Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Generation of spin in single cholesterol molecules on gold
Sujoy
Karan†
* and
Richard
Berndt
*
Institut für Experimentelle und Angewandte Physik, Christian-Albrechts-Universität zu Kiel, 24098 Kiel, Germany. E-mail: berndt@physik.uni-kiel.de
First published on 1st March 2016
Abstract
Compact islands of cholesterol on Au(111) were investigated with scanning tunneling microscopy at ∼5 K. Single molecules have been switched among several states, three of which exhibit a sharp spectroscopic feature at the Fermi level. This feature signals the presence of a localized spin and suggests that the molecule may be controllably switched between paramagnetic and diamagnetic states.
Most materials investigated in spintronics are metallic compounds and semiconductors.1,2 Recently efforts have been made to establish organic spintronics,3 one reason being that weak spin–orbit and hyperfine interactions in organic molecules tend to imply long spin coherence times.4 A few stable organic radicals have been shown to preserve their spin on metal surfaces.5,6 Moreover, electron transfer from a substrate can induce a magnetic moment in organic complexes.7 Here we report indication that a spin can be generated and quenched in cholesterol, a closed shell molecule, on the Au(111) surface. A scanning tunneling microscope (STM) was used to create radical cations from the pristine adsorbed molecules via hole injection from the tip into the molecule.
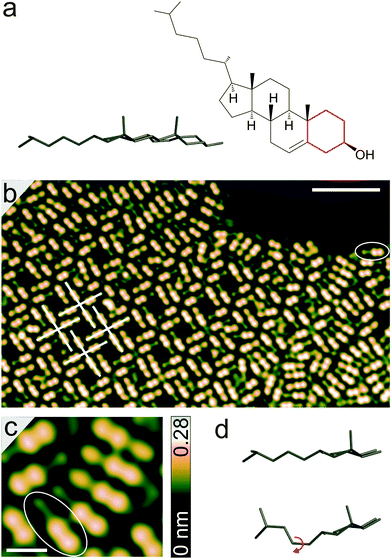
Cholesterol (Fig. 1a) consists of four fused hydrocarbon rings (denoted tetra-cyclic unit below) linked to an isooctyl chain, with a hydroxyl group attached to the cyclohexane ring (highlighted in red) at the far end. A pseudo-three-dimensional schematic viewed across the plane of the hydrocarbon rings shows that two methyl groups attached to the hydrocarbon rings protrude from the plane of the tetra-cyclic unit and the octyl chain. It may be interesting to note that the molecule is an essential component of cell membranes, plays a vital role in lipid metabolism, and contributes to the functions of living organism through signal transduction.8–10
Cholesterol was sublimated onto Au(111) at room temperature in ultra-high vacuum. Experiments were carried out with a STM operated at ∼5 K. Electrochemically etched tungsten wires were used as tips, treated in situ by indenting them into the Au substrate.
Fig. 1b shows an image of a molecular island formed upon deposition on Au(111). The ellipse drawn on the image indicates a single molecule (also see Fig. 1c). The tetra-cyclic units of the molecules appear to lie flat on the surface with the methyl groups, attached to hydrocarbon rings, in upright orientation. This apparently causes the dumbbell shaped protrusions appearing higher (apparent height ∼0.21 nm) than the octyl chains (discussed in detail below). Incommensurate adsorption of the molecules with respect to atomic lattice of the substrate leads to slight variations from molecule to molecule.
Much of the surface is cluttered with molecules (as in the right part of Fig. 1b), although some short-range order is seen with strings of molecules arranged in parallel or antiparallel. In addition, rhombic pores surrounded by cruciform patterns (examples marked in white in Fig. 1b, acute angle ∼84°) are frequently observed within the agglomerates. These arrangements involve a head-to-head orientation of the molecules which is presumably caused by intermolecular hydrogen bonds among hydroxyl groups. Previous studies of cholesterol monolayers at a variety of liquid–solid interfaces have also shown the molecules to be oriented head-to-head in pairs, owing to hydrogen bonds between the hydroxyl groups.11–14 Parallel arrangements of dimers have also been reported and attributed to interaction through the tetra-cyclic units.13 This is consistent with our observations of molecules being mostly grouped in parallel, although in small domains.
