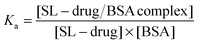Open Access Article
Open Access ArticleEPR studies of intermolecular interactions and competitive binding of drugs in a drug–BSA binding model†
Y.
Akdogan
*a,
M.
Emrullahoglu
b,
D.
Tatlidil
a,
M.
Ucuncu
b and
G.
Cakan-Akdogan
c
aMaterials Science and Engineering Department, İzmir Institute of Technology, 35430 Urla, İzmir, Turkey. E-mail: yasarakdogan@iyte.edu.tr
bDepartment of Chemistry, İzmir Institute of Technology, 35430 Urla, İzmir, Turkey
cDokuz Eylul University, İzmir Biomedicine and Genome Center, İzmir, Turkey
First published on 25th July 2016
Abstract
Understanding intermolecular interactions between drugs and proteins is very important in drug delivery studies. Here, we studied different binding interactions between salicylic acid and bovine serum albumin (BSA) using electron paramagnetic resonance (EPR) spectroscopy. Salicylic acid was labeled with a stable radical (spin label) in order to monitor its mobilized (free) or immobilized (bound to BSA) states. In addition to spin labeled salicylic acid (SL-salicylic acid), its derivatives including SL-benzoic acid, SL-phenol, SL-benzene, SL-cyclohexane and SL-hexane were synthesized to reveal the effects of various drug binding interactions. EPR results of these SL-molecules showed that hydrophobic interaction is the main driving force. Whereas each of the two functional groups (–COOH and –OH) on the benzene ring has a minute but detectable effect on the drug–protein complex formation. In order to investigate the effect of electrostatic interaction on drug binding, cationic BSA (cBSA) was synthesized, altering the negative net charge of BSA to positive. The salicylic acid loading capacity of cBSA is significantly higher compared to that of BSA, indicating the importance of electrostatic interaction in drug binding. Moreover, the competitive binding properties of salicylic acid, ibuprofen and aspirin to BSA were studied. The combined EPR results of SL-salicylic acid/ibuprofen and SL-ibuprofen/salicylic acid showed that ibuprofen is able to replace up to ∼83% of bound SL-salicylic acid, and salicylic acid can replace only ∼14% of the bound SL-ibuprofen. This indicates that ∼97% of all salicylic acid and ibuprofen binding sites are shared. On the other hand, aspirin replaces only ∼23% of bound SL-salicylic acid, and salicylic acid replaces ∼50% of bound SL-aspirin, indicating that ∼73% of all salicylic acid and aspirin binding sites are shared. These results show that EPR spectroscopy in combination with the spin labeling technique is a very powerful method to investigate drug binding dynamics in detail.
Introduction
Electron paramagnetic resonance (EPR) spectroscopy is a highly selective and sensitive method to locally probe paramagnetic molecules (spin labels or spin probes). The spin labeling technique makes EPR spectroscopy a powerful method for studying interactions between spin labeled drugs and proteins.1–3 Although spin labeled drugs have been used in the studies of drug mechanisms for a long time, the number of EPR studies is still not sufficient in the field of drug delivery.4–7 The most common exogenous stable free radicals, Tempo (2,2,6,6-tetramethylpiperidine-1-oxyl) based nitroxide radicals, are used as spin labels. Information on the spin label dynamics can be obtained by analyzing the line shape of the continuous wave (cw) EPR spectrum at room temperature.8 The rotational dynamics of spin labeled molecules determines the line shape of the EPR spectrum. Immobilized drugs (bound) have broad signals coming from restricted rotational motion. On the other hand, mobilized drugs (free) have three sharp line signals coming from freely rotational motion.8,9Different Tempo based nitroxide radicals are commercially available with various functional groups. These spin labels can be covalently attached to drugs via their appropriate functional groups. Therefore, we are able to monitor drugs in a protein solution using EPR spectroscopy.
Salicylic acid, acetyl salicylic acid (aspirin) and ibuprofen are non-steroidal anti-inflammatory drugs that can bind to bovine serum albumin (BSA) efficiently with 104–106 M−1 binding constants.10,11 Serum albumin is the main binding protein in blood plasma for a large number of drugs. Mostly, drugs bind to serum albumin at one of two primary sites (site I and site II).12 Therefore, co-administration of drugs causes a competition for the binding sites of serum albumin. A drug with a higher affinity can replace a bound drug with a lower affinity to serum albumin. Therefore, the level of active (free) drug changes in the presence of a second drug, which is crucial to the therapeutic effect of drugs.13
Our study consists of two parts. In the first part, intermolecular interactions between salicylic acid and BSA were studied. In the second part, we showed that EPR spectroscopy can be used as an alternative technique to study the competitive binding of drugs to serum albumin.
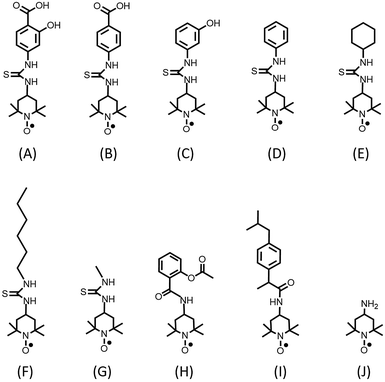
Usually drug/protein binding is a reversible process. For reversible binding, the drug binds to a protein with several weak chemical bonds such as electrostatic interactions, hydrogen bonds, hydrophobic forces and van der Waals interactions.14 Here, the effects of these intermolecular interactions on the binding process were studied using synthesized SL-salicylic acid, SL-benzoic acid, SL-phenol, SL-benzene, SL-cyclohexane and SL-hexane by EPR spectroscopy (Fig. 1).
