emergence of the corresponding resonance from the aqua species were monitored as a function of time. By following the disappearance of the H6 resonance of the Me2bpy ligand for [Ru(tpy)(Me2bpy)Cl]+ (9.71 ppm) and Rubb7-Cl (9.52 ppm) and the emergence of the corresponding resonance of the aqua complexes (9.52 and 9.24 ppm, respectively), it was determined that 50% aquation is achieved in 90 minutes for [Ru(tpy)(Me2bpy)Cl]+ and 210 minutes for Rubb7-Cl (see Fig. 3).Reaction of [Ru(tpy)(Me2bpy)(D2O)]2+ with nucleotides
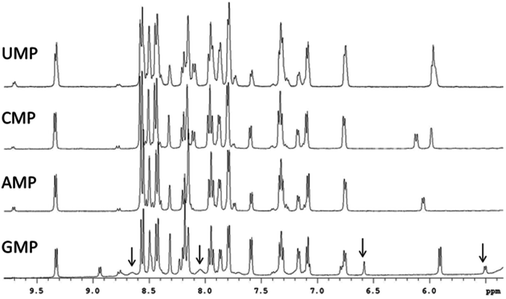
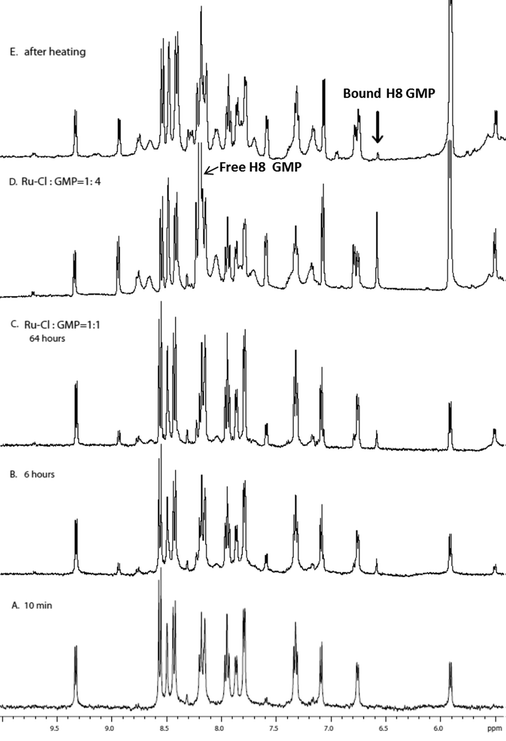
The covalent binding of [Ru(tpy)(Me2bpy)(D2O)]2+ with each of the di-anions of guanosine monophosphate (5′-GMP), cytidine monophosphate (5′-CMP), adenosine monophosphate (5′-AMP) and uridine monophosphate (5′-UMP) was studied. After a 24-hour incubation of [Ru(tpy)(Me2bpy)(D2O)]2+ with each nucleotide, the 1H NMR spectrum was recorded and the spectra are shown in Fig. 4. Addition of AMP, CMP and UMP to [Ru(tpy)(Me2bpy)(D2O)]2+ only resulted in the slight broadening of the nucleotide resonances without the emergence of new peaks, suggesting a weak reversible association between the ruthenium complex and the nucleotides. However, 24 hours after the addition of GMP to [Ru(tpy)(Me2bpy)(D2O)]2+ distinct differences in the NMR spectrum were observed. The absence of a resonance at 9.71 ppm (H6 of Me2bpy of the chlorido species) indicated that the equilibrium had shifted towards the aqua complex. Furthermore, a small second set of broad peaks emerged, indicating the formation of a covalently-bound adduct. Based upon the relative integrals of the sugar H1′ of the free GMP (5.90 ppm) and ruthenium complex-bound GMP (5.50 ppm), approximately 35% of the GMP covalently bound [Ru(tpy)(Me2bpy)(D2O)]2+ in 24 hours.Given the observed preferential binding of [Ru(tpy)(Me2bpy)(D2O)]2+ with GMP, further studies were only carried out with GMP. Fig. 5 shows the 1H NMR spectrum of [Ru(tpy)(Me2bpy)(D2O)]2+ with added GMP as a function of time, and after the addition of further GMP after 64 hours. The H8 of GMP was tentatively assigned by heating the sample at 50 °C for 16 hours. The H8 of GMP will slowly exchange with a deuterium in the D2O solvent at 50 °C; e.g. see the reduction in intensity of the H8 of the free GMP in Fig. 5D and E. Consequently, the singlet resonance at 6.56 ppm could be assigned to the H8 of bound GMP.
