Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effect of Zn2+ ions on the assembly of amylin oligomers: insight into the molecular mechanisms†
Vered
Wineman-Fisher
ab and
Yifat
Miller
*ab
aDepartment of Chemistry, Ben-Gurion University of the Negev, P.O. Box 653, Be'er Sheva 84105, Israel. E-mail: ymiller@bgu.ac.il; Fax: +972-86428709; Tel: +972-86428705
bIlse Katz Institute for Nanoscale Science and Technology, Ben-Gurion University of the Negev, Beér-Sheva 84105, Israel
First published on 6th July 2016
Abstract
Amylin is an endocrine hormone and is a member of the family of amyloid peptides and proteins that emerge as potential scaffolds by self-assembly processes. Zn2+ ions can bind to amylin peptides to form self-assembled Zn2+–amylin oligomers. In the current work the binding sites of Zn2+ ions in the self-assembled amylin oligomers at various concentrations of zinc have been investigated. Our results yield two conclusions. First, in the absence of Zn2+ ions polymorphic states (i.e. various classes of amylin oligomers) are obtained, but when Zn2+ ions bind to amylin peptides to form Zn2+–amylin oligomers, the polymorphism is decreased, i.e. Zn2+ ions bind only to specific classes of amylin. At low concentrations of Zn2+ ions the polymorphism is smaller than at high concentrations. Second, the structural features of the self-assembled amylin oligomers are not affected by the presence of Zn2+ ions. This study proposes new molecular mechanisms of the self-assembly of Zn2+–amylin oligomers.
1. Introduction
The family of amyloid peptides and amyloid proteins that are related to amyloidogenic diseases are self-assembled either from synthetic peptides or globular proteins.1–5 Amyloid assembly is a multistep process, starting from an isolated monomer of amyloid, followed by aggregation into oligomers and fibrils,6 and it has become the subject of intensive research in soft matter and materials science.7,8 The family of amyloid peptides and amyloid proteins emerge as potential scaffolds and biomaterials for the growth of mammalian cells.9 Spectroscopic and calorimetric approaches have been employed to characterize the stability of aggregated amyloids and elucidate the complex mechanism of fibril polymerization and depolymerization.10Metal ions are used to modulate the in vitro formation of protein nano-objects for creating functional materials.7,8 Metal ions also affect amyloid aggregation, promoting precipitation and fibrillogenesis, and alter the morphology of aggregates.11 Transition metals such as Cu2+, Fe3+ and Zn2+ are particularly interesting as they are found at high concentrations in the core and periphery of senile amyloid plaque deposits.12 It was shown that Cu2+, Fe3+ and Zn2+ promote the aggregation of α-synuclein in the brain of patients with Parkinson's disease (PD).13–15 The role of metal ions in other amyloid peptides and proteins with regard to amyloid aggregation is well-documented. Cu2+ and Zn2+ ions that have been found in the brain of Creutzfeldt–Jacob patients showed induction of prion aggregates.16–19 Extensive studies have shown that the contribution of Cu2+ and Zn2+ ions to the pathology of Alzheimer's disease (AD) is by promoting Aβ aggregation.20–22 Furthermore, the characterization of the structures of Cu2+–Aβ and Zn2+–Aβ oligomers and fibrils was extensively investigated both by experimental and computational studies.22–25
Amylin, which is also known as human islet amyloid polypeptide (hIAPP), is an endocrine hormone that consists of a 37-residue sequence and is a member of the family of amyloid peptides and proteins. In vitro studies have shown that Cu2+ ions inhibit the formation of amylin fibrils,26,27 and prevent the formation of the β-sheet structures of amylin that are identified as oligomers and fibrils.28 Yet, studying the role of Zn2+ in amylin aggregation is particularly of greater interest than the role of Cu2+ ions. While Cu2+ ions do not emerge from the pancreas together with amylin, it is known that amylin and Zn2+ ions are stored and packed inside the β-cell granules in the pancreatic islets29,30 and enter the blood stream together.29 In the pathological state of type 2 diabetes (T2D), amylin peptides aggregate to form various fibrillary structures.31,32 Furthermore, it is well-known that during the aggregation of amylin, toxic species of amylin oligomers are involved with the progressive process of β-cell destruction in T2D.28,33–35 It is also known that amylin and Zn2+ ions have a crucial role in glycemic regulation, but interruption in their delicate balance may cause the aggregation of amylin and thus the progression of T2D.33,34 Therefore, amylin peptides can interact with Zn2+ ions to form similar amyloid plaque deposits as other amyloids. Yet, the mechanisms and chemistry through which Zn2+ ions interact with the toxic amylin oligomers are still elusive.
In vitro studies have shown that Zn2+ ions play a role in promoting amylin aggregation30,36,37 and it was suggested that Zn2+ ions bind only to His18 in the amylin monomer.37,38 Ramamoorthy and coworkers proposed that Zn2+ ions have a complex effect on amylin aggregation because the Zn2+ ions have different effects depending on the various stages of amylin aggregation. At an early stage of aggregation (40 min) and at high concentrations of Zn2+ ions (10 mM), the Zn2+ ions induce the formation of large sizes of Zn2+–amylin aggregates compared with the absence of Zn2+ ions. However, at an early stage of aggregation (40 min) and at low concentrations of Zn2+ ions (100 μM), the Zn2+ ions promote the formation of even larger sizes of Zn2+–amylin aggregates than those that are formed at high Zn2+ concentrations.39 At the later stage of aggregation in which amylin fibrils are formed, the Zn2+ ions inhibit fibril formation at low concentrations, but the Zn2+ ions accelerate the formation of fibrils at higher concentrations.37,39,40
Despite the extensive work done previously, there are still questions that remain to be solved. How Zn2+ ions bind to amylin oligomers at the atomic resolution, i.e. what are the coordination binding sites? How Zn2+ ions affect the structure and stability of amylin oligomers? and what are the possible zinc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratios in amylin oligomers? This work is the first study that is aimed to address these important questions.
amylin ratios in amylin oligomers? This work is the first study that is aimed to address these important questions.
To address these issues, we applied our previous amylin oligomers,41 which were derived from the structural ssNMR models31 and from the crystal structural models.32 We have shown that there are four different amylin oligomers that are derived from the experimental studies that vary by the orientation of the residues along the sequence of the β-arch structure of amylin oligomers. Herein, we present a first study that investigates the specific binding sites in the various amylin oligomers, considering His18 as a potential residue that binds Zn2+ ions, as previously proposed by ssNMR.39,40 In order to provide insight into the effect of Zn2+ ions on the structure and the stability of amylin oligomers, we compare the structural characterization of Zn2+–amylin oligomers versus our previous amylin oligomers (in the absence of Zn2+ ions). Finally, we examine various Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratios, which have been proposed by experimental studies.39,40 Our molecular dynamics (MD) simulations revealed that Zn2+ ions selectively bind to specific classes of amylin oligomers thus reducing the polymorphism, but within these classes of Zn2+–amylin oligomers, polymorphic states were characterized due to the various concentrations of Zn2+ ions.
amylin ratios, which have been proposed by experimental studies.39,40 Our molecular dynamics (MD) simulations revealed that Zn2+ ions selectively bind to specific classes of amylin oligomers thus reducing the polymorphism, but within these classes of Zn2+–amylin oligomers, polymorphic states were characterized due to the various concentrations of Zn2+ ions.