In the second part of the study, we used EPR spectroscopy to make an analysis of the competitive binding of ibuprofen–salicylic acid and aspirin–salicylic acid to BSA. Several techniques have been used to study the competitive binding of drugs to serum albumin including gel filtration techniques, equilibrium dialysis, NMR spectroscopy and fluorescence spectroscopy.15–18 Kuchimanchi et al. studied the binding of cosalane to BSA in the presence of salicylic acid by a gel filtration technique.15 Sakai et al. observed the competitive interaction between indoxyl sulfate and dansyl-L-asparagine to the N-conformer of human serum albumin (HSA) by equilibrium dialysis.16 Cui et al. analysed the competitive binding of ibuprofen and salicylic acid to HSA by NMR relaxation measurements.17 The observed 1H relaxation rate of drugs was regarded as a weighted average of that of the free and bound forms of drugs. Among the techniques used in drug binding studies, fluorescence spectroscopy has been used frequently. The presence of intrinsic fluorescence active sites in serum albumin (tryptophan, tyrosine and phenylalanine) makes the fluorescence spectroscopy technique very convenient for drug binding studies. Also, external site marker fluorescence probes for binding sites (warfarin, dansylamide, dansyl-L-asparagine, ibuprofen, etc.) have been used to characterize binding sites and their affinity towards drugs.18–21
As an alternative technique, we used cw EPR spectroscopy to monitor bound and unbound (free) drugs simultaneously at physiological albumin concentrations. The calculated areas under the bound and free signals give their concentrations separately. Therefore, the change in the bound and free concentrations of a drug can be monitored easily in the presence of another type of drug. This provides information about the association constants of drugs and also their shared binding sites.
Experimental
Reagents
Bovine serum albumin (fatty acid free), Tempo-4-amino (97%), 4-amino salicylic acid (99%), 4-amino benzoic acid (99%), 3-amino phenol (98%), phenyl isothiocyanate (97%), cyclohexyl isothiocyanate (98%), hexyl isothiocyanate (95%), ibuprofen (98%), acetyl salicylic acid (99%), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, 98%), hydrochloric acid (37%, 12.2 M), dichloromethane (99.8%), trimethylamine (99.5%), ethyl acetate (99.5%), methanol (99.8%) were purchased from Sigma-Aldrich. Thiophosgene (99%) was purchased from Merck.Synthesis of SL-molecules
The mechanisms of synthesis of SL-molecules and their EPR spectra in buffer solutions are shown in Fig. S1 and S2 (ESI†), respectively.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol, 15
methanol, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 13
1, 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 12
1, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10
1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 46%.
1). The yield is 46%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 4
ethyl acetate 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 25%.
1). The yield is 25%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 4
ethyl acetate 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 37%.
1). The yield is 37%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 4
ethyl acetate 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 55%.
1). The yield is 55%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 4
ethyl acetate 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 25%.
1). The yield is 25%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 4
ethyl acetate 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 28%.
1). The yield is 28%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 3
ethyl acetate 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 32%.
1). The yield is 32%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio under argon and stirred for 18 hours at 25 °C. SL-ibuprofen was purified with column chromatography (hexane
1 ratio under argon and stirred for 18 hours at 25 °C. SL-ibuprofen was purified with column chromatography (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 5
ethyl acetate 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 69%.
1). The yield is 69%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio under argon and stirred for 18 hours at 25 °C. SL-aspirin was purified by column chromatography (hexane
1 ratio under argon and stirred for 18 hours at 25 °C. SL-aspirin was purified by column chromatography (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 4
ethyl acetate 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2.3
1 and 2.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The yield is 58%.
1). The yield is 58%.
Preparation of cHSA
Cationization of BSA was achieved by applying a procedure that has been reported previously.23 BSA (50 mg, 0.15 mmol) was dissolved in 5 mL ethylenediamine aqueous solution (50 mM, pH: 4.75) at 25 °C. After that N-(3-dimethylaminopropyl)-N′-ethylcarbodiimidehydrochloride (410 mg, 2.1 mmol) was introduced. After stirring for 2 hours at 25 °C, the reaction was stopped by adding acetate-buffer (360 μL, 4 M, pH: 4.75). The colorless solution was concentrated by ultrafiltration (Amicon Ultra 50 kDa MWCO), washed three times with acetate-buffer (100 mM, pH: 4.75) and five times with Milli-Q water. The yield of the experiment was 95%. A Malvern dynamic light scattering (DLS) Nano-ZS instrument was used for zeta potential measurements.Binding of spin labeled molecules to BSA and to cBSA
Aqueous solutions of BSA and cBSA at different concentrations were prepared in 0.1 M phosphate buffer (pH 7.4). One of the most common buffers used in biological reactions is phosphate buffer (pKa = 6.8). The buffering range of phosphate buffer (pH = pKa ± 1) matches the pH of mammalian cells (pH = 7.35–7.45) with a lack of toxicity. Also, phosphoric acid and dihydrogen phosphate ions are found in the blood at very low concentrations.24 The SL-molecule stock solution (1 M) was prepared in DMSO. The desired ratio of spin labeled molecules to BSA was obtained with the addition of the appropriate amount of spin labeled molecule solution to the protein solution with constant stirring. The concentrations (0.5–0.7 mM) of used drugs are similar to the typical concentrations of drugs in the blood plasma. In the literature, Morra et al.25 reported that the serum concentrations of salicylic acid were between 0.2 mM and 0.66 mM, and Janssen et al.26 showed that the mean plasma concentration of ibuprofen was 0.4 mM. The DMSO concentration was kept below 1% (v/v). The solution was allowed to equilibrate at 37 °C for 10 min for EPR measurements.EPR measurements
A CMS 8400 (Adani) benchtop spectrometer provided with a TE102 resonator cavity was used for all X-band EPR measurements at a microwave frequency of ∼9.4 GHz at 25 °C. Measurements were performed in quartz capillary sample tubes. The shown EPR spectra were normalized to the intensity of the high field line of the unbound spin labeled molecules. All spectra were simulated using the Matlab-based Easyspin 4.5.5 software package.27Results and discussion
In this study, we tried to mimic in vivo conditions such as the physiological concentrations of albumin, the pH value of blood, buffer and also the body temperature. We studied mainly the physiological concentrations of albumin (0.5–0.7 mM). The pH value was kept constant at 7.4 just like the pH value of blood using phosphate buffer. Phosphate buffer is a nontoxic buffer and its buffering range is appropriate for the pH value of blood. Finally, we mixed the drug and albumin at a body temperature of 37 °C about 10 minutes. Therefore, the experimental conditions are close to the conditions in vivo.Analysis of bound and free drugs by EPR spectroscopy
Cw EPR spectroscopy allows us to obtain bound and free spin labeled dugs simultaneously in the physiological range of albumin (0.5–0.7 mM). In solution, analysis of the EPR spectral line shape reveals the rotational behaviour of the spin labeled molecules. The rotational motion of molecules is strongly affected by macromolecule binding. Therefore, rotational correlation time increases from the ps to μs range depending on the immobilization strength.
