Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sequence-specific recognition of methylated DNA by an engineered transcription activator-like effector protein†
Shogo
Tsuji
,
Shiroh
Futaki
and
Miki
Imanishi
*
Institute for Chemical Research, Kyoto University, Uji, Kyoto 611-0011, Japan. E-mail: imiki@scl.kyoto-u.ac.jp
First published on 16th November 2016
Abstract
A 5mC-selective TALE-repeat was created by screening a TALE repeat library containing randomized amino acids at repeat variable diresidues and their neighboring residues. The new repeat showed high 5mC discrimination ability. An artificial TALE containing the new repeat activated an endogenous gene in a genomic methylation status-dependent manner.
DNA methylation is an important epigenetic marker that regulates gene expression, chromatin remodeling, and genome stability.1 DNA methylation status changes dynamically during development, cell cycle, and disease.2–4 In mammals, it occurs mainly at the cytosine base of CpG dinucleotides to produce 5-methylcytosine (5mC). To understand individual biological functions of locus-specific 5mC, many different 5mC detection methods have been developed. However, sequence-specific 5mC detection methods in living cells, which allow us to understand the biological roles of methylation status at individual cytosine residues, are lacking. Bisulfite sequencing is one of the most powerful existing identification methods.5 This method reveals the genomic methylation status at single base resolution, but requires extraction of genomic DNA from cells. Thus, it is not applicable to living cells. Another approach is using anti-5mC antibodies or 5mC binding proteins to examine global DNA methylation levels.6–9 Although these approaches can observe the overall methylation status directly at the cellular level, antibodies or proteins employed in these methods are neither sequence-selective nor contain sequence constraints. Tools that recognize 5mC at a specific site without sequence constraints are needed.
Transcription activator-like effectors (TALEs) have attracted broad attention as designable DNA-binding scaffolds.10–12 Their DNA-binding specificity is determined by a series of tandem repeats of, typically, 34 highly conserved amino acids. Each repeat recognizes one target base. These repeats contain variable diresidues at positions 12 and 13, called repeat variable diresidues (RVD), which define the base preference of a repeat (Fig. 1A). Owing to the simple one-to-one base recognition of each repeat, TALEs can be readily designed to target specific DNA sequences by simply modifying the RVDs. An RVD recognizing 5mC but not C would provide useful TALEs that discriminate methylation status with designable sequence-specificity.
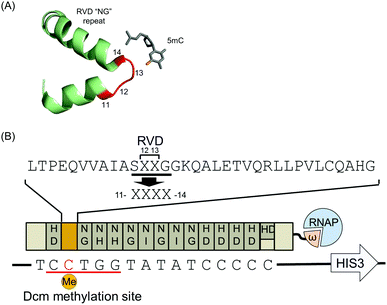 | ||
| Fig. 1 Schematic representation of the bacterial one-hybrid screening for 5mC-specific TALE repeats. (A) DNA recognition mode of a TALE repeat. The structure (PDB: 4GJP) shows an interaction between the RVD “NG” repeat and 5mC. Four amino acid residues (11–14), including RVD, are shown in red. (B) The TALE-ω fusion protein targets the sequence containing Dcm methylated cytosine (red) on the promoter of the HIS3 reporter. The TALE contains 14.5 repeats with RVDs “NG”, “HD”, “NI”, and “NH” for T, C, A, and G recognition, respectively. Target DNA sequences in the reporter vector are aligned to the TALE repeats. Four amino acid residues (11–14) of repeat 2 were randomized and are shown as XXXX. | ||
The commonly used RVD “NG” (Asn-Gly), specific for the thymine nucleobase, also binds to 5mC because of its structural similarity to thymine (Fig. 1A).13,14 Recently, using the RVD “