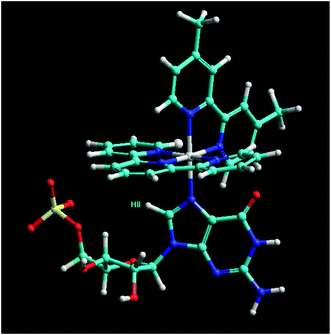
The assignment of the proton resonances from the [Ru(tpy)(Me2bpy)GMP] adduct was determined from DQFCOSY and NOESY spectra (data not shown). In the NOESY spectrum of the aromatic to sugar H1′ region, strong NOE cross peaks were observed between the tpy H6 and H6′′, H5 and H5′′, H4 and H4′′, H3 and H3′′ and H3′ and H5′, indicating the tpy protons were in slow exchange (on the NMR time scale) between two forms. Due to rotation around the Ru–N7 bond, it is possible that [Ru(tpy)(Me2bpy)GMP] complex may exist in two conformers that are in slow exchange. An NOE cross peak between the H8 and sugar H1′ of the bound GMP at 6.56/5.50 ppm provided further support for the assignment of the H8 of the bound GMP. Based on the NMR analysis, the binding site of ruthenium complex is most likely at the N7 of the GMP, given the large shift observed for the GH8 resonance. A molecular model of the ruthenium complex-GMP adduct is shown in Fig. 6. In the model, the GMP H8 is positioned directly below the tpy aromatic rings, which may explain the unusual upfield shift observed for the resonance upon N7-metallation.
The time-course 1H NMR spectra of the reaction between the dinuclear complex Rubb7-Cl and GMP also showed the emergence of resonances for the ruthenium complex-GMP adduct (data not shown). A singlet at 6.65 ppm could be assigned to the H8 from bound GMP, and new broad peaks in the 8.7–8.9 ppm region were possibly from the bound metal complex. These observations were similar to those from the reaction between [Ru(tpy)(Me2bpy)Cl]+ and GMP, described above, suggesting that Rubb7-Cl interacts with GMP by forming a covalent bond.
Antimicrobial activity
The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values for the Rubbn-Cl complexes against S. aureus, MRSA, E. coli and P. aeruginosa were determined and are summarised in Table 1. All Rubbn-Cl complexes showed significant activity against both the Gram-negative and Gram-positive strains, with the Gram-negative strain P. aeruginosa being the least susceptible. Overall, Rubb12-Cl showed the best activity across the Rubbn-Cl series. Compared to Rubb12-Cl, Rubb16-Cl was similarly active against the two Gram-positive strains, but showed considerably lower activity against the Gram-negative strains. On the other hand, Rubb7-Cl was less active than Rubb12-Cl against the Gram-positive strains, but equally as active as Rubb12-Cl against the Gram-negative bacteria. As the MBC values were ≤2 × MIC, it is concluded that all the Rubbn-Cl complexes are bactericidal, rather than bacteriostatic. The Rubbn-Cl complexes generally showed similar or better activity than the Rubbn and Cl-Rubbn-Cl complexes, with Rubb12-Cl exhibiting as good an activity profile as any of the other previously-reported ruthenium complexes linked by the bbn ligand, including the tri- and tetra-nuclear inert complexes.18,24| Complexes | S. aureus | MRSA | E. coli | P. aeruginosa | ||||
|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| Δ-Rubb7-Cl | 5.6 | 11.2 | 11.2 | 22.4 | 0.7 | 0.7 | 11.2 | 22.4 |
| Δ-Rubb12-Cl | 0.7 | 0.7 | 0.7 | 1.4 | 0.7 | 0.7 | 11.2 | 11.2 |
| Δ-Rubb16-Cl | 0.7 | 0.7 | 1.4 | 1.4 | 2.7 | 2.7 | 43.2 | 43.2 |
| ΔΔ-Rubb7 | 10.7 | 21.3 | 10.7 | 21.3 | 10.7 | 10.7 | 85.3 | >85 |
| ΔΔ-Rubb12 | 0.7 | 1.3 | 0.7 | 1.3 | 1.3 | 1.3 | 20.4 | 20.4 |
| ΔΔ-Rubb16 | 0.6 | 0.6 | 0.6 | 1.2 | 1.2 | 1.2 | 9.8 | 9.8 |
| Gentamicin | 0.4 | 0.8 | 28 | >200 | 0.8 | 0.8 | 1.6 | 3.2 |
Over the four bacteria examined in this study, Rubb7-Cl and Rubb12-Cl showed slightly better antimicrobial activities compared to their inert analogues, Rubb7 and Rubb12. Conversely, Rubb16-Cl displayed slightly lower activity than Rubb16. Interestingly, Rubb7-Cl and Rubb12-Cl show better activities against the two Gram-negative species E. coli and P. aeruginosa than Rubb7 and Rubb12, respectively. Particularly noteworthy is the activity of Rubb7-Cl against E. coli compared to the Gram-positive species S. aureus and MRSA: Rubb7-Cl is eight- and sixteen-fold more active towards E. coli than against S. aureus and MRSA, respectively. This preferential activity towards a Gram-negative species is very unusual for metal-based antimicrobial agents.