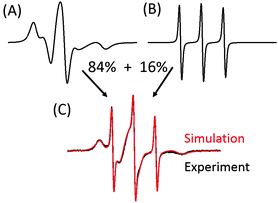
Fig. 2 shows the EPR spectrum of 0.6 mM SL-salicylic acid in buffered solution of 0.6 mM BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with the corresponding simulation. The simulations of bound and free (unbound) SL-salicylic acid were added with the calculated proportions giving the experimental result. At room temperature, protein bound spin labeled drugs show broad outer hyperfine signals due to restricted rotational motion (10 ns), and free spin labeled drugs show three sharp signals (0.07 ns), signatures of the freely tumbling motion of radicals. The area under each signal (by double integration of EPR spectrum) of the simulated EPR spectra of bound and unbound SL-salicylic acid reveals the ratio of bound to unbound drugs, e.g. the ratio is ∼5
1) with the corresponding simulation. The simulations of bound and free (unbound) SL-salicylic acid were added with the calculated proportions giving the experimental result. At room temperature, protein bound spin labeled drugs show broad outer hyperfine signals due to restricted rotational motion (10 ns), and free spin labeled drugs show three sharp signals (0.07 ns), signatures of the freely tumbling motion of radicals. The area under each signal (by double integration of EPR spectrum) of the simulated EPR spectra of bound and unbound SL-salicylic acid reveals the ratio of bound to unbound drugs, e.g. the ratio is ∼5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of 0.6 mM SL-salicylic acid/BSA (1
1 of 0.6 mM SL-salicylic acid/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution.
1) solution.
Intermolecular interactions of SL-salicylic acid and BSA
Hydrophobic interactions, electrostatic interactions, hydrogen bonding and van der Waals interactions determine the extent of binding of drugs to serum albumin.14 Here, we study the effect of these intermolecular interactions on the binding of salicylic acid to albumin individually by EPR spectroscopy. In our previous study, we showed that SL-salicylic acid binds to albumin by 80% at physiological concentrations.3 Salicylic acid has a phenyl group and two functional groups, a hydroxyl group and a carboxylic acid group. At pH 7.4 salicylic acid carries a negative charge on the carboxylate group (pKa = 2.97) and contains a hydrogen bond forming group (hydroxyl group). The aromatic group of salicylic acid may interact with other aromatic rings in albumin forming pi–pi stacking in addition to hydrophobic interactions.Fig. 1 shows chemical structures of spin labeled molecules used in this study. SL-salicylic acid has two functional groups (–COOH and –OH) but SL-benzoic acid and SL-phenol have only the –COOH group and the –OH group, respectively. Although SL-benzene and SL-cyclohexane do not have these functional groups, SL-benzene conserves aromaticity but SL-cyclohexane does not.
Therefore, we can control the pi–pi stacking effect on the formation of a drug-albumin complex by comparing the results of SL-benzene and SL-cyclohexane. The polarity of these molecules is also different. Salicylic acid with two functional groups is expected to be more polar than others. Benzene without two functional groups must be less polar than salicylic acid, benzoic acid and phenol. The polarity of cyclohexane is similar to benzene but it does not have aromaticity. In addition to them, we also synthesized SL-hexane which is more hydrophobic than the others.28
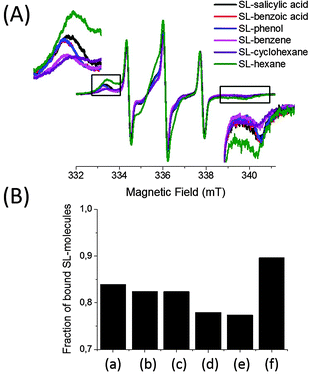
The uptake of spin labeled molecules by BSA is shown in Fig. 3(A). The EPR spectra are normalized to the intensity of the high field line of the unbound molecule which is the least affected signal by the presence of bound signals of SL-molecules. Simulations of each spectrum of 0.6 mM SL-salicylic acid, SL-benzoic acid, SL-phenol, SL-benzene and SL-cyclohexane in 0.6 mM BSA solutions showed that these molecules extensively bind to albumin with bound fractions of 0.84–0.76 (Fig. 3(B)). Since each molecule has a hydrophobic fragment, such as a benzene ring and a cyclohexane ring, these hydrophobic fragments are thought to cause the main effect on the formation of a strong similar molecule–albumin complex (0.84–0.76). In order to study the hydrophobic effect on protein binding, we also measured the SL-hexane/BSA complex. Most of the SL-hexane molecules are bound to BSA with a fraction of bound SL-hexane as 0.90. Since SL-hexane is more hydrophobic than the others with its aliphatic hexyl chain, the fraction of bound SL-hexane is higher than those of other SL-molecules.
Nevertheless, these molecules have different bound to unbound molecule ratios with BSA because of their structural differences. Although SL-salicylic acid, SL-benzoic acid, SL-phenol and SL-benzene all have an aromatic benzene ring, the number of functional groups on the molecules is different. SL-salicylic acid binds to BSA slightly more than both SL-benzoic acid and SL-phenol, and evidently more than SL-benzene. The fractions of bound SL-salicylic acid, SL-benzoic acid, SL-phenol, and SL-benzene are 0.84, 0.82, 0.82 and 0.77, respectively. The presence of –COOH and –OH groups on the benzene ring increases the bound ratio slightly in the order of SL-benzene < SL-phenol ≈ SL-benzoic acid < SL-salicylic acid. This indicates that the presence of a negative charge carrying group (–COOH) and hydrogen bond forming groups (–COOH and –OH) promotes albumin binding.
Another structural factor which is known to promote protein binding is pi–pi stacking. Comparison of SL-benzene/BSA and SL-cyclohexane/BSA spectra shows that SL-benzene and SL-cyclohexane bind to BSA very similarly. The fractions of bound SL-benzene and SL-cyclohexane are 0.77 and 0.76, respectively. Since their polarities are similar28 and pi–pi stacking is only possible for SL-benzene and not for SL-cyclohexane, these results show that the presence of a pi–pi stacking interaction between SL-benzene and albumin does not have considerable effect on protein binding.