2. Experimental section
Molecular dynamics (MD) simulations protocol
We constructed various models of Zn2+–amylin octamers with different Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratios by using Accelerys Discovery Studio software.42 Since the His18 residues in all constructed models bind Zn2+ ions, all His18 residues were deprotonated. MD simulations of the solvated models were performed in the NPT ensemble using the NAMD package43 with the CHARMM27 force field.44,45 The models were energy minimized and explicitly solvated in a TIP3P water box46,47 with a minimum distance of 15 Å from each edge of the box. Each water molecule within 2.5 Å of the models was removed. Counter ions were added at random locations to neutralize the models' charge. The Langevin piston method43,48,49 with a decay period of 100 fs and a damping time of 50 fs was used to maintain a constant pressure of 1 atm. A temperature of 330 K was controlled using a Langevin thermostat with a damping coefficient of 10 ps−1.43 The short-range van der Waals interactions were calculated using the switching function, with a twin range cut-off of 10.0 and 12.0 Å. Long-range electrostatic interactions were calculated using the particle mesh Ewald method with a cutoff of 12.0 Å.48,50 The equations of motion were integrated using the leapfrog integrator with a step of 1 fs. The solvated systems were energy minimized for 2000 conjugated gradient steps, where the hydrogen bonding distance between the β-sheets in each oligomer was fixed in the range 2.2–2.5 Å. The counter ions and water molecules were allowed to move. The hydrogen atoms were constrained to the equilibrium bond using the SHAKE algorithm.51 The minimized solvated systems were energy minimized for 5000 additional conjugate gradient steps and 20
amylin ratios by using Accelerys Discovery Studio software.42 Since the His18 residues in all constructed models bind Zn2+ ions, all His18 residues were deprotonated. MD simulations of the solvated models were performed in the NPT ensemble using the NAMD package43 with the CHARMM27 force field.44,45 The models were energy minimized and explicitly solvated in a TIP3P water box46,47 with a minimum distance of 15 Å from each edge of the box. Each water molecule within 2.5 Å of the models was removed. Counter ions were added at random locations to neutralize the models' charge. The Langevin piston method43,48,49 with a decay period of 100 fs and a damping time of 50 fs was used to maintain a constant pressure of 1 atm. A temperature of 330 K was controlled using a Langevin thermostat with a damping coefficient of 10 ps−1.43 The short-range van der Waals interactions were calculated using the switching function, with a twin range cut-off of 10.0 and 12.0 Å. Long-range electrostatic interactions were calculated using the particle mesh Ewald method with a cutoff of 12.0 Å.48,50 The equations of motion were integrated using the leapfrog integrator with a step of 1 fs. The solvated systems were energy minimized for 2000 conjugated gradient steps, where the hydrogen bonding distance between the β-sheets in each oligomer was fixed in the range 2.2–2.5 Å. The counter ions and water molecules were allowed to move. The hydrogen atoms were constrained to the equilibrium bond using the SHAKE algorithm.51 The minimized solvated systems were energy minimized for 5000 additional conjugate gradient steps and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 heating steps at 250 K, with all atoms being allowed to move. Then, the system was heated from 250 to 330 K for 300 ps and equilibrated at 330 K for 300 ps. All simulations were run for 50 ns at 330 K. These conditions were applied to all of the Zn2+–amylin octamers. The temperature of 330 K (which is higher that room/body temperature) has been chosen in order to investigate the stability of the Zn2+–amylin oligomers. Obviously, Zn2+–amylin oligomers that are stable at this temperature will be stable also at lower temperatures. The timescale of 50 ns has been examined for all simulated Zn2+–amylin octamers to ensure that this timescale is sufficient for these systems (see the Results and discussion section).
000 heating steps at 250 K, with all atoms being allowed to move. Then, the system was heated from 250 to 330 K for 300 ps and equilibrated at 330 K for 300 ps. All simulations were run for 50 ns at 330 K. These conditions were applied to all of the Zn2+–amylin octamers. The temperature of 330 K (which is higher that room/body temperature) has been chosen in order to investigate the stability of the Zn2+–amylin oligomers. Obviously, Zn2+–amylin oligomers that are stable at this temperature will be stable also at lower temperatures. The timescale of 50 ns has been examined for all simulated Zn2+–amylin octamers to ensure that this timescale is sufficient for these systems (see the Results and discussion section).
Analysis details
We examined the structural stability of the studied models by following the changes in the number of hydrogen bonds between β-strands, with the hydrogen bond cut-off being set to 2.5 Å. We further computed the root-mean square deviations (RMSDs), root-mean square fluctuations (RMSFs) and monitored the change in the inter-sheet distance (Cα backbone–backbone distance) in the core domain of all of the simulated Zn2+–amylin octamers. We estimated the core domain, as we previously estimated for amylin oligomers.41 In all the simulated Zn2+–amylin octamers that are based on model A2, the core domain was defined as the distance between residue 12 and residue 30. In all the simulated Zn2+–amylin octamers that are based on model A3, the distance between residue 12 and residue 29 was defined as the core domain. Finally, we estimated the average number of water molecules around each side-chain Cβ carbon within 4 Å for each simulated Zn2+–amylin octamer.Generalized born method with molecular volume (GBMV)
To obtain the relative conformational energies of the previous simulated amylin octamers41 and the current simulated Zn2+–amylin octamers, the trajectories of the last 5 ns of each simulated Zn2+–amylin octamer were first extracted from the explicit MD simulation excluding the water molecules. The solvation energies of all systems were calculated using the GBMV.52,53 In the GBMV calculations, the dielectric constant of water was set to 80. The hydrophobic solvent-accessible surface area (SASA) term factor was set to 0.00592 kcal mol−1 Å−2. Each conformer was minimized using 1000 cycles, and the conformational energy was evaluated by grid-based GBMV.A total of 1500 conformations (500 conformations for each of the 3 previous conformers41) were used to construct the energy landscape of the simulated amylin octamer that are based on model A2 (models M2, M2D and M5) and to evaluate the conformer probabilities by using Monte Carlo (MC) simulations. Similarly, a total of 1500 conformations were used: 500 conformations for each of the 2 examined conformers of the simulated Zn2+–amylin octamers that are based on model A2 with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (models R1 and R2) and one examined conformer simulated Zn2+–amylin octamer that is based on model A3 with the Zn2+
2 (models R1 and R2) and one examined conformer simulated Zn2+–amylin octamer that is based on model A3 with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (model S1). Finally, we used 1000 conformations, 500 conformations for each of the 2 examined conformers: one for the simulated Zn2+–amylin octamer that is based on model A2 with the Zn2+
2 (model S1). Finally, we used 1000 conformations, 500 conformations for each of the 2 examined conformers: one for the simulated Zn2+–amylin octamer that is based on model A2 with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (model R3) and the second for the simulated Zn2+–amylin octamer that is based on model A3 with the Zn2+
4 (model R3) and the second for the simulated Zn2+–amylin octamer that is based on model A3 with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (model S3).
4 (model S3).