Cytotoxicity against eukaryotic cells and selective activity
To further evaluate the potential of the ruthenium complexes as antimicrobial agents, an understanding of their cytotoxicity towards mammalian cells is necessary. The cytotoxicities of the ruthenium complexes against the kidney cell lines BHK and HEK-293 were determined for incubation times of 24 hours. As there were no differences in the cytotoxicities (within experimental error) between the Δ-Rubbn-Cl and the rac-Rubbn-Cl complexes in preliminary experiments, the rac-Rubbn-Cl complexes were used to determine the 24-hour IC50 values. The 24 h-IC50 values of the Rubbn-Cl and Rubbn complexes are summarised in Table 2.| Complexes | IC50 | SI | ||||
|---|---|---|---|---|---|---|
| BHK | HEK-293 | BHK vs. S. aureus | BHK vs. E. coli | HEK-293 vs. S. aureus | HEK-293 vs. E. coli | |
| Rac-Rubb7-Cl | 238.1 ± 14.8 | 69.1 ± 5.6 | 42.5 | 340.1 | 12.3 | 98.7 |
| Rac-Rubb12-Cl | 47.4 ± 0.9 | 58.7 ± 2.1 | 67.7 | 67.7 | 83.9 | 83.9 |
| Rac-Rubb16-Cl | 22.6 ± 2.3 | 41.1 ± 1.9 | 32.3 | 8.4 | 58.7 | 15.2 |
| ΔΔ-Rubb12 | 70.5 ± 26.4 | 50.9 ± 19.9 | 100.7 | 54.2 | 72.7 | 39.2 |
| ΔΔ-Rubb16 | 29.8 ± 1.1 | 21.0 ± 10.8 | 49.7 | 24.8 | 35 | 17.5 |
Generally, the complexes with a longer linking chain were more toxic to the cells. Among the rac-Rubbn-Cl complexes, Rubb16-Cl was the most toxic complex towards BHK and HEK-293 cells. In contrast, Rubb7-Cl was nontoxic (>200 μM) towards BHK but showed moderate cytotoxicity towards HEK-293, while Rubb12-Cl showed similar cytotoxicity towards both cell lines. These observations suggest that the linking chain length plays an important role in their cytotoxicities. The inert complexes ΔΔ-Rubb12 and ΔΔ-Rubb16 also showed the same trend with ΔΔ-Rubb16 being more toxic than ΔΔ-Rubb12.
The good antimicrobial activity of the ruthenium complexes suggests they may have potential as antimicrobial agents. However, to be clinically useful as antimicrobial agents, it is desirable that the compounds exhibit significantly greater toxicity towards bacterial cells than mammalian cells. Table 2 also summarises the selectivity indices (SI, 24 h-IC50/MIC) between the BHK and HEK-293 cell lines and the Gram-positive bacterium S. aureus and the Gram-negative species E. coli. In general, the SI values demonstrated that all the ruthenium complexes were more toxic against bacterial cells than eukaryotic cells, with the SI values ranging from 8–340. Noticeably, Rubb7-Cl displayed the highest SI values for both BHK and HEK-293 cells against E. coli. Of particular note, the SI value of 340 between BHK and E. coli was at least five-times higher than with the other complexes. By contrast, Rubb12-Cl showed similar selectivity towards S. aureus and E. coli, compared to both kidney cell lines. The inert complex Rubb12 displayed a better selectivity for S. aureus than E. coli against both kidney cell lines. Taken together, Rubb12-Cl exhibited the best overall selectivity (but only marginally better than Rubb12), while Rubb7-Cl showed the best selectivity towards the Gram-negative E. coli.