Furthermore, the release profiles of SL-salicylic acid and its spin labeled derivatives (SL-benzoic acid, SL-phenol and SL-benzene) from BSA were studied by EPR spectroscopy (Fig. S3, ESI†). 0.5 mM SL-molecules/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solutions were placed in 3 mL D-tube dialyzers (MWCO 6–8 kDa) and at pre-fixed times 7 μL samples were removed and measured by EPR spectroscopy. Each time both bound and unbound SL-molecules in albumin solution were detected simultaneously. The EPR line intensities of the SL-molecules coming from both the bound and free fractions decrease gradually. Therefore, the bound fractions of SL-molecules do not change with time in BSA solutions. At 37 °C the relative diffusion rates of SL-salicylic acid and its derivatives are very similar in the SL-molecules/BSA (1
1) solutions were placed in 3 mL D-tube dialyzers (MWCO 6–8 kDa) and at pre-fixed times 7 μL samples were removed and measured by EPR spectroscopy. Each time both bound and unbound SL-molecules in albumin solution were detected simultaneously. The EPR line intensities of the SL-molecules coming from both the bound and free fractions decrease gradually. Therefore, the bound fractions of SL-molecules do not change with time in BSA solutions. At 37 °C the relative diffusion rates of SL-salicylic acid and its derivatives are very similar in the SL-molecules/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution (Fig. S3, ESI†). This indicates that the release profiles of SL-salicylic acid and its spin labeled derivatives (SL-benzoic acid, SL-phenol and SL-benzene) from BSA are comparable because of having similar binding interactions with BSA.
1) solution (Fig. S3, ESI†). This indicates that the release profiles of SL-salicylic acid and its spin labeled derivatives (SL-benzoic acid, SL-phenol and SL-benzene) from BSA are comparable because of having similar binding interactions with BSA.
Taking the EPR results together, we conclude that hydrophobic interaction is the main force between salicylic acid and BSA. The presence of two functional groups (–COOH and –OH) on the benzene ring has a minute but detectable effect on the formation of drug–protein complexes.
As a control experiment, SL-methyl was synthesized and a mixture of SL-methyl and BSA solution was measured by EPR spectroscopy (Fig. S4, ESI†). Since methyl has a very low hydrophobicity compared to hexane, SL-methyl did not bind to albumin and only three sharp signals coming from free SL-methyl were obtained. Moreover, Tempo-4-amino was also mixed with BSA and measured by EPR spectroscopy (Fig. S4, ESI†). Again, only three sharp signals were observed, signatures of free radicals.
Comparison of SL-salicylic acid binding to BSA and to cBSA
Cationic albumin was prepared in order to observe the effect of the net charge of albumin on drug binding. Albumin cationization was achieved by following the procedure that has been previously described.23 The negative net charge (−19 ± 2) on the BSA was converted into a positive net charge (+23 ± 3) after the reaction of BSA with ethylenediamine at pH 4.75. The uptake of 0.6 mM SL-salicylic acid by 0.6 mM cBSA was monitored by EPR spectroscopy (Fig. 4(A)). The bound fraction of SL-salicylic acid increases from 0.84 to 0.89 in BSA and in cBSA, respectively. An obvious increase in the bound fraction of SL-salicylic acid can be explained by the presence of electrostatic attraction between SL-salicylic acid and cBSA.23,29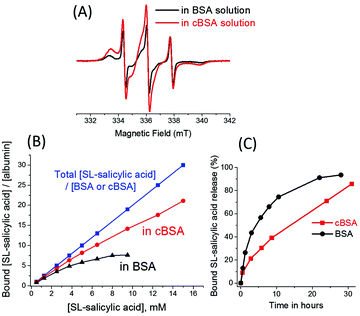 | ||
| Fig. 4 (A) Cw EPR spectra of 0.6 mM SL-salicylic acid in BSA solution (black) and in cBSA solution (red) at a drug/albumin ratio of 1. (B) The concentration ratio of bound SL-salicylic acid to BSA saturates (black) with increasing SL-salicylic acid concentration in a constant 0.5 mM BSA solution. The concentration ratio of bound SL-salicylic acid to cBSA increases (red) with increasing SL-salicylic acid concentration in a constant 0.5 mM cBSA solution. (C) The bound SL-salicylic acid release profiles from BSA and from cBSA with time at 37 °C. The data of SL-salicylic acid/BSA in (B) and (C) were taken from ref. 3. | ||
In a previous study, it has been shown that a maximum of 7 SL-salicylic acid molecules can bind to each BSA molecule in the physiological concentration range.3 In order to find out the the loading capacity of cBSA, a series of SL-salicylic acid samples with different concentrations from 0.5 mM to 15 mM were added to 0.5 mM cBSA solution (Fig. S5, ESI†). In the normalized EPR spectra, the bound fraction of SL-salicylic acid decreases with increasing SL-salicylic acid concentration (Fig. S5, ESI†). Yet, the number of bound SL-salicylic acid per cBSA increases regularly and does not reach a saturation point under the experimental conditions (up to 15 mM SL-salicylic acid) (Fig. 4(B)). This indicates that cBSA at physiological concentrations has numerous binding sites for SL-salicylic acid. If we compare the bound fractions of SL-salicylic acid in BSA and in cBSA solutions, SL-salicylic acid forms a saturated complex with BSA at a molar ratio of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 but each cBSA can load ∼20 SL-salicylic acid at a [SL-salicylic acid]/[cBSA] molar ratio of 30
1 but each cBSA can load ∼20 SL-salicylic acid at a [SL-salicylic acid]/[cBSA] molar ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The –COOH and –OH functional groups collectively provide a partial negative charge on SL-salicylic acid. Therefore, SL-salicylic acid can bind to cBSA with a higher efficiency compared to its binding to anionic BSA. Repelling of the negatively charged drug by anionic BSA can be explained in terms of a heterogeneous charge distribution on the BSA molecule. The larger negative domain of the protein hinders the anionic drug binding and also prevents it electrostatically from the attractive cationic domain.30 Furthermore, theoretical studies of polyelectrolyte (PE) binding onto net-neutral Janus nanospheres (JNSs), with two equally but oppositely charged hemispheres, support the drug binding behaviour of BSA.31 PE–JNS adsorption at low salt concentration is carried out with PE attraction by the oppositely charged hemisphere and PE repulsion by the similarly charged hemisphere.
1. The –COOH and –OH functional groups collectively provide a partial negative charge on SL-salicylic acid. Therefore, SL-salicylic acid can bind to cBSA with a higher efficiency compared to its binding to anionic BSA. Repelling of the negatively charged drug by anionic BSA can be explained in terms of a heterogeneous charge distribution on the BSA molecule. The larger negative domain of the protein hinders the anionic drug binding and also prevents it electrostatically from the attractive cationic domain.30 Furthermore, theoretical studies of polyelectrolyte (PE) binding onto net-neutral Janus nanospheres (JNSs), with two equally but oppositely charged hemispheres, support the drug binding behaviour of BSA.31 PE–JNS adsorption at low salt concentration is carried out with PE attraction by the oppositely charged hemisphere and PE repulsion by the similarly charged hemisphere.