In the first step, one conformation of conformer i and one conformation of conformer j were randomly selected. Then, the Boltzmann factor was computed as e−(Ej − Ei)/KT, where Ei and Ej are the conformational energies evaluated using the GBMV calculations for conformations i and j, respectively, K is the Boltzmann constant and T is the absolute temperature (298 K used here). If the value of the Boltzmann factor was larger than the random number, then the move from conformation i to conformation j was allowed. After 1 million steps, the conformations ‘visited’ for each conformer were counted.
3. Results and discussion
Zn2+ ions selectively bind to specific classes of amylin oligomers
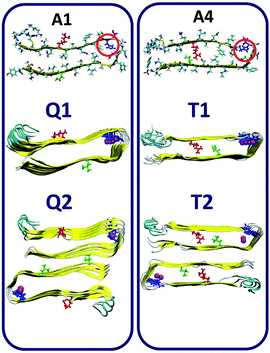
It has been shown by Ramamoorthy's group that Zn2+ ions in amylin solution lead to the formation of oligomers and fibrils.37,39 Therefore, the Zn2+ ions induce the formation of the cross-β structure of amylin and thus our initial constructed models here are based on the amylin oligomers and fibril-like structures with their self-assembled cross-β structure. Previously,41 we generated eight various molecular structural amylin cross-β octamers of single- and double-layer conformations applying models A1–A4 as ‘building blocks’ from Tycko's31 and Eiesnberg's32 coordinates (Fig. 1). These four models share the same sequence of 37 residues of amylin, but they differ in the orientations of the residues both along the backbone of the two β-strands and the U-turn of the β-arch structure of amylin. A particular difference that is related to this study is the location of His18 in the U-turn domain of amylin. In models A1 and A4, His18 is located inside the core domain, while in models A2 and A3 His18 is located outside the core domain (Fig. 1).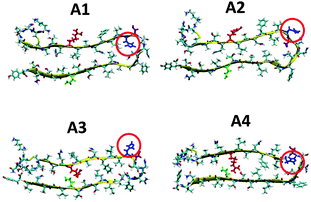 | ||
| Fig. 1 Representation of the molecular models of the fibril-like structures of amylin that differ in the orientations of the residues along the β-arch and the turn domain. The models were extracted from experimental studies31,32 and from our previous work.41 Residue colors: Arg11, red; His18, blue; Asn19, purple; and Asn31, green. | ||
We first examined the binding site of Zn2+ ions in amylin oligomers that are based on these four models A1–A4 for single- and double-layer conformations. The Zn2+ ions were interacted with His18, since it was suggested that Zn2+ ions bind to His18 in the amylin monomer.37,38 We constructed various conformations with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
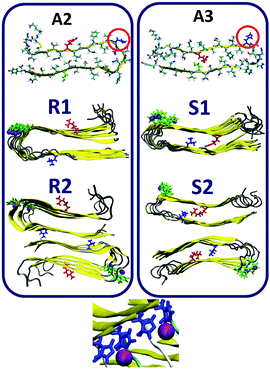
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 models Q1, Q2, T1 and T2 (Fig. 2) and R1, R2, S1 and S2 (Fig. 3). Table 1 summarizes these various conformations of Zn2+–amylin oligomers. The single- and the double-layer conformations that were constructed from models A1 (models Q1 and Q2) and A4 (models T1 and T2), in which His18 is located in the core domain, illustrated unstable Zn2+–amylin oligomers. During minimization, the Zn2+ ions escaped upon interaction with His18. We suggest that the steric effects inside the core domain in the β-arch domain of the amylin oligomers do not allow Zn2+ ions to interact with His18 residues (Fig. S1, ESI†). We therefore propose that Zn2+ ions do not bind to amylin oligomers that are based on models A1 and A4 and thus decrease the polymorphism, which is strongly reflected in the aggregation of amylin with the absence of Zn2+ ions, as we previously demonstrated for amylin oligomers.41
2 models Q1, Q2, T1 and T2 (Fig. 2) and R1, R2, S1 and S2 (Fig. 3). Table 1 summarizes these various conformations of Zn2+–amylin oligomers. The single- and the double-layer conformations that were constructed from models A1 (models Q1 and Q2) and A4 (models T1 and T2), in which His18 is located in the core domain, illustrated unstable Zn2+–amylin oligomers. During minimization, the Zn2+ ions escaped upon interaction with His18. We suggest that the steric effects inside the core domain in the β-arch domain of the amylin oligomers do not allow Zn2+ ions to interact with His18 residues (Fig. S1, ESI†). We therefore propose that Zn2+ ions do not bind to amylin oligomers that are based on models A1 and A4 and thus decrease the polymorphism, which is strongly reflected in the aggregation of amylin with the absence of Zn2+ ions, as we previously demonstrated for amylin oligomers.41
| Model | Constructed from A type model | Conformation | Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio amylin ratio |
|---|---|---|---|
| Q1 | A1 | Single layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| Q2 | A1 | Double layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| T1 | A4 | Single layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| T2 | A4 | Double layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| R1 | A2 | Single layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| R2 | A2 | Double layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| S1 | A3 | Single layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| S2 | A3 | Double layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| R3 | A2 | Single layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
| S3 | A3 | Single layer | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
In models A2 and A3 His18 is located outside the core domain of the β-arch. The single- and the double-layer conformations of models R1 and R2, which are based on model A2, and the single-layer conformation model S1, which is based on model A3, demonstrated that during minimization and MD simulations the Zn2+ ions were not escaped from the binding site. Surprisingly, in the double-layer conformation model S2, which is based on model A3, the Zn2+ ions escaped from the binding site after 5 ns of simulations (Fig. S2, ESI†). Finally, for the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, only single-layer conformation model R3 is based on model A2 and single-layer conformation model S3 is allowed to bind Zn2+ ions within this ratio. Table 1 summarizes these two conformations of Zn2+–amylin oligomers with the ratio of 1
4, only single-layer conformation model R3 is based on model A2 and single-layer conformation model S3 is allowed to bind Zn2+ ions within this ratio. Table 1 summarizes these two conformations of Zn2+–amylin oligomers with the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. We therefore focused our simulations and analysis only on models R1, R2 and S1 for the Zn2+
4. We therefore focused our simulations and analysis only on models R1, R2 and S1 for the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and R3 and S3 for the ratio of 1
2 and R3 and S3 for the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.
4.