Closer inspection of Fig. 1c reveals two distinct adsorption geometries of the molecules. While the octyl chains of some molecules are imaged as elliptical protrusions with apparent heights of ∼0.16 nm with respect to the substrate, others appear lower (∼0.1 nm). This difference may be understood from the flexibility of the octyl chains, which is also observed in crystal structures.15 The chains may adapt their geometries to maximize the van der Waals interaction with the substrate and to reduce the steric repulsion within a molecule as well as with its neighbors.16 The pattern surrounded by an ellipse in Fig. 1c is consistent with a fully extended anti form of the octyl chain (Fig. 1d), which is the energetically most favored structure.15 In this form, the methyl end-groups of the octyl chain are located at heights similar to that of methylenes and the chain as a whole remains at a height below the methyl groups attached to the tetra-cyclic unit. The other pattern presumably corresponds to a conformation of the chain involving one of the C–C bonds in gauche form (marked with curved red arrow in Fig. 1d), which leads to an upright orientation of one of the methyl end-groups and to an elliptical protrusion in STM images.
Each of the molecules may be switched to a number of states by applying voltage pulses to the center of the tetra-cyclic unit (red crosses in Fig. 2a). A particular state was obtained by changing the sample voltage to V ∼ −2.5 V from a value of approximately ∼−1 V with the current-feedback disabled and subsequently monitoring the current I. V was changed back to its original value when an abrupt change of I, which typically occurred on a time scale of seconds, signaled a change of the molecule. This procedure had the effect to freeze the manipulated state. The images in Fig. 2b–d illustrate a sequence of switching events. Fig. 2b presents the result of pulses to the molecules indicated with crosses in Fig. 2a. The pulses switched the molecules to two states denoted A and B in Fig. 2b. A few other molecules in the island changed to state A as well. Inelastic scattering of electrons propagating through the surface may induce these changes.17,18 Moreover, it is conceivable that a geometrical change of a molecule is transmitted via lateral forces within the close-packed islands.
 | ||
| Fig. 2 (a–d) STM images showing a sequence of switching events. The manipulations were effected at V = −2.5 V with disabled current-feedback. The positions of the applied pulses are marked with red crosses. Different states are marked with arrows. (e) dI/dV spectra recorded over the center of the tetra-cyclic rings of different molecular states. The bottom two spectra were recorded after and before the manipulation of the molecule in state C in Fig. 2c. For clarity, the spectra are shifted vertically (top to bottom: by 4, 2, 1, 0, and −1 nS). The data were recorded using a lock-in amplifier by adding a sinusoidal modulation of 2 mVrms at 985 Hz to the sample voltage. Before opening the current feedback, the STM was operated at V = −100 mV and I = 100 pA. Red curves represent fits of Frota functions (details in the text) to the measured spectra. | ||
Fig. 2c shows two other states, which are denoted C and D below, of the molecules that resulted from another four pulses applied at the positions marked in Fig. 2b (red crosses). In addition, a few molecules were also switched to state A by those pulses.
The tetra-cyclic unit of state A appears as a rounded trapezium with an apparent height similar to that of pristine molecules. In state B, one end of these rings appears higher than the other with a sharp depression between them. C and D appear quite similar to each other with the cyclohexane ring (red in Fig. 1a) at the far end of the tetra-cyclic unit being slightly higher (∼0.1 nm) in C than in D. The octyl chains remain essentially unchanged in all states. Molecules were found to be stable for hours at ∼5 K in any of these manipulated states, although spontaneous conversions of octyl chains from gauche to anti form (vide supra) were often observed while imaging at slightly elevated voltages |V| > 0.5 V. The substantial increase in apparent height at the far end of the tetra-cyclic units appears to indicate a conformational change of the cyclohexane ring. Although the tetra-cyclic unit is planar, the cyclohexane ring can adopt other conformations in crystal structures.15
Fig. 2d recorded after Fig. 2c demonstrates that the switching may be reversed by additional pulses applied in a manner similar to that of one mentioned before. While state C (Fig. 2c) was switched back to the pristine state, state D was changed to A. Conversely, one of the molecules in state A was switched to state D. Furthermore, another B state was prepared from a pristine molecule (cross mark in Fig. 2c).