The liver cell line HepG2 was chosen to determine the cytotoxicities of the ruthenium complexes in liver cells. The cytotoxicities of the ruthenium complexes against HepG2 cells were determined after a 24-hour incubation, and the results are summarised in Table 3. The HepG2 cells were more susceptible than the kidney cells to Rubb7-Cl and Rubb12-Cl, but were more resistant to Rubb16-Cl. Interestingly, and unlike what was observed with the BHK and HEK-293 cells, the cytotoxicities of the Rubbn-Cl complexes decreased with increasing chain length in the bbn ligand. Surprisingly, Rubb7-Cl was the most cytotoxic of all the ruthenium complexes assayed against the HepG2 cells, with the 24 h-IC50 being 3.7 μM. The comparative selectivities between the two kidney cell lines and the HepG2 cell line were calculated for each of the ruthenium complexes and the results are summarised in Table 3. Rubb7-Cl clearly exhibited significantly higher toxicity towards the HepG2 cell line − compared to the two kidney cell lines − than did the other ruthenium complexes. Although the relative difference was much smaller, Rubb12-Cl was the only other complex to show some preferential toxicity to the HepG2 cells.
| Complexes | IC50 | SX | |
|---|---|---|---|
| HepG2 | BHK vs. HepG2 | HEK-293 vs. HepG2 | |
| Rac-Rubb7-Cl | 3.7 ± 0.7 | 64.4 | 18.7 |
| Rac-Rubb12-Cl | 24.3 ± 3.9 | 2.0 | 2.4 |
| Rac-Rubb16-Cl | 52.0 ± 2.9 | 0.4 | 0.8 |
| ΔΔ-Rubb12 | 61.7 ± 5.5 | 1.1 | 0.8 |
| ΔΔ-Rubb16 | 41.5 ± 2.9 | 0.7 | 0.5 |
Cellular localisation of rac-Rubbn-Cl complexes in HepG2 cells
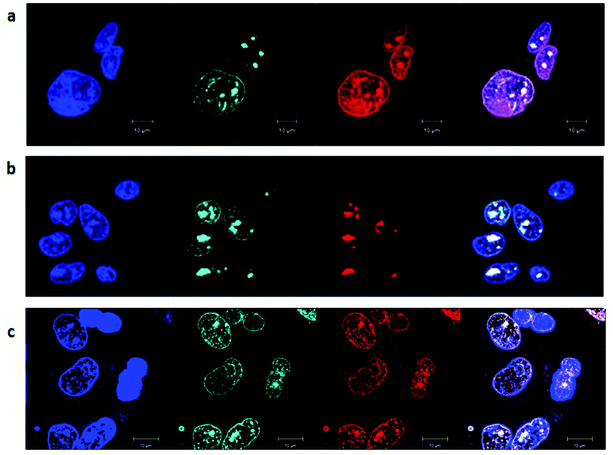
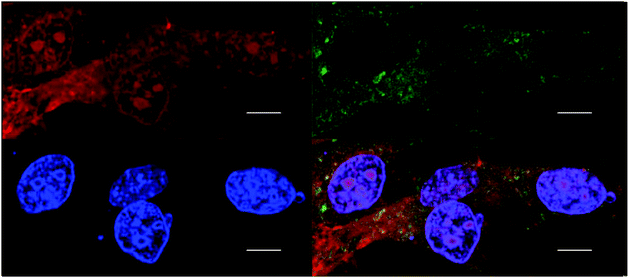
Cellular localisation patterns are important in unravelling the mechanism of cytotoxicity for any drug. Therefore, the intrinsic phosphorescence properties of the ruthenium complexes were used to study their cellular localisation by laser-scanning confocal microscopy. To achieve this, the ruthenium complex phosphorescence patterns were overlaid with the fluorescence patterns of DNA/RNA/mitochondria-specific stains in co-labelling experiments.As shown in Fig. 7, the rac-Rubbn-Cl complexes showed similar nucleolus localisation to that previously reported for the inert Rubbn complexes.22 The nucleolus of HepG2 cells were stained by Rubbn-Cl and SYTO 9. In addition to SYTO 9-stained rRNA, the most toxic complex Rubb7-Cl also showed localisation with DAPI-stained DNA. By contrast, Rubb12-Cl and Rubb16-Cl overlaid more with SYTO 9-stained rRNA in the nucleolus, and less with DAPI-stained DNA. In particular, Rubb12-Cl was exclusively localised in the nucleolus. These results suggest that Rubb12-Cl and Rubb16-Cl have greater selectivity for rRNA, compared to DNA, than does Rubb7-Cl.