Examining the variety of Zn2+ coordination modes in amylin oligomers
In numerous biological systems Zn2+ ions bind to ligands, such as amino acids and water molecules. Since it is known that Zn2+ ions bind His18 in the amylin monomer,37 it is great of interest to investigate the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio in Zn2+–amylin oligomers. To examine the possible Zn2+
amylin ratio in Zn2+–amylin oligomers. To examine the possible Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio in Zn2+–amylin oligomers, we constructed various Zn2+
amylin ratio in Zn2+–amylin oligomers, we constructed various Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratios of octamers that are based on models A2 and A3. There is a diversity of coordination modes of Zn2+ ions in proteins. It has been suggested that Zn2+ ions can bind to 4–6 amino acids or/and water molecules in proteins.54 Yet, it has been suggested that Zn2+ ions prefer the tetrahedral coordination mode, i.e. 4 amino acids/water molecules in proteins.54–56 Previously Ramamoorthy's group suggested that Zn2+ ions bind to 3, 4 and 6 helical amylin monomeric structure in His18.39,40 Yet, Ramammorthy's group also suggested that Zn2+ ions induce the formation of amylin fibrils, i.e. cross-β structures of amylin. Therefore, the coordination mode of Zn2+ ions in the self-assembled cross-β structure of amylin oligomers is still unknown. To address this issue, we examined various Zn2+
amylin ratios of octamers that are based on models A2 and A3. There is a diversity of coordination modes of Zn2+ ions in proteins. It has been suggested that Zn2+ ions can bind to 4–6 amino acids or/and water molecules in proteins.54 Yet, it has been suggested that Zn2+ ions prefer the tetrahedral coordination mode, i.e. 4 amino acids/water molecules in proteins.54–56 Previously Ramamoorthy's group suggested that Zn2+ ions bind to 3, 4 and 6 helical amylin monomeric structure in His18.39,40 Yet, Ramammorthy's group also suggested that Zn2+ ions induce the formation of amylin fibrils, i.e. cross-β structures of amylin. Therefore, the coordination mode of Zn2+ ions in the self-assembled cross-β structure of amylin oligomers is still unknown. To address this issue, we examined various Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratios of 1
amylin ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. S3, ESI†), 1
1 (Fig. S3, ESI†), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (models R1, R2 and S1) and 1
2 (models R1, R2 and S1) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (models R3 and S3). The ratio of 1
4 (models R3 and S3). The ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 has not been examined here and ongoing work is performed in our group to examine this ratio. Finally, the ratio of 1
3 has not been examined here and ongoing work is performed in our group to examine this ratio. Finally, the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 may be applicable to be examined for the six helical monomeric structures of amylin,39,40 but this ratio cannot be applicable in the cross-β structure of amylin oligomers that are studied here.
6 may be applicable to be examined for the six helical monomeric structures of amylin,39,40 but this ratio cannot be applicable in the cross-β structure of amylin oligomers that are studied here.
We examined the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for both single and double-layer conformations that are based on models A2 and A3. Not surprisingly, the Zn2+ ions were escaped from the binding site, probably due to the electrostatic repulsions between the Zn2+ ions (Fig. S3, ESI†). Furthermore, it is more likely that the Zn2+
1 for both single and double-layer conformations that are based on models A2 and A3. Not surprisingly, the Zn2+ ions were escaped from the binding site, probably due to the electrostatic repulsions between the Zn2+ ions (Fig. S3, ESI†). Furthermore, it is more likely that the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein ratio or the Zn2+
protein ratio or the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 is not possible when the Zn2+ ion binds to only one residue, such as His18 in amylin, and our simulations provided support to this hypothesis.
1 is not possible when the Zn2+ ion binds to only one residue, such as His18 in amylin, and our simulations provided support to this hypothesis.
As seen in Fig. 3, we also examined the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 both for single- and double layer conformations that are based on models A2 (models R1 and R2) and A3 (model S1). Our simulations suggest that each Zn2+ ion binds to two Nε atoms (each of two His18 residues of two amylin monomers) in these models, and four O atoms of water molecules complete the coordination mode to six coordination by forming an octahedral molecular geometry (Fig. S4 and Table S1, ESI†). Interestingly, the ratio of Zn2+
2 both for single- and double layer conformations that are based on models A2 (models R1 and R2) and A3 (model S1). Our simulations suggest that each Zn2+ ion binds to two Nε atoms (each of two His18 residues of two amylin monomers) in these models, and four O atoms of water molecules complete the coordination mode to six coordination by forming an octahedral molecular geometry (Fig. S4 and Table S1, ESI†). Interestingly, the ratio of Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Aβ of 1
Aβ of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 has also been illustrated by Miller et al. for Zn2+–Aβ oligomers; however, the completion of coordination with water molecules has not been studied for these oligomers.25 As noted in the previous section, surprisingly, in the double-layer conformation model S2, which is based on model A3, the Zn2+ ions escaped from the binding site after 5 ns of the simulations (Fig. S2, ESI†).
2 has also been illustrated by Miller et al. for Zn2+–Aβ oligomers; however, the completion of coordination with water molecules has not been studied for these oligomers.25 As noted in the previous section, surprisingly, in the double-layer conformation model S2, which is based on model A3, the Zn2+ ions escaped from the binding site after 5 ns of the simulations (Fig. S2, ESI†).
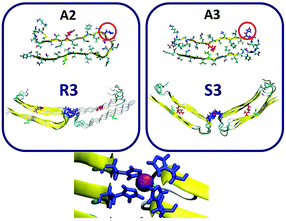
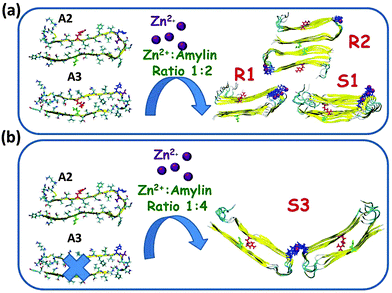
Finally, we examined the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 for models that are based on model A2 (model R3) and model A4 (model S3). In each one of models R3 and S3, two single-layer conformations were joined together by Zn2+ ions (Fig. 4). These two models, R3 and S3, exhibited stable Zn2+–amylin oligomers. Each Zn2+ ion in these two models binds to Nε atoms (each of four His18 residues of four amylin monomers) and two water molecules (Fig. S5 and Table S2, ESI†). The Zn2+
4 for models that are based on model A2 (model R3) and model A4 (model S3). In each one of models R3 and S3, two single-layer conformations were joined together by Zn2+ ions (Fig. 4). These two models, R3 and S3, exhibited stable Zn2+–amylin oligomers. Each Zn2+ ion in these two models binds to Nε atoms (each of four His18 residues of four amylin monomers) and two water molecules (Fig. S5 and Table S2, ESI†). The Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Aβ ratio of 1
Aβ ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 has also been shown by Miller et al. for Zn2+–Aβ oligomers, but the completion of the coordination with water molecules has not been shown.25 It should be noted that the ratio of 1
4 has also been shown by Miller et al. for Zn2+–Aβ oligomers, but the completion of the coordination with water molecules has not been shown.25 It should be noted that the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 cannot allow the formation of double-layer conformations of Zn2+–amylin oligomers in the well-organized cross-β structure.
4 cannot allow the formation of double-layer conformations of Zn2+–amylin oligomers in the well-organized cross-β structure.
Polymorphic states of Zn2+–amylin oligomers depend on the concentration of Zn2+ ions
We have previously shown that amylin oligomers exhibit polymorphic states, i.e. there are several conformations of amylin oligomers with similar energies and similar populations.41 It was previously suggested that Zn2+ ions increase the polymorphism in Aβ oligomers.25,57 Herein, we showed that because Zn2+ ions cannot bind to all polymorphic amylin oligomers, the Zn2+ ions decrease polymorphism in amylin oligomers. Yet, within the structures of Zn2+–amylin oligomers, it is of great interest to examine whether binding Zn2+ ions to amylin oligomers may also lead to polymorphism. To investigate this issue, we estimated the conformational energies of Zn2+–amylin oligomer models with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, which are based on model A2: models R1, R2 and the model that is based on A3: model S1. We further estimated the conformational energies of Zn2+–amylin oligomer models with the Zn2+
2, which are based on model A2: models R1, R2 and the model that is based on A3: model S1. We further estimated the conformational energies of Zn2+–amylin oligomer models with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 for the model that is based on A2 (model R3) and models that are based on A3 (model S3). The values of the averaged conformational energies are listed in Table S3 (ESI†). Finally, we estimated the relative conformational energies values of the equivalent amylin oligomers: models M2, M2d and M5 from our previous study.41 The values of the averaged conformational energies are also listed in Table S3 (ESI†).