Differential conductance (dI/dV) spectra of all states of the molecule are presented in Fig. 2e. Pristine molecules (second spectrum from the bottom) and molecules in state A have featureless spectra in an energy range close to the Fermi level (V = 0). The spectra of the other states surprisingly show a zero-bias resonance. In spatially resolved measurements the resonance was found all over the molecules except the very end of the octyl chains. Slight variations of the resonance-width were observed from different manipulated molecules, presumably due to small differences in their local environments. The reversibility of the switching observed in the STM images in Fig. 2 along with the spectra recorded on a molecule after manipulation (bottom spectrum in Fig. 2e) show that dissociation of the molecule is not the cause of the zero-bias resonance.
The small width of the resonance close to the Fermi level, in particular in state D (full width half maximum ≈12 meV), is difficult to reconcile with an electronic single particle excitation. As an alternative interpretation a low-energy vibrational excitation should be considered.19,20 The asymmetry of the lines, which is particularly obvious in states B and C, however, does not favor such a scenario. We therefore attribute the resonance to a Kondo effect. This is a many-body effect in which a localized spin is screened by the conduction electrons of the substrate. It has been observed from various magnetic atoms and molecules on surfaces.21–28 Organic radicals on surfaces were also shown to exhibit this effect.5,6,29–31 The corresponding line-shape may be described by a Frota function, which has recently been used to model the Kondo fingerprints of a range of systems.32–35 Using a generalized Frota function,34 which allows for an asymmetry parameter, our experimental data is reproduced very well (red curves in Fig. 2e).36
The Kondo effect requires the presence of a localized spin. Indeed, no such effect is observed from pristine cholesterol on Au(111) as expected for a closed-shell molecule. State A may tentatively be attributed to a geometrical isomer because its electronic structure is unchanged in dI/dV spectra over a wider voltage range (−0.5–0.5 V, data not shown). No significant differences between pristine molecules and state A were found in these data. We propose that states B–D are due to the formation of radical cation. The possibility of generating stable radical cations has indeed been reported for organic biomolecules such as retinoic acid,37,38 whereas the creation of an anion may dissociate the molecule through deprotonation.39 A cationic state is also consistent with the experimental observation that these states are obtained upon electron transfer from the molecule to the tip at elevated bias voltage. It may be speculated that the extracted electron stems from the endocyclic double bond, similar to the case of retinoic acid.37,38 At the low temperature of the experiment the cationic state may be stabilized by a concomitant change of the molecular geometry. A closely related mechanism has been reported from charged metal atoms on insulators.40,41
In conclusion, single cholesterol molecules have been reversibly switched on a Au(111) substrate among several states, three of which exhibit a sharp spectroscopic feature at the Fermi level. It is attributed to the creation of a radical cation carrying a localized spin. The multistability of the molecule with paramagnetic and diamagnetic states that may be controllably switched may make it a candidate for spintronic applications. It may also be interesting to look for spin effects in cholesterol in a biochemical environment.
Financial support of the Deutsche Forschungsgemeinschaft (DFG) via Sonderforschungsbereich 677 is acknowledged.
References
- C. Chappert, A. Fert and F. N. Van Dau, Nat. Mater., 2007, 6, 813 CrossRef CAS PubMed.
- D. D. Awshalom and M. M. Flatté, Nat. Phys., 2007, 3, 153 CrossRef.
- T. Sugawara and M. M. Matsushita, J. Mater. Chem., 2009, 19, 1738–1753 RSC.