BHK cells incubated with Rubb7-Cl at a concentration of 100 μM (approx. 50% of the IC50) for 20 hours are shown in Fig. 8. The Rubb7-Cl phosphorescence is observed throughout the cytoplasm and in the nucleus; however, within the nucleus the phosphorescence was mainly in the nucleolus, with little overlay with the DAPI-stained DNA. When BHK cells were incubated with Rubb7-Cl at a concentration of 50 μM for 20 hours, almost no phosphorescence was observed in the nucleus, with only relatively weak accumulation of the ruthenium complex in the cytoplasm (see Fig. 9).
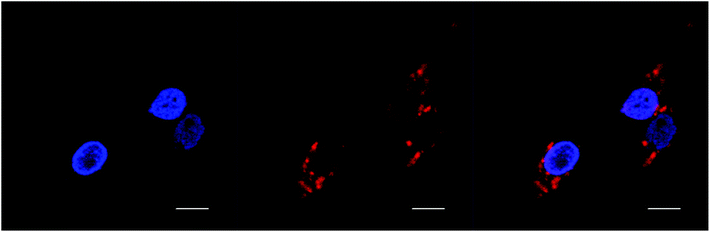 | ||
| Fig. 9 DAPI (blue) and rac-Rubb7-Cl (red; 50 μM; 20 hour incubation) localisation in BHK cells. Scale bar = 10 μm. | ||
Discussion
We have previously demonstrated that the dinuclear ruthenium complexes Rubbn and Cl-Rubbn-Cl have good antimicrobial activity.18,25 As would be expected, significant differences in activity are observed upon changes to the total charge, charge separation and lipophilicity of the ruthenium complexes. To further investigate the interplay between these parameters, we have in this study synthesised and analysed the antimicrobial activities of the Rubbn-Cl series of complexes. The results demonstrate that the Rubbn-Cl complexes also have good antimicrobial activity and are bactericidal, with the most active complex against both Gram-positive and Gram-negative strains being Rubb12-Cl. Similarly to what was observed for the Rubbn complexes, the Rubbn-Cl complexes were considerably more toxic to bacterial cells than towards eukaryotic cells. With the exception of P. aeruginosa, Rubb12-Cl was 35- to 84-times more toxic to the bacteria used in this study than to BHK, HEK-293 or HepG2 cells. However, and perhaps of greatest interest, was the observed toxicity profile of Rubb7-Cl. Rubb7-Cl was eight- to sixteen-fold more active against E. coli than the Gram-positive species, and even more significantly, Rubb7-Cl was vastly less toxic towards BHK cells but considerably more toxic towards HepG2 cells than Rubb12-Cl, Rubb16-Cl, Rubb12 and Rubb16.The preferential activity of Rubb7-Cl towards Gram-negative E. coli over the Gram-positive species S. aureus and MRSA is very unusual for a metal-based antimicrobial agent. A few metal complexes have shown higher activity against Gram-negative than Gram-positive bacteria. For example, two helicate-like dinuclear iron complexes showed two-fold higher activity against E. coli than S. aureus,35 and a [Ru2L3]4+ triply-stranded helicate complex also showed higher activity towards E. coli than S. aureus, but the activities were modest.36 However, most metal complexes have shown greater antimicrobial activity against Gram-positive species than Gram-negative species.9–11,18–25,37,38
The Rubbn-Cl complexes also showed a different pattern of activities to the Rubbn and Cl-Rubbn-Cl complexes. Against the Gram-positive bacteria, Rubb12-Cl and Rubb16-Cl were of equal activity and considerably more active than Rubb7-Cl. However, for the Gram-negative species, Rubb7-Cl and Rubb12-Cl were of equal activity and were considerably more active than Rubb16-Cl. For the Rubbn and Cl-Rubbn-Cl complexes, the same pattern of activities was observed for all bacteria. For the Rubbn complexes, the order of the activities across the four bacteria was Rubb16 ≥ Rubb12 > Rubb7; while for the Cl-Rubbn-Cl series, Cl-Rubb12-Cl > Cl-Rubb7-Cl ≥ Cl-Rubb16-Cl across the bacteria. Although the differences are relatively small, Rubb12-Cl showed the best activity profile against the four bacterial species used in this study compared to any of the bbn-linked ruthenium complexes that have been previously reported.18–25