4 for the model that is based on A2 (model R3) and models that are based on A3 (model S3). The values of the averaged conformational energies are listed in Table S3 (ESI†). Finally, we estimated the relative conformational energies values of the equivalent amylin oligomers: models M2, M2d and M5 from our previous study.41 The values of the averaged conformational energies are also listed in Table S3 (ESI†).
The averaged conformational energies and the distribution of the last 500 conformations for these models are in seen in Fig. 5. One can see that with the absence of Zn2+ ions the energy difference between amylin oligomers that are based on model A2 (model M2 and M2d) and the amylin oligomer that is based on model A3 (model M5) is in the range of 200–320 kcal mol−1. However, these are not the only models of amylin oligomers, as we previously shown, amylin oligomers are polymorphic and there are at least 12 different amylin oligomers.41 When adding Zn2+ ions with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, the energy difference between the models that are based on model A2 (model R3) and the model that is based on model A3 (model S3) is ∼250 kcal mol−1. Therefore, we suggest that adding relatively low concentrations of Zn2+ ions decrease polymorphism and yield a relatively high preference of one model over the other model. In this case, model R3 is highly preferred over model S3. However, at higher concentrations, i.e. in the Zn2+
4, the energy difference between the models that are based on model A2 (model R3) and the model that is based on model A3 (model S3) is ∼250 kcal mol−1. Therefore, we suggest that adding relatively low concentrations of Zn2+ ions decrease polymorphism and yield a relatively high preference of one model over the other model. In this case, model R3 is highly preferred over model S3. However, at higher concentrations, i.e. in the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1
amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the energy difference between the models that are based on model A2 (models R1 and R2) and the model that is based on A3 (model S1) is ∼100 kcal mol−1. Thus, at high concentrations of Zn2+ ions, the polymorphic states are increased compared with low concentration of Zn2+ ions, and models that are based on both A2 and A3 are preferred. Obviously, the polymorphism in such a case is relatively low compared with the case with the absence of Zn2+ ions.
2, the energy difference between the models that are based on model A2 (models R1 and R2) and the model that is based on A3 (model S1) is ∼100 kcal mol−1. Thus, at high concentrations of Zn2+ ions, the polymorphic states are increased compared with low concentration of Zn2+ ions, and models that are based on both A2 and A3 are preferred. Obviously, the polymorphism in such a case is relatively low compared with the case with the absence of Zn2+ ions.
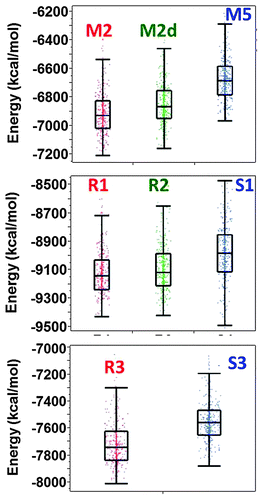 | ||
Fig. 5 Distributions of the conformational energy values (obtained from the GBMV calculations52,53) of the simulated models of amylin oligomers M2, M5 and M5d (obtained from our previous study), simulated models of Zn2+–amylin oligomers with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1 amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 – models R1, R2, and S1 and simulated models of Zn2+–amylin oligomers with the Zn2+ 2 – models R1, R2, and S1 and simulated models of Zn2+–amylin oligomers with the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio of 1 amylin ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 – models R3 and S3. 4 – models R3 and S3. | ||
Previously, it was suggested by Miller et al.58 that Aβ oligomers present polymorphic states and that metal ions increase the polymorphism, as previously shown for Zn2+–Aβ1–42 oligomers.25 Interestingly, we found that in amylin the Zn2+ ions may decrease or increase the polymorphism within the classes of amylin oligomers due to the concentrations of Zn2+, i.e., due to the Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin ratio.
amylin ratio.
Zn2+ ions do not change the structural features of amylin oligomers
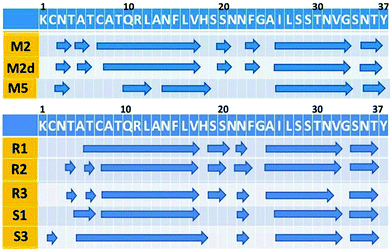
A previous study by Ramamoorthy's group has shown that both in the absence and in the presence of Zn2+ ions, amylin peptides are self-assembled into fibrillar structures, i.e. forming cross-β structures, and that the structural features are similar.39 To investigate the structural features of our simulated Zn2+–amylin oligomer models, we first examined whether the fibril-like structures are well-packed (Fig. S6, ESI†). We further estimated the percentage of hydrogen bonds along the cross-β structure that are retained throughout the MD simulations (Fig. S7, ESI†). Finally, we computed the root-mean square deviations (RMSDs) (Fig. S8, ESI†). These analyses show that all simulated models are well-packed with cross-β fibrillar structures, as suggested by the experimental observation.39 Furthermore, the RMSD analyses show that all simulated models were structurally converged in the last 10 ns of the simulations.To focus in more detail on the structural stability, we performed analysis of the secondary structure of the simulated amylin oligomers. According to dihedral angles ψ and φ analysis, the secondary structure of the Zn2+–amylin oligomers is similar to our previous equivalent amylin oligomers (Fig. 6). As seen in the secondary structure of the studied models, the two β-strand domains (residues 8–17 and residues 26–37) retained the stability, similarly to the equivalent amylin oligomer models that we previously studied.41 Therefore, we propose that the Zn2+ ions do not change the formation of stable cross-β structures of amylin, as previously suggested by experimental observations.39
Zn2+ ions affect the turn domains of specific classes of amylin oligomers
As noted above, the Zn2+ ions that bind to amylin oligomers in models that are based on models A2 and A3 in this study for both the ratios of Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin of 1
amylin of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 have no significant effect on the secondary structure of the turn domain (Fig. 6). The secondary structure of the Zn2+–amylin oligomers demonstrates a similar secondary structure to the equivalent amylin oligomers.
4 have no significant effect on the secondary structure of the turn domain (Fig. 6). The secondary structure of the Zn2+–amylin oligomers demonstrates a similar secondary structure to the equivalent amylin oligomers.