- S. Pramanik, C.-G. Stefanita, S. Patibandla, S. Bandyopadhyay, K. Garre, N. Harth and M. Cahay, Nature, 2007, 2, 216 CAS.
- J. Liu, H. Isshiki, K. Katoh, T. Morita, B. K. Breedlove, M. Yamashita and T. Komeda, J. Am. Chem. Soc., 2013, 135, 651–658 CrossRef CAS PubMed.
- S. Müllegger, M. Rashidi, M. Fattinger and R. Koch, J. Phys. Chem. C, 2013, 117, 5718–5721 Search PubMed.
- M. Garnica, D. Stradi, S. Barja, F. Calleja, C. Diaz, M. Alcami, N. Martin, A. L. Vázquez de Parga, F. Martin and R. Miranda, Nat. Phys., 2013, 9, 368–374 CrossRef CAS.
- P. L. Yeagle, Biochim. Biophys. Acta, 1985, 882, 267–287 CrossRef.
- D. Needham and R. S. Nunn, Biophys. J., 1990, 58, 997–1009 CrossRef CAS PubMed.
- M. Doxastakis, A. K. Sum and J. J. de Pablo, J. Phys. Chem. B, 2005, 109, 24173–24181 CrossRef CAS PubMed.
- B. M. Craven, Nature, 1976, 260, 727–729 CrossRef CAS PubMed.
- H. S. Shieh, L. G. Hoard and C. E. Nordman, Nature, 1977, 267, 287–289 CrossRef PubMed.
- M. Hibino and H. Tsuchiya, Langmuir, 2014, 30, 6852–6857 CrossRef CAS PubMed.
- S. Sek, S. Xu, M. Chen, G. Szymanski and J. Lipkowski, J. Am. Chem. Soc., 2008, 130, 5736–5743 CrossRef CAS PubMed.
- W. L. Duax, J. F. Griffin, D. C. Rohrer and C. M. Weeks, Lipids, 1980, 15, 783–792 CrossRef CAS PubMed.
- N. Hauptmann, K. Scheil, T. G. Gopakumar, F. L. Otte, C. h. Schütt, R. Herges and R. Berndt, J. Am. Chem. Soc., 2013, 135, 8814–8817 CrossRef CAS PubMed.
- P. Maksymovych, D. B. Dougherty, X.-Y. Zhu and J. T. Yates, Jr., Phys. Rev. Lett., 2007, 99, 016101 CrossRef PubMed.
- T. G. Gopakumar, F. Matino, H. Naggert, A. Bannwarth, F. Tuczek and R. Berndt, Angew. Chem., Int. Ed., 2012, 51, 6262 CrossRef CAS PubMed.
- M. Pivetta, M. Ternes, F. Patthey and W.-D. Schneider, Phys. Rev. Lett., 2007, 99, 126104 CrossRef PubMed.
- W. A. Hofer, G. Teobaldi and N. Lorente, Nanotechnology, 2008, 19, 305701 CrossRef PubMed.
- J. Li, W.-D. Schneider, R. Berndt and B. Delley, Phys. Rev. Lett., 1998, 80, 2893 CrossRef CAS.
- V. Madhavan, W. Chen, T. Jamneala, M. F. Crommie and N. S. Wingreen, Science, 1998, 280, 567 CrossRef CAS PubMed.
- U. G. E. Perera, H. J. Kulik, V. Iancu, L. G. G. V. Dias da Silva, S. E. Ulloa, N. Marzari and S.-W. Hla, Phys. Rev. Lett., 2010, 105, 106601 CrossRef CAS PubMed.
- A. Mugarza, R. Robles, C. Krull, R. Korytár, N. Lorente and P. Gambardella, Phys. Rev. B: Condens. Matter Mater. Phys., 2012, 85, 155437 CrossRef.
- E. Minamitani, N. Tsukahara, D. Matsunaka, Y. Kim, N. Takagi and M. Kawai, Phys. Rev. Lett., 2012, 109, 086602 CrossRef PubMed.