Cytotoxicity assays against the kidney cell lines BHK and HEK-293 and the liver cell line HepG2 were carried out to estimate the toxicity of the Rubbn-Cl complexes towards mammalian organ cells. In BHK and HEK-293 cells, the 24 h-IC50 values for the Rubbn-Cl series decreased with the increasing number of methylene groups in the bbn ligand. A similar trend was observed for the Rubbn complexes. These results suggest that cellular uptake is the key parameter, at least to a first approximation, with the uptake being correlated to the lipophilicity of the ruthenium complex. However, in HepG2 cells the least lipophilic complex Rubb7-Cl exhibited the highest cytotoxicity while the most lipophilic complex Rubb16-Cl was the least toxic. Hence, it is probable that while lipophilicity is important, at least in terms of cellular uptake, there are other factors to be considered. Compared with healthy cells, the cancer cell outer membrane leaflet contains three- to nine-times more negatively-charged lipids and greater levels of negatively-charged O-glycosylated mucins.39–41 In contrast, in non-cancerous cells the membrane is largely occupied by zwitterionic phospholipids, with negligible or weak negative charge.42,43 In addition, the increased number of microvilli on cancer cells, which lead to an increase in cell surface area, may also enhance their susceptibility.44,45 Therefore, the cellular uptake of the polycationic ruthenium complexes may be less affected by lipophilicity in cancer cell lines compared with healthy cells.
Intracellular localisation could also be an important aspect of cytotoxicity. Confocal microscopy was used to determine the cellular localisation of the Rubbn-Cl complexes in HepG2 cells. The Rubbn-Cl complexes preferentially accumulated in the nucleolus, while significant DNA binding was also observed at higher concentrations. The preference for rRNA is consistent with our previous studies on the localisation of Rubb12 in BHK cells22 and Rubb16 in the ribosomes of E. coli.21 However, the Rubbn-Cl complexes showed differences in nuclear localisation in HepG2 cells based upon the length of the alkyl chain in the bbn linking ligand. The least lipophilic complex Rubb7-Cl appeared to co-localise to a higher degree with DNA, while the other two complexes, particularly Rubb12-Cl, showed greater accumulation in the nucleolus. The effect of lipophilicity on the cellular localisation of ruthenium complexes has been previously reported. Lincoln, Nordén and co-workers found the length of an alkyl chain in a dppz-based complex (dppz = dipyrido[3,2-a:2′,3′-c]phenazine) had a significant effect on the localisation pattern – the least lipophilic complex was found to stain nuclear DNA, the most lipophilic complex preferably stained cellular membranes, whereas the derivative of intermediate lipophilicity selectively stained the RNA-rich nucleoli.46 Furthermore, Thomas and co-workers demonstrated that the dinuclear complex [{Ru(phen)2}2{μ-tpphz}]4+ (where tpphz = tetrapyridophenazine) localised in the nucleus, while the more lipophilic 4,7-diphenyl-1,10-phenanthroline analogue [{Ru(DIP)2}2{μ-tpphz}]4+ localised in the endoplasmic reticulum.47,48 While other factors are yet to be examined, the greater level of DNA localisation in HepG2 by Rubb7-Cl, compared to Rubb12-Cl and Rubb12, could be related to its higher cytotoxicity against this cell line. Consistent with this proposal was the observed low level of DNA binding, compared to that in the nucleolus and cytoplasm, by Rubb7-Cl in BHK cells at concentrations considerably higher than the 24 h-IC50 determined against HepG2 cells.
In conclusion, a new class of dinuclear ruthenium complexes, Rubbn-Cl, has been synthesised and characterised. These ruthenium complexes exhibited good antimicrobial activities; and interestingly, showed relatively better activity towards Gram-negative bacteria, (compared to Gram-positive species) than previously reported ruthenium complexes linked by the bbn ligand. In addition, the Rubbn-Cl complexes were considerably less toxic to eukaryotic cells, compared to bacteria, with Rubb7-Cl showing striking differences in cytotoxicity between the eukaryotic cell lines. It is possible that Rubb7-Cl could become a new lead compound for metal-based anticancer or antimicrobial agents.