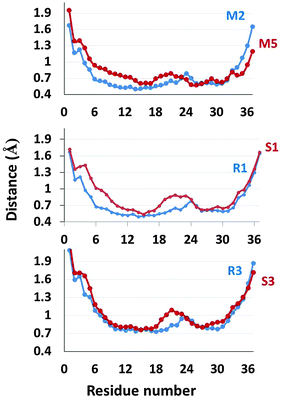
Since the Zn2+ ions bind to His18 that is located in the turn domain of the β-arch of the self-assembled amylin, it is important to examine whether the Zn2+ ions affect the morphologies of this domain. To examine the fluctuations of the simulated amylin oligomers (M2, M2d and M5) and the equivalent simulated Zn2+–amylin oligomers, we computed the root-mean square fluctuations (RMSFs) for each residue of the amylin oligomers (Fig. 7). We previously have shown that the turn domain of all amylin oligomer models fluctuates relatively more than the two β-strands domains.41 Specifically, the turn domain of model M2 (which are based on model A2) illustrated relatively less fluctuations than that of model M5 (which is based on model A3) (Fig. S7, ESI†). Herein, one can see a similar scenario for both ratios of Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin of 1
amylin of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Fig. 7). In models that are based on model A2 (models R1, R2 and R3), the turn domain fluctuates less compared with the turn domain in models that are based on model A3 (models S1 and S3).
4 (Fig. 7). In models that are based on model A2 (models R1, R2 and R3), the turn domain fluctuates less compared with the turn domain in models that are based on model A3 (models S1 and S3).
Interestingly, these fluctuations more likely affect the inter-sheet distance (Cα backbone–backbone distance) in the core domain of amylin oligomers. The inter-sheet distances of models R1 and R3 demonstrate relatively smaller values than those of models S1 and S3 (Fig. S6, ESI†). Indeed, one can see from Fig. 7 that the fluctuations of residues in the N-terminal domain of model S1 are relatively large compared to those of model R1. Therefore, it indicates that the cross-β structures of models R1 and R3 are well packed compared to those of models S1 and S3.
Finally, the turn domains in models that are based on model A2 are more solvated than models that are based on model A3 for both ratios of Zn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) amylin of 1
amylin of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Fig. S9, ESI†). One can see that the turn domain of model S1 is more solvated than that of model R1 (and R2) and the turn domain of model S3 is more solvated than that of model R3. Interestingly, the solvation of the turn domain is in accordance with the fluctuation of the turn domain (Fig. 7). When the turn domain fluctuates more, it is more exposed to solvent and therefore it is more solvated.
4 (Fig. S9, ESI†). One can see that the turn domain of model S1 is more solvated than that of model R1 (and R2) and the turn domain of model S3 is more solvated than that of model R3. Interestingly, the solvation of the turn domain is in accordance with the fluctuation of the turn domain (Fig. 7). When the turn domain fluctuates more, it is more exposed to solvent and therefore it is more solvated.
4. Conclusions
Amylin aggregation is associated with β-cell death in T2D. The role of Zn2+ in T2D is controversial. It is known that patients with T2D have zinc deficiency in β-cells.59 One of the fundamental questions is what is the effect of zinc deficiency in β-cell granules on the development of T2D, such as on the aggregation of amylin. Previously, it was suggested that amylin and Zn2+ ions have a crucial role in glycemic regulation, but interruption in their delicate balance may cause aggregation of amylin and thus the progression of T2D.33,34 It has been suggested that Zn2+ ions interact with amylin peptides and lead to the self-assembly of amylin peptides into fibrillar structures, via the oligomeric toxic species.39 In this study we investigated the interactions of the Zn2+ ions within the amylin peptides at the atomic resolution in the toxic oligomeric states of amylin. Specifically, we investigated the effect of these interactions on the self-assembly of amylin. Our results led to two main conclusions. First, while polymorphic states were obtained in the absence of Zn2+ ions, i.e. various classes of amylin oligomers were present,41 in the absence of Zn2+ ions only specific classes of amylin oligomers were obtained. We therefore suggest that Zn2+ ions decrease amylin polymorphism. Yet, the level of polymorphic states depends on the concentration of the Zn2+ ions. Fig. 8 illustrates the various mechanisms proposed for the formation of amylin oligomers at different concentrations of Zn2+ ions. At high concentrations of Zn2+ ions, various polymorphic states of Zn2+–amylin oligomers are obtained, while at low concentrations of Zn2+ ions there is high preference for one Zn2+–amylin oligomer. Second, Zn2+ ions do not affect the structural features of the self-assembly of amylin, as previously shown by experimental measurements.39 Yet, in several classes of amylin oligomers there are slight fluctuations in the turn domain of amylin, due to the interactions with the Zn2+ ions. It was found that the turn domain in the amylin peptide is crucial for seeding and for self-assembly of amylin into oligomers.60–64 It was also proposed that at an early stage of amylin oligomerization in solution, amylin forms interactions between His18 and Tyr37, i.e. interactions between the turn domain (in which His18 is located and can bind Zn2+ ions) and the C-terminal of amylin (residue Tyr37).38 We therefore suggest that interactions of the Zn2+ ions can affect the initiation of the process of amylin aggregation. Herein, we have shown that stable Zn2+–amylin oligomers were less solvated, and thus more prone to aggregate. Yet, future experimental work is needed to approve this interesting finding from our simulations.Acknowledgements
This research was supported by the Israel Science Foundation (grant No. 532/15) and partly by the FP7-PEOPLE-2011-CIG, research grant no. 303741. All simulations were performed using the high-performance computational facilities of the Miller Lab in BGU HPC computational center. The support of the BGU HPC computational center staff is greatly appreciated.References
- M. N. Bongiovanni, D. B. Scanlon and S. L. Gras, Functional fibrils derived from the peptide TTR1-cycloRGDfK that target cell adhesion and spreading, Biomaterials, 2011, 32, 6099–6110 CrossRef CAS PubMed.
- G. A. Silva, C. Czeisler, K. L. Niece, E. Beniash, D. A. Harrington, J. A. Kessler and S. I. Stupp, Selective differentiation of neural progenitor cells by high-epitope density nanofibers, Science, 2004, 303, 1352–1355 CrossRef CAS PubMed.
- C. Li, J. Adamcik and R. Mezzenga, Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties, Nat. Nanotechnol., 2012, 7, 421–427 CrossRef CAS PubMed.
- N. P. Reynolds, M. Charnley, R. Mezzenga and P. G. Hartley, Engineered lysozyme amyloid fibril networks support cellular growth and spreading, Biomacromolecules, 2014, 15, 599–608 CrossRef CAS PubMed.
- N. P. Reynolds, K. E. Styan, C. D. Easton, Y. Li, L. Waddington, C. Lara, J. S. Forsythe, R. Mezzenga, P. G. Hartley and B. W. Muir, Nanotopographic surfaces with defined surface chemistries from amyloid fibril networks can control cell attachment, Biomacromolecules, 2013, 14, 2305–2316 CrossRef CAS PubMed.
- S. I. Cohen, S. Linse, L. M. Luheshi, E. Hellstrand, D. A. White, L. Rajah, D. E. Otzen, M. Vendruscolo, C. M. Dobson and T. P. Knowles, Proliferation of amyloid-beta42 aggregates occurs through a secondary nucleation mechanism, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 9758–9763 CrossRef CAS PubMed.
- S. Bolisetty, J. J. Vallooran, J. Adamcik and R. Mezzenga, Magnetic-responsive hybrids of Fe3O4 nanoparticles with beta-lactoglobulin amyloid fibrils and nanoclusters, ACS Nano, 2013, 7, 6146–6155 CrossRef CAS PubMed.