The net charge of albumin also affects the drug release. For the releasing study, 0.5 mM SL-salicylic acid/cBSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and 0.5 mM SL-salicylic acid/BSA (1
1) and 0.5 mM SL-salicylic acid/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solutions were placed in 3 mL D-tube dialyzers (MWCO 6–8 kDa) and at prefixed times 7 μL samples were removed and measured by EPR spectroscopy (Fig. S6, ESI†). At 37 °C the relative diffusion rate of SL-salicylic acid in the SL-salicylic acid/cBSA (1
1) solutions were placed in 3 mL D-tube dialyzers (MWCO 6–8 kDa) and at prefixed times 7 μL samples were removed and measured by EPR spectroscopy (Fig. S6, ESI†). At 37 °C the relative diffusion rate of SL-salicylic acid in the SL-salicylic acid/cBSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution is slower than the diffusion rate of SL-salicylic acid in the SL-salicylic acid/BSA (1
1) solution is slower than the diffusion rate of SL-salicylic acid in the SL-salicylic acid/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution (Fig. 4(C)). This can be explained by the relatively strong attractive forces between cBSA and SL-salicylic acid. Attractive electrostatic forces between cationic albumin and anionic SL-salicylic acid slow down the SL-salicylic acid release. For example, only 30% of bound SL-salicylic acid is released from c BSA after 6 hours, but in the same time interval 57% of bound SL-salicylic acid is released from BSA.
1) solution (Fig. 4(C)). This can be explained by the relatively strong attractive forces between cBSA and SL-salicylic acid. Attractive electrostatic forces between cationic albumin and anionic SL-salicylic acid slow down the SL-salicylic acid release. For example, only 30% of bound SL-salicylic acid is released from c BSA after 6 hours, but in the same time interval 57% of bound SL-salicylic acid is released from BSA.
Binding of SL-ibuprofen, SL-aspirin and SL-salicylic acid to BSA
In addition to salicylic acid, we also studied ibuprofen and aspirin by EPR spectroscopy. Spin labels were attached to the –COOH groups of ibuprofen and aspirin (Fig. 1). Fig. 5(A) shows the comparison of the EPR spectra of 0.5 mM SL-ibuprofen, SL-aspirin and SL-salicylic acid in 0.5 mM BSA solutions. Most of the SL-ibuprofen is bound to BSA with a fraction of bound SL-ibuprofen as 0.93. But, the bound fractions of SL-salicylic acid and SL-aspirin are 0.79 and 0.76, respectively which are lower than the bound fraction of ibuprofen (Fig. 5(B)). The bound fractions of SL-drugs depend on the serum albumin concentration (at a drug![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA ratio of 1). Therefore, the bound fractions of SL-salicylic acid are 0.84 and 0.79 in 0.6 mM and 0.5 mM BSA solutions, respectively.
BSA ratio of 1). Therefore, the bound fractions of SL-salicylic acid are 0.84 and 0.79 in 0.6 mM and 0.5 mM BSA solutions, respectively.
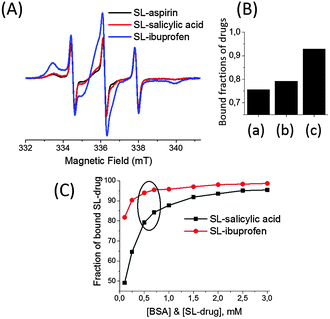 | ||
Fig. 5 (A) Cw EPR spectra of 0.5 mM SL-drugs in 0.5 mM BSA solution: SL-aspirin (black), SL-salicylic acid (red) and SL-ibuprofen (blue). (B) Fractions of bound SL-drugs obtained from simulations of spectra from (A): (a) SL-aspirin, (b) SL-salicylic acid, and (c) SL-ibuprofen. (C) Fractions of bound SL-salicylic acid and SL-ibuprofen obtained from simulations of EPR spectra of SL-drug/BSA solutions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The concentrations of SL-drug and BSA decrease from 3.0 mM to 0.1 mM for both drugs. The circle shows the concentrations of BSA in the physiological range (0.5–0.7 mM BSA). The data for SL-salicylic acid/BSA in (C) were taken from ref. 3. 1). The concentrations of SL-drug and BSA decrease from 3.0 mM to 0.1 mM for both drugs. The circle shows the concentrations of BSA in the physiological range (0.5–0.7 mM BSA). The data for SL-salicylic acid/BSA in (C) were taken from ref. 3. | ||
In a previous study, we showed that the extent of SL-salicylic acid/BSA conjugation decreases sharply especially below the physiological concentrations (Fig. 5(C)).3 Therefore, we emphasized the importance of the physiological concentration of albumin for drug delivery. Fig. S7 (ESI†) shows the EPR spectra of SL-ibuprofen in BSA solution at a SL-ibuprofen/BSA ratio of 1. For the concentrated solution (3.0 mM of SL-ibuprofen and BSA), the bound fraction of SL-ibuprofen is 0.99. When the mixture of SL-ibuprofen/BSA solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) is diluted with buffer, the fraction of bound SL-ibuprofen decreases only from 0.99 to 0.82 at 3.0 mM and 0.1 mM concentrations, respectively (Fig. 5(C)). As a result, a significant amount of SL-ibuprofens is still bound to BSA in the diluted system. On the other hand, dilution causes a strong release of SL-salicylic acid from BSA, the fraction of bound SL-salicylic acid decreases from 0.95 to 0.50 at 3.0 mM and 0.1 mM concentrations, respectively.3
1) is diluted with buffer, the fraction of bound SL-ibuprofen decreases only from 0.99 to 0.82 at 3.0 mM and 0.1 mM concentrations, respectively (Fig. 5(C)). As a result, a significant amount of SL-ibuprofens is still bound to BSA in the diluted system. On the other hand, dilution causes a strong release of SL-salicylic acid from BSA, the fraction of bound SL-salicylic acid decreases from 0.95 to 0.50 at 3.0 mM and 0.1 mM concentrations, respectively.3
The drug–albumin association constants can also be calculated from the EPR results. Salicylic acid, aspirin and ibuprofen bind to BSA via noncovalent bonding therefore an equilibrium exists between the bound and free states of the drugs:
| SL-drug + BSA ⇔ SL-drug/BSA complex |
where [SL-drug] is the concentration of the free drug, [BSA] is the concentration of free protein, and [SL-drug/BSA complex] is the concentration of the drug bound to the protein at equilibrium. The free BSA and free drug concentrations are assumed to be equal by accepting that one BSA takes up one SL-drug in the mixture of drug
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) albumin with a 1
albumin with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar concentration ratio.