Acknowledgements
F. R. K. acknowledges the award of a Visiting Professorship from UNSW Canberra.References
- J. H. Powers, Clin. Microbiol. Infect., 2004, 10(Suppl 4), 23 CrossRef PubMed.
- E. Wong and C. M. Giandomenico, Chem. Rev., 1999, 99, 2451 CrossRef CAS PubMed.
- B. Nordén, P. Lincoln, B. Akerman and E. Tuite, Met. Ions Biol. Syst., 1996, 33, 177 Search PubMed.
- C. Metcalfe and J. A. Thomas, Chem. Soc. Rev., 2003, 32, 215 RSC.
- B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565 RSC.
- M. R. Gill and J. A. Thomas, Chem. Soc. Rev., 2012, 41, 3179 RSC.
- A. D. Richards, A. Rodger, M. J. Hannon and A. Bolhuis, Int. J. Antimicrob. Agents, 2009, 33, 469 CrossRef CAS PubMed.
- N. S. Ng, P. Leverett, D. E. Hibbs, Q. Yang, J. C. Bulanadi, M. J. Wu and J. R. Aldrich-Wright, Dalton Trans., 2013, 42, 3196 RSC.
- F. P. Dwyer, E. C. Gyarfas, W. P. Rogers and J. H. Koch, Nature, 1952, 170, 190 CrossRef CAS PubMed.
- F. P. Dwyer, I. K. Reid, A. Shulman, G. M. Laycock and S. Dixson, Aust. J. Exp. Biol. Med. Sci., 1969, 47, 203 CrossRef CAS PubMed.
- A. Bolhuis, L. Hand, J. E. Marshall, A. D. Richards, A. Rodger and J. Aldrich-Wright, Eur. J. Pharm. Sci., 2011, 42, 313 CrossRef CAS PubMed.
- C. S. Devi, D. A. Kumar, S. S. Singh, N. Gabra, N. Deepika, Y. P. Kumar and S. Satyanarayana, Eur. J. Med. Chem., 2013, 64, 410 CrossRef PubMed.
- K. A. Kumar, K. L. Reddy, S. Vidhisha and S. Satyanarayana, Appl. Organomet. Chem., 2009, 23, 409 CrossRef CAS.
- N. H. Khan, N. Pandya, R. I. Kureshy, S. H. R. Abdi, S. Agrawal, H. C. Bajaj, J. Pandya and A. Gupte, Spectrochim. Acta, Part A, 2009, 74, 113 CrossRef PubMed.
- P.-L. Lam, G.-L. Lu, K.-M. Hon, K.-W. Lee, C.-L. Ho, X. Wang, J. C.-O. Tang, K.-H. Lam, R. S.-M. Wong, S. H.-L. Kok, Z.-X. Bian, H. Li, K. K.-H. Lee, R. Gambari, C.-H. Chui and W.-Y. Wong, Dalton Trans., 2014, 43, 3949 RSC.
- N. Thilagavathi, A. Manimaran and C. Jayabalakrishnan, J. Coord. Chem., 2010, 63, 1252 CrossRef CAS.
- C. S. Allardyce, P. J. Dyson, D. J. Ellis, P. A. Salter and R. Scopelliti, J. Organomet. Chem., 2003, 668, 35 CrossRef CAS.
- F. Li, Y. Mulyana, M. Feterl, J. M. Warner, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 5032 RSC.
- F. Li, M. Feterl, Y. Mulyana, J. M. Warner, J. G. Collins and F. R. Keene, J. Antimicrob. Chemother., 2012, 67, 2686 CrossRef CAS PubMed.
- F. Li, M. Feterl, J. M. Warner, F. R. Keene and J. G. Collins, J. Antimicrob. Chemother., 2013, 68, 2825 CrossRef CAS PubMed.
- F. Li, E. J. Harry, A. L. Bottomley, M. D. Edstein, G. W. Birrell, C. E. Woodward, F. R. Keene and J. G. Collins, Chem. Sci., 2014, 5, 685 RSC.
- X. Li, A. K. Gorle, T. D. Ainsworth, K. Heimann, C. E. Woodward, J. G. Collins and F. R. Keene, Dalton Trans., 2015, 44, 3594 RSC.