- T. P. Knowles and M. J. Buehler, Nanomechanics of functional and pathological amyloid materials, Nat. Nanotechnol., 2011, 6, 469–479 CrossRef CAS PubMed.
- C. Zhong, T. Gurry, A. A. Cheng, J. Downey, Z. Deng, C. M. Stultz and T. K. Lu, Strong underwater adhesives made by self-assembling multi-protein nanofibres, Nat. Nanotechnol., 2014, 9, 858–866 CrossRef CAS PubMed.
- T. Ikenoue, Y. H. Lee, J. Kardos, H. Yagi, T. Ikegami, H. Naiki and Y. Goto, Heat of supersaturation-limited amyloid burst directly monitored by isothermal titration calorimetry, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6654–6659 CrossRef CAS PubMed.
- S. S. Leal, H. M. Botelho and C. M. Gomes, Metal ions as modulators of protein conformation and misfolding in neurodegeneration, Coord. Chem. Rev., 2012, 256, 2253–2270 CrossRef CAS.
- A. I. Bush, The metallobiology of Alzheimer's disease, Trends Neurosci., 2003, 26, 207–214 CrossRef CAS PubMed.
- A. Prakash, K. Bharti and A. B. Majeed, Zinc: indications in brain disorders, Fundam. Clin. Pharmacol., 2015, 29, 131–149 CrossRef CAS PubMed.
- C. G. Dudzik, E. D. Walter and G. L. Millhauser, Coordination features and affinity of the Cu(2)+ site in the alpha-synuclein protein of Parkinson's disease, Biochemistry, 2011, 50, 1771–1777 CrossRef CAS PubMed.
- M. Bisaglia, I. Tessari, S. Mammi and L. Bubacco, Interaction between alpha-synuclein and metal ions, still looking for a role in the pathogenesis of Parkinson's disease, NeuroMol. Med., 2009, 11, 239–251 CrossRef CAS PubMed.
- F. Stellato, A. Spevacek, O. Proux, V. Minicozzi, G. Millhauser and S. Morante, Zinc modulates copper coordination mode in prion protein octa-repeat subdomains, Eur. Biophys. J., 2011, 40, 1259–1270 CrossRef CAS PubMed.
- A. A. Valiente-Gabioud, V. Torres-Monserrat, L. Molina-Rubino, A. Binolfi, C. Griesinger and C. O. Fernandez, Structural basis behind the interaction of Zn(2)(+) with the protein alpha-synuclein and the Abeta peptide: a comparative analysis, J. Inorg. Biochem., 2012, 117, 334–341 CrossRef CAS PubMed.
- E. Gaggelli, F. Bernardi, E. Molteni, R. Pogni, D. Valensin, G. Valensin, M. Remelli, M. Luczkowski and H. Kozlowski, Interaction of the human prion PrP (106-126) sequence with copper(II), manganese(II), and zinc(II): NMR and EPR studies, J. Am. Chem. Soc., 2005, 127, 996–1006 CrossRef CAS PubMed.
- E. D. Walter, D. J. Stevens, M. P. Visconte and G. L. Millhauser, The prion protein is a combined zinc and copper binding protein: Zn2+ alters the distribution of Cu2+ coordination modes, J. Am. Chem. Soc., 2007, 129, 15440–15441 CrossRef CAS PubMed.
- D. R. Brown and H. Kozlowski, Biological inorganic and bioinorganic chemistry of neurodegeneration based on prion and Alzheimer diseases, Dalton Trans., 2004, 1907–1917 RSC.
- J. W. Karr and V. A. Szalai, Cu(II) binding to monomeric, oligomeric, and fibrillar forms of the Alzheimer's disease amyloid-beta peptide, Biochemistry, 2008, 47, 5006–5016 CrossRef CAS PubMed.
- S. Parthasarathy, F. Long, Y. Miller, Y. Xiao, D. McElheny, K. Thurber, B. Ma, R. Nussinov and Y. Ishii, Molecular-level examination of Cu2 binding structure for amyloid fibrils of 40-residue Alzheimer's β by solid-state NMR spectroscopy, J. Am. Chem. Soc., 2011, 133, 3390–3400 CrossRef CAS PubMed.
- S. C. Drew, C. L. Masters and K. J. Barnham, Alanine-2 carbonyl is an oxygen ligand in Cu2 coordination of Alzheimer's disease amyloid-β peptide-relevance to N-terminally truncated forms, J. Am. Chem. Soc., 2009, 131, 8760–8761 CrossRef CAS PubMed.
- C. Hureau and P. Faller, Aβ-mediated ROS production by Cu ions: structural insights, mechanisms and relevance to Alzheimer's disease, Biochimie, 2009, 91, 1212–1217 CrossRef CAS PubMed.
- Y. Miller, B. Ma and R. Nussinov, Zinc ions promote Alzheimer Abeta aggregation via population shift of polymorphic states, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9490–9495 CrossRef CAS PubMed.
- D. Drago, S. Bolognin and P. Zatta, Role of metal ions in the Aβ oligomerization in Alzheimer's disease and in other neurological disorders, Curr. Alzheimer Res., 2008, 5, 500–507 CrossRef CAS PubMed.
- B. Ward, K. Walker and C. Exley, Copper(II) inhibits the formation of amylin amyloid in vitro, J. Inorg. Biochem., 2008, 102, 371–375 CrossRef CAS PubMed.
- K. Jeong, W. Y. Chung, Y. S. Kye and D. Kim, Cu(II) cyclen cleavage agent for human islet amyloid peptide, Bioorg. Med. Chem., 2010, 18, 2598–2601 CrossRef CAS PubMed.
- D. Li, S. Chen, E. A. Bellomo, A. I. Tarasov, C. Kaut, G. A. Rutter and W. H. Li, Imaging dynamic insulin release using a fluorescent zinc indicator for monitoring induced exocytotic release (ZIMIR), Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 21063–21068 CrossRef CAS PubMed.
- P. Westermark, Z. C. Li, G. T. Westermark, A. Leckstrom and D. F. Steiner, Effects of beta cell granule components on human islet amyloid polypeptide fibril formation, FEBS Lett., 1996, 379, 203–206 CrossRef CAS PubMed.
- S. Luca, W.-M. Yau, R. Leapman and R. Tycko, Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR, Biochemistry, 2007, 46, 13505–13522 CrossRef CAS PubMed.
- J. J. W. Wiltzius, S. A. Sievers, M. R. Sawaya, D. Cascio, D. Popov, C. Riekel and D. Eisenberg, Atomic structure of the cross-β spine of islet amyloid polypeptide (amylin), Protein Sci., 2008, 17, 1467–1474 CrossRef CAS PubMed.
- J. Janson, R. H. Ashley, D. Harrison, S. McIntyre and P. C. Butler, The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles, Diabetes, 1999, 48, 491–498 CrossRef CAS PubMed.
- R. A. Ritzel, J. J. Meier, C. Y. Lin, J. D. Veldhuis and P. C. Butler, Human islet amyloid polypeptide oligomers disrupt cell coupling, induce apoptosis, and impair insulin secretion in isolated human islets, Diabetes, 2007, 56, 65–71 CrossRef CAS PubMed.