1 molar concentration ratio.
At physiological concentrations of albumin (0.5–0.7 mM), the calculated Ka values of SL-ibuprofen are about 3.8–5.6 × 105 M−1, while for SL-salicylic acid and SL-aspirin, the Ka values are about 3.6–4.8 × 104 M−1, and 2.6–3.3 × 104 M−1, respectively. EPR results show that the binding affinity of SL-ibuprofen to BSA is ∼10 times higher than the binding affinities of SL-salicylic acid and SL-aspirin to BSA.
Ibuprofen is a more hydrophobic molecule compared to aspirin and salicylic acid; therefore it is not surprising that the more hydrophobic ibuprofen molecule could bind to BSA higher than the binding of aspirin and salicylic acid to BSA.
EPR studies of competitive binding of drugs with BSA
In the literature, interactions between small molecules and biomacromolecules have been extensively studied by X-ray crystallography.32–34 Structures of several albumin-drug complexes have also been resolved by X-ray crystallography.35 A 3D structure of the albumin crystal revealed that the main drug binding sites in BSA site I and site II are located in the hydrophobic cavities of sub-domains IIA and IIIA, respectively.12,35 Yang et al. showed 3D structures of both aspirin and salicylic acid bind to HSA at site I.36 The phenyl group of salicylic acid has mostly hydrophobic interactions with the surrounding aminoacids in site I. Ghuman et al.35 crystallized an ibuprofen–HSA complex and showed the binding sites of ibuprofen as mainly site II and also to a small extent site I.Furthermore, fluorescence studies showed that salicylic acid primarily binds to site I as well as to a smaller extent to site II of BSA.20 Site II is the main binding site for ibuprofen and aspirin.19 Also, aspirin binds to BSA to a small extent in site I.19
In addition to these drug high binding sites of BSA, several low affinity binding sites are also available in BSA.17 Drugs may also compete for these low affinity binding sites in BSA.
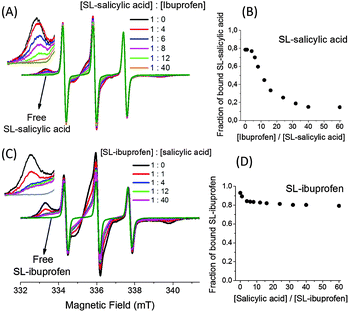
Fig. 6(A) shows the EPR spectra of the SL-salicylic acid/BSA complex before and after the addition of ibuprofen with different concentrations. At 0.5 mM SL-salicylic acid/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the bound fraction of SL-salicylic acid is 0.79. After the addition of ibuprofen, first the bound fraction of SL-salicylic acid does not change (up to 4
1), the bound fraction of SL-salicylic acid is 0.79. After the addition of ibuprofen, first the bound fraction of SL-salicylic acid does not change (up to 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, ibuprofen
1, ibuprofen![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SL-salicylic acid) but then it decreases sharply to 0.14 (Fig. 6(B)). This shows that ibuprofen first binds to empty sites and then additional ibuprofens displace the bound SL-salicylic acid from BSA. Since the association constant of ibuprofen is higher than that of salicylic acid, ibuprofen can easily replace the salicylic acid. Combining the knowledge from the literature and our results, one can speculate that SL-salicylic acid occupies mainly site I and to a small extent site II. Ibuprofen primarily binds to the empty side of site II and then replaces SL-salicylic acid from site II and also most of the SL-salicylic acid from site I.19,20,35 In contrast, we monitored the state of SL-ibuprofen in a solution of BSA in the presence of different amounts of salicylic acid (Fig. 6(C)). At 0.5 mM SL-ibuprofen/BSA (1
SL-salicylic acid) but then it decreases sharply to 0.14 (Fig. 6(B)). This shows that ibuprofen first binds to empty sites and then additional ibuprofens displace the bound SL-salicylic acid from BSA. Since the association constant of ibuprofen is higher than that of salicylic acid, ibuprofen can easily replace the salicylic acid. Combining the knowledge from the literature and our results, one can speculate that SL-salicylic acid occupies mainly site I and to a small extent site II. Ibuprofen primarily binds to the empty side of site II and then replaces SL-salicylic acid from site II and also most of the SL-salicylic acid from site I.19,20,35 In contrast, we monitored the state of SL-ibuprofen in a solution of BSA in the presence of different amounts of salicylic acid (Fig. 6(C)). At 0.5 mM SL-ibuprofen/BSA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the bound fraction of SL-ibuprofen is 0.93. After the addition of 0.5 mM and 2.0 mM salicylic acid, an obvious decrease in the bound fraction of SL-ibuprofen from 0.93 to 0.89 and to 0.84, respectively, is observed.
1), the bound fraction of SL-ibuprofen is 0.93. After the addition of 0.5 mM and 2.0 mM salicylic acid, an obvious decrease in the bound fraction of SL-ibuprofen from 0.93 to 0.89 and to 0.84, respectively, is observed.
However, the bound fraction of SL-ibuprofen does not change excessively after the addition of salicylic acid; e.g., the bound fraction of SL-ibuprofen reduces only to 0.80 in the presence of 30 mM salicylic acid (Fig. 6(D)). This result correlates with the preceding result; the bound fraction of SL-ibuprofen only reduces from 0.93 to 0.80 because salicylic acid with a lower association constant cannot replace ibuprofen easily. Salicylic acid displaces only a part of bound ibuprofens from the very limited sites which have low Ka values for ibuprofen and high Ka values for salicylic acid, probably site I.20
We can also find out the percentage of binding sites shared by salicylic acid and ibuprofen. Since additional ibuprofen (30 mM) replaces 83% of bound salicylic acid (0.5 mM), the remaining 17% of bound salicylic acid must either have a higher association constant with BSA or have different binding sites with respect to ibuprofen. To solve the remaining 17% of the bound salicylic acid situation, we used the data obtained from SL-ibuprofen/salicylic acid measurements. The addition of 30 mM salicylic acid to a 0.5 mM SL-ibuprofen/albumin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution replaces only 14% of bound SL-ibuprofen. It means 14% of SL-ibuprofen initially occupies the places in which salicylic acid has a higher association constant compared to that of ibuprofen. Therefore, 97% of all sites of salicylic acid and ibuprofen are shared (83% + 14%).