- T. Komeda, H. Isshiki, J. Liu, Y.-F. Zhang, N. Lorente, K. Katoh, B. K. Breedlove and M. Yamashita, Nat. Commun., 2011, 2, 217 CrossRef PubMed.
- A. DiLullo, S.-H. Chang, N. Baadji, K. Clark, J.-P. Klöckner, M.-H. Prosenc, S. Sanvito, R. Wiesendanger, G. Hoffmann and S.-W. Hla, Nano Lett., 2012, 12, 3174 CrossRef CAS PubMed.
- S. Karan, D. Jacob, M. Karolak, C. h. Hamann, Y. Wang, A. Weismann, A. I. Lichtenstein and R. Berndt, Phys. Rev. Lett., 2015, 115, 016802 CrossRef PubMed.
- Y.-h. Zhang, S. Kahle, T. Herden, C. Stroh, M. Mayor, U. Schlickum, M. Ternes, P. Wahl and K. Kern, Nat. Commun., 2013, 4, 2110 Search PubMed.
- R. Requist, S. Modesti, P. P. Baruselli, A. Smogunov, M. Fabrizio and E. Tosatti, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 69–74 CrossRef CAS PubMed.
- T. Choi, S. Bedwani, A. Rochefort, C.-Y. Chen, A. J. Epstein and J. A. Gupta, Nano Lett., 2010, 10, 4175–4180 CrossRef CAS PubMed.
- Y.-S. Fu, S.-H. Ji, X. Chen, X.-C. Ma, R. Wu, C.-C. Wang, W.-H. Duan, X.-H. Qiu, B. Sun, P. Zhang, J.-F. Jia and Q.-K. Xue, Phys. Rev. Lett., 2007, 99, 256601 CrossRef PubMed.
- H. Prüser, M. Wenderoth, P. E. Dargel, A. Weismann, R. Peters, T. Pruschke and R. G. Ulbrich, Nat. Phys., 2011, 7, 203 CrossRef.
- H. Prüser, M. Wenderoth, A. Weismann and R. G. Ulbrich, Phys. Rev. Lett., 2012, 108, 166604 CrossRef PubMed.
- D.-J. Choi, M. V. Rastei, P. Simon and L. Limot, Phys. Rev. Lett., 2012, 108, 266803 CrossRef PubMed.
- From the fits Kondo temperatures of ∼22,∼125, and ∼125 K are obtained for states D, B, and C, respectively. It should be noted that these values may deviate from the actual Kondo temperatures.29.
- K. Li, M. Wang, T. Wang, D. Sun, R. Zhu, X. Sun, X. Wu and S.-L. Wang, Photochem. Photobiol., 2013, 89, 1064–1070 CrossRef CAS PubMed.
- S. Karan, N. Li, Y. Zhang, Y. He, I.-P. Hong, H. Song, J.-T. Lü, Y. Wang, L. Peng, K. Wu, G. S. Michelitsch, R. J. Maurer, K. Diller, K. Reuter, A. Weismann and R. Berndt, Phys. Rev. Lett., 2016, 116, 027201 CrossRef PubMed.
- L. Xu and N. A. Porter, Free Radical Res., 2015, 49, 835–849 CrossRef CAS PubMed.
- W. Steurer, J. Repp, L. Gross, I. Scivetti, M. Persson and G. Meyer, Phys. Rev. Lett., 2015, 114, 036108 CrossRef PubMed.
- M. Sterrer, T. Risse, U. M. Pozzoni, L. Giordano, M. Heyde, H.-P. Rust, G. Pacchioni and H.-J. Freund, Phys. Rev. Lett., 2007, 98, 096107 CrossRef PubMed.
Footnote |
| † Present address: Institut für Experimentelle und Angewandte Physik, Universität Regensburg, 93053 Regensburg, Germany. E-mail: sujoy.karan@physik.uni-regensburg.de |
| This journal is © the Owner Societies 2016 |