- F. Li, J. G. Collins and F. R. Keene, Chem. Soc. Rev., 2015, 44, 2529 RSC.
- A. K. Gorle, M. Feterl, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2014, 43, 16713 RSC.
- M. Pandrala, F. Li, M. Feterl, Y. Mulyana, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2013, 42, 4686 RSC.
- Y. Mulyana, G. Collins and R. Keene, J. Inclusion Phenom. Macrocyclic Chem., 2011, 71, 371 CrossRef CAS.
- J. L. Morgan, C. B. Spillane, J. A. Smith, D. P. Buck, J. G. Collins and F. R. Keene, Dalton Trans., 2007, 4333 RSC.
- P. A. Adcock, F. R. Keene, R. S. Smythe and M. R. Snow, Inorg. Chem., 1984, 23, 2336 CrossRef CAS.
- X. Hua and A. von Zelewsky, Inorg. Chem., 1995, 34, 5791 CrossRef CAS.
- D. J. States, R. A. Haberkorn and D. J. Ruben, J. Magn. Reson., 1982, 48, 286 CAS.
- H. R. Hypercube Inc., Gainesville, FL, USA, 2005.
- British Society for Antimicrobial Chemotherapy Working Party, J. Antimicrob. Chemother., 1991, 27, 22 CrossRef.
- M. Motyl, K. Dorso, J. Barrett and R. Giacobbe, in Current Protocols in Pharmacology, John Wiley & Sons, Inc., 2005, pp. 13A.3.1–13A.3.22 Search PubMed.
- J. O'Brien, I. Wilson, T. Orton and F. Pognan, Eur. J. Biochem., 2000, 267, 5421 CrossRef.
- S. E. Howson, A. Bolhuis, V. Brabec, G. J. Clarkson, J. Malina, A. Rodger and P. Scott, Nat. Chem., 2011, 4, 31 CrossRef PubMed.
- S. V. Kumar, W. K. C. Lo, H. J. L. Brooks and J. D. Crowley, Inorg. Chim. Acta, 2015, 425, 1 CrossRef CAS.
- N. S. Ng, P. Leverett, D. E. Hibbs, Q. Yang, J. C. Bulanadi, M. Jie Wu and J. R. Aldrich-Wright, Dalton Trans., 2013, 42, 3196 RSC.
- A. Srishailam, Y. P. Kumar, N. M. D. Gabra, P. V. Reddy, N. Deepika, N. Veerababu and S. Satyanarayana, J. Fluoresc., 2013, 23, 897 CrossRef CAS PubMed.
- J. S. Mader and D. W. Hoskin, Expert Opin. Inves. Drugs, 2006, 15, 933 CrossRef CAS PubMed.
- N. Papo and Y. Shai, Cell. Mol. Life Sci., 2005, 62, 784 CrossRef CAS PubMed.
- R. F. Zwaal and A. J. Schroit, Blood, 1997, 89, 1121 CAS.
- I. L. Ang, T. C. W. Poon, P. B. S. Lai, A. T. C. Chan, S.-M. Ngai, A. Y. Hui, P. J. Johnson and J. J. Y. Sung, J. Proteome Res., 2006, 5, 2691 CrossRef CAS PubMed.
- S. Riedl, D. Zweytick and K. Lohner, Chem. Phys. Lipids, 2011, 164, 766 CrossRef CAS PubMed.
- N. Papo and Y. Shai, Biochemistry, 2003, 42, 9346 CrossRef CAS PubMed.
- S. C. Chan, L. Hui and H. M. Chen, Anticancer Res., 1998, 18, 4467 CAS.
- M. Matson, F. R. Svensson, B. Nordén and P. Lincoln, J. Phys. Chem. B, 2011, 115, 1706 CrossRef CAS PubMed.
- M. R. Gill, J. Garcia-Lara, S. J. Foster, C. Smythe, G. Battaglia and J. A. Thomas, Nat. Chem., 2009, 1, 662 CrossRef CAS PubMed.
- M. R. Gill, D. Cecchin, M. H. Walker, R. S. Mulla, G. Battaglia, C. Smythe and J. A. Thomas, Chem. Sci., 2013, 4, 4512 RSC.
Footnote |
| † Current address: School of Chinese Medicine, Hong Kong Baptist University, Hong Kong, China. |
| This journal is © The Royal Society of Chemistry 2016 |