1) solution replaces only 14% of bound SL-ibuprofen. It means 14% of SL-ibuprofen initially occupies the places in which salicylic acid has a higher association constant compared to that of ibuprofen. Therefore, 97% of all sites of salicylic acid and ibuprofen are shared (83% + 14%).
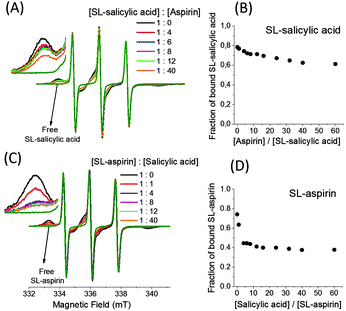
We also investigate the competitive interaction of salicylic acid and aspirin. Fig. 7 shows the EPR spectra of 0.5 mM SL-salicylic acid/aspirin (Fig. 7(A)) and also SL-aspirin/salicylic acid (Fig. 7(C)) mixtures in 0.5 mM BSA solutions. First, in the SL-salicylic acid/aspirin mixtures, aspirin concentration increases from 0.5 mM to 30 mM. The fraction of bound SL-salicylic acid decreases gradually from 0.79 to 0.61 by increasing the ratio of aspirin/SL-salicylic acid from 0 to 60 (Fig. 7(B)).
Since the calculated association constant of SL-salicylic acid is slightly higher than that of SL-aspirin, it is expected that aspirin would not replace the SL-salicylic acid completely in BSA. The small amount of replacement of SL-salicylic acid by aspirin can be explained by the weak binding ability of SL-salicylic acid at site II and the higher binding ability of aspirin at site II.19,20 The remaining high proportion of SL-salicylic acid locates probably at site I.19 Second, in the SL-aspirin/salicylic acid mixtures, the addition of salicylic acid rapidly reduces the bound fraction of 0. 5 mM SL-aspirin from 0.76 to 0.44 after the addition of 2 mM salicylic acid. It means that the added salicylic acid directly replaces some SL-aspirin but additional salicylic acid (up to 30 mM) only reduces the bound fraction of SL-aspirin to 0.38 (Fig. 7(D)). This correlates with the above results; salicylic acid replaces SL-aspirin easily at site I but some of the SL-aspirin located at site II is still bound.19,20
The percentage of binding sites shared by salicylic acid and aspirin can be calculated from the above results. Since additional aspirin (30 mM) replaces only 23% of the bound salicylic acid (0.5 mM), the remaining 77% of the bound salicylic acid must either have a higher association constant with BSA or have different binding sites with respect to aspirin. The addition of 30 mM salicylic acid to 0.5 mM SL-aspirin/albumin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution replaces 50% of the bound SL-aspirin. It means 50% of the bound SL-aspirin initially occupies the places where salicylic acid has a higher association constant compared to that of aspirin. Therefore, 73% of all sites of salicylic acid and aspirin are shared (23% + 50%).
1) solution replaces 50% of the bound SL-aspirin. It means 50% of the bound SL-aspirin initially occupies the places where salicylic acid has a higher association constant compared to that of aspirin. Therefore, 73% of all sites of salicylic acid and aspirin are shared (23% + 50%).
The extent of drug transportation is important to determine the dosage of drugs. In the body, drugs can be transported either as free floating molecules or as protein bound molecules. The concentration of free drugs is used to determine the effect of a drug rather than the protein bound concentration. Instead, a protein bound drug can escape from fast metabolism, which is crucial for sustained drug delivery applications. Our EPR results suggested that when two typical non-steroidal anti-inflammatory drugs, ibuprofen and salicylic acid, are co-administered, mostly ibuprofen binds to albumin and salicylic acid floats in the blood. Therefore, salicylic acid is expected to affect faster or/and be metabolized faster than ibuprofen.
Conclusions
In this study, we have presented the use of EPR spectroscopy and the spin labeling (SL) technique in the field of drug binding. Labeling drugs with stable nitroxide based Tempo radicals allows monitoring of the state of drugs in protein solution since bound and free spin labeled drugs have different EPR signals. Moreover, the areas under these signals give us the concentration of the bound and free drugs individually.In the first part of the study, we emphasized the intermolecular interactions between salicylic acid and BSA. Therefore, we have synthesized different derivatives of SL-salicylic acid. In these derivatives, the functional groups on the salicylic acid and its aromaticity were changed one by one. EPR measurements of these SL-molecules in BSA solution showed that hydrophobic interaction is the main interaction among the intermolecular interactions. While the bound fractions of these molecules are very close to each other (0.76–0.84), the EPR technique is able to differentiate the minute effects of structural variation on protein binding. Moreover, the importance of electrostatic interactions was determined using cationic BSA (cBSA) instead of anionic BSA. SL-salicylic acid with negative charge binds more strongly to cBSA than to BSA, and the SL-salicylic acid loading capacity of cBSA becomes greater than that of BSA.
In the second part, we used EPR spectroscopy as an alternative technique to study the competitive binding of drugs to BSA. At physiological concentrations of albumin (0.5–0.7 mM), the association constants of SL-drugs/BSA calculated from EPR spectra are 3.8–5.6 × 105 M−1 (SL-ibuprofen), 3.6–4.8 × 104 M−1 (SL-salicylic acid), and 2.6–3.3 × 104 M−1 (SL-aspirin). Since SL-ibuprofen has ten times higher binding affinity to BSA compared to the binding affinity of SL-salicylic acid, additional ibuprofens replace ∼83% of the bound SL-salicylic acid. However, additional salicylic acid replaces only ∼14% of the bound SL-ibuprofen. Altogether, ∼97% of all binding sites of salicylic acid and ibuprofen are shared. On the other hand, the calculated association constants of aspirin and salicylic acids with BSA are very close to each other. Therefore, additional salicylic acids replace only ∼50% of the bound SL-aspirin, and additional aspirin replace only ∼23% of the bound SL-salicylic acid. This shows that ∼73% of all binding sites of salicylic acid and aspirin are shared.