- S. Sakagashira, H. J. Hiddinga, K. Tateishi, T. Sanke, T. Hanabusa, K. Nanjo and N. L. Eberhardt, S20G mutant amylin exhibits increased in vitro amyloidogenicity and increased intracellular cytotoxicity compared to wild-type amylin, Am. J. Pathol., 2000, 157, 2101–2109 CrossRef CAS PubMed.
- F. Bellia and G. Grasso, The role of copper(II) and zinc(II) in the degradation of human and murine IAPP by insulin-degrading enzyme, J. Mass Spectrom., 2014, 49, 274–279 CrossRef CAS PubMed.
- J. R. Brender, K. Hartman, R. P. Nanga, N. Popovych, R. de la Salud Bea, S. Vivekanandan, E. N. Marsh and A. Ramamoorthy, Role of zinc in human islet amyloid polypeptide aggregation, J. Am. Chem. Soc., 2010, 132, 8973–8983 CrossRef CAS PubMed.
- L. Wei, P. Jiang, W. Xu, H. Li, H. Zhang, L. Yan, M. B. Chan-Park, X. W. Liu, K. Tang, Y. Mu and K. Pervushin, The molecular basis of distinct aggregation pathways of islet amyloid polypeptide, J. Biol. Chem., 2011, 286, 6291–6300 CrossRef CAS PubMed.
- J. R. Brender, J. Krishnamoorthy, G. M. Messina, A. Deb, S. Vivekanandan, C. La Rosa, J. E. Penner-Hahn and A. Ramamoorthy, Zinc stabilization of prefibrillar oligomers of human islet amyloid polypeptide, Chem. Commun., 2013, 49, 3339–3341 RSC.
- S. Salamekh, J. R. Brender, S. J. Hyung, R. P. Nanga, S. Vivekanandan, B. T. Ruotolo and A. Ramamoorthy, A two-site mechanism for the inhibition of IAPP amyloidogenesis by zinc, J. Mol. Biol., 2011, 410, 294–306 CrossRef CAS PubMed.
- V. Wineman-Fisher, Y. Atsmon-Raz and Y. Miller, Orientations of residues along the beta-arch of self-assembled amylin fibril-like structures lead to polymorphism, Biomacromolecules, 2015, 16, 156–165 CrossRef CAS PubMed.
- http://accelrys.com/products/discovery-studio/ .
- L. Kalé, R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan and K. Schulten, NAMD2: greater scalability for parallel molecular dynamics, J. Comput. Phys., 1999, 151, 283–312 CrossRef.
- B. R. Brooks, R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan and M. Karplus, CHARMM: A program for macromolecular energy, minimization, and dynamics calculations, J. Comput. Chem., 1983, 4, 187–217 CrossRef CAS.
- A. D. MacKerell, M. Feig and C. L. Brooks, Improved treatment of the protein backbone in empirical force fields, J. Am. Chem. Soc., 2004, 126, 698–699 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- M. W. Mahoney and W. L. Jorgensen, A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions, J. Chem. Phys., 2000, 112, 8910–8922 CrossRef CAS.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, A smooth particle mesh Ewald method, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- K. Tu, D. J. Tobias and M. L. Klein, Constant pressure and temperature molecular dynamics simulation of a fully hydrated liquid crystal phase dipalmitoylphosphatidylcholine bilayer, Biophys. J., 1995, 69, 2558–2562 CrossRef CAS PubMed.
- A. D. MacKerell Jr, D. Bashford, M. Bellott, R. L. Dunbrack Jr, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo and S. Ha, All-atom empirical potential for molecular modeling and dynamics studies of proteins, J. Phys. Chem. B, 1998, 102, 3586–3616 CrossRef PubMed.
- P. Jungwirth and D. J. Tobias, Molecular structure of salt solutions: a new view of the interface with implications for heterogeneous atmospheric chemistry, J. Phys. Chem. B, 2001, 105, 10468–10472 CrossRef CAS.
- M. S. Lee, M. Feig, F. R. Salsbury and C. L. Brooks, New analytic approximation to the standard molecular volume definition and its application to generalized Born calculations, J. Comput. Chem., 2003, 24, 1348–1356 CrossRef CAS PubMed.
- M. S. Lee, F. R. Salsbury Jr and C. L. Brooks III, Novel generalized Born methods, J. Chem. Phys., 2002, 116, 10606–10614 CrossRef CAS.
- K. Patel, A. Kumar and S. Durani, Analysis of the structural consensus of the zinc coordination centers of metalloprotein structures, Biochim. Biophys. Acta, 2007, 1774, 1247–1253 CrossRef CAS PubMed.
- I. L. Alberts, K. Nadassy and S. J. Wodak, Analysis of zinc binding sites in protein crystal structures, Protein science: a publication of the Protein Society, 1998, 7, 1700–1716 CrossRef CAS PubMed.
- M. Laitaoja, J. Valjakka and J. Janis, Zinc coordination spheres in protein structures, Inorg. Chem., 2013, 52, 10983–10991 CrossRef CAS PubMed.
- Y. Miller, H. M. Botelho and R. Nussinov, Metal binding sites in amyloid oligomers: Complexes and mechanisms, Coord. Chem. Rev., 2012, 256, 2245–2252 CrossRef CAS.
- Y. Miller, B. Ma and R. Nussinov, Polymorphism in Alzheimer Abeta amyloid organization reflects conformational selection in a rugged energy landscape, Chem. Rev., 2010, 110, 4820–4838 CrossRef CAS PubMed.
- W. Maret, Zinc and human disease, Met. Ions Life Sci., 2013, 13, 389–414 Search PubMed.
- J. R. Brender, U. H. Durr, D. Heyl, M. B. Budarapu and A. Ramamoorthy, Membrane fragmentation by an amyloidogenic fragment of human Islet Amyloid Polypeptide detected by solid-state NMR spectroscopy of membrane nanotubes, Biochim. Biophys. Acta, 2007, 1768, 2026–2029 CrossRef CAS PubMed.
- S. Gilead and E. Gazit, The role of the 14–20 domain of the islet amyloid polypeptide in amyloid formation, Exp. Diabetes Res., 2008, 2008, 256954 CrossRef PubMed.
- E. T. Jaikaran, C. E. Higham, L. C. Serpell, J. Zurdo, M. Gross, A. Clark and P. E. Fraser, Identification of a novel human islet amyloid polypeptide beta-sheet domain and factors influencing fibrillogenesis, J. Mol. Biol., 2001, 308, 515–525 CrossRef CAS PubMed.
- D. Milardi, M. F. Sciacca, M. Pappalardo, D. M. Grasso and C. La Rosa, The role of aromatic side-chains in amyloid growth and membrane interaction of the islet amyloid polypeptide fragment LANFLVH, Eur. Biophys. J., 2011, 40, 1–12 CrossRef CAS PubMed.
- L. A. Scrocchi, K. Ha, Y. Chen, L. Wu, F. Wang and P. E. Fraser, Identification of minimal peptide sequences in the (8–20) domain of human islet amyloid polypeptide involved in fibrillogenesis, J. Struct. Biol., 2003, 141, 218–227 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6cp04105a |
| This journal is © the Owner Societies 2016 |