Thus, applications of cw EPR spectroscopy in combination with the spin labeling technique can provide information on the intermolecular interactions in drug/protein complexes and on the details of competitive binding of drugs.
Acknowledgements
This work was financially supported by Turkish Scientific and Technological Research Council (Tubitak) via 2232 Program under grant 114C082.References
- G. Martini and L. Ciani, Phys. Chem. Chem. Phys., 2009, 11, 211 RSC.
- S. Kempe, H. Metz and K. Mader, Eur. J. Pharm. Biopharm., 2010, 74, 55 CrossRef CAS PubMed.
- D. Tatlidil, M. Ucuncu and Y. Akdogan, Phys. Chem. Chem. Phys., 2015, 17, 22678 RSC.
- N. Rapoport and L. Pitina, J. Pharm. Sci., 1998, 87, 321 CrossRef CAS PubMed.
- H. Omote and M. K. Al-Shawi, J. Biol. Chem., 2002, 277, 45688 CrossRef CAS PubMed.
- A. Besheer, K. M. Wood, N. A. Peppas and K. Mader, J. Controlled Release, 2006, 111, 73 CrossRef CAS PubMed.
- H. Dugas, C. Spino and M. Quellette, Can. J. Chem., 1983, 61, 2540 CrossRef CAS.
- W. L. Hubbell, D. S. Cafiso and C. Altenbach, Nat. Struct. Biol., 2000, 7, 735 CrossRef CAS PubMed.
- Y. Akdogan, M. J. N. Junk and D. Hinderberger, Biomacromolecules, 2011, 12, 1072 CrossRef CAS PubMed.
- S. Nafisi, G. B. Sadeghi and A. Panahyab, J. Photochem. Photobiol., B, 2011, 105, 198 CrossRef CAS PubMed.
- Z.-M. Li, C.-W. Wei, Y. Zhang, D.-S. Wang and Y.-N. Liu, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2011, 879, 1934 CrossRef CAS PubMed.
- G. Sudlow, D. J. Birkett and D. N. Wade, Mol. Pharmacol., 1975, 11, 824 CAS.
- A. Sulkowska, B. Bojka, J. Rownicka and W. W. Sulkowski, Biopolymers, 2006, 81, 472 CrossRef PubMed.
- A. Kar, Essentials of Biopharmaceutics and Pharmacokinetics, Elsevier, 2010 Search PubMed.
- K. R. Kuchimanchi, M. S. Ahmed, T. P. Johnston and A. K. Mitra, J. Pharm. Sci., 2001, 90, 659 CrossRef CAS PubMed.
- T. Sakai, K. Yamasaki, T. Sako, U. K. Hansen, A. Suenaga and M. Otagiri, Pharm. Res., 2001, 18, 520 CrossRef CAS.
- Y. F. Cui, G. Y. Bai, C. G. Li, C. H. Ye and M. L. Liu, J. Pharm. Biomed. Anal., 2004, 34, 247 CrossRef CAS PubMed.
- X. Yu, Y. Yang, S. Lu, Q. Yao, H. Liu, X. Li and P. Yi, Spectrochim. Acta, Part A, 2011, 83, 322 CrossRef CAS PubMed.
- Y. Ni, R. Zhu and S. Kokot, Analyst, 2011, 136, 4794 RSC.
- Y. Ni, S. Su and S. Kokot, Anal. Chim. Acta, 2006, 580, 206 CrossRef CAS PubMed.
- F. Moreno and J. G. Jimenez, Chem.-Biol. Interact., 1999, 121, 237 CrossRef CAS PubMed.
- S. Park, B. L. Hayes, F. Marankan, D. C. Mulhearn, L. Wanna, A. D. Mesecar, B. D. Santarsiero, M. E. Johnson and D. L. Venton, J. Med. Chem., 2003, 46, 936 CrossRef CAS PubMed.
- Y. Akdogan, Y. Wu, K. Eisele, M. Schaz, T. Weil and D. Hinderberger, Soft Matter, 2012, 8, 11106 RSC.
- B. Srilakshmi, Nutrition Science, New Age International Publishers, 2006 Search PubMed.
- P. Morra, W. R. Bartle, S. E. Walker, S. N. Lee, S. K. Bowles and R. A. Reeves, Ann. Pharmacother., 1996, 30, 935 CAS.
- G. M. Janssen and J. F. Venema, J. Int. Med. Res., 1985, 13, 68 CAS.
- S. Stoll and A. Schweiger, J. Magn. Reson., 2006, 178, 42 CrossRef CAS PubMed.
- E. Heftmann, Chromatography, Elsevier, 1992 Search PubMed.
- S. T. Moerz and P. Huber, J. Phys. Chem. C, 2014, 119, 27072 Search PubMed.
- C. L. Cooper, A. Goulding, A. B. Kayitmazer, S. Ulrich, S. Stoll, S. Turksen, S. Yusa, A. Kumar and P. L. Dubin, Biomacromolecules, 2006, 7, 1025 CrossRef CAS PubMed.
- S. J. de Carvalho, R. Metzler and A. G. Cherstvy, Phys. Chem. Chem. Phys., 2014, 16, 15539 RSC.
- Z. Zhang, V. Martiny, D. Lagorce, Y. Ikeguchi, E. Alexov and M. A. Miteva, PLoS One, 2014, 9, e110884 Search PubMed.
- L. M. Stevers, C. V. Lam, S. F. R. Leysen, F. A. Meijer, D. S. van Scheppingen, R. M. J. M. de Vries, G. W. Carlile, L. G. Milroy, D. Y. Thomas, L. Brunsveld and C. Ottmann, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E1152 CrossRef CAS PubMed.
- Z. Zhang, S. Witham, M. Petukh, G. Moroy, M. Miteva, Y. Ikeguchi and E. Alexov, J. Am. Med. Inform. Assoc., 2013, 20, 643 CrossRef PubMed.
- J. Ghuman, P. A. Zunszain, I. Petitpas, A. A. Bhattacharya, M. Otagiri and S. Curry, J. Mol. Biol., 2005, 353, 38 CrossRef CAS PubMed.
- F. Yang, C. Bian, L. Zhu, G. Zhao, Z. Huang and M. Huang, J. Struct. Biol., 2007, 157, 348 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6cp04137j |
| This journal is © the Owner Societies 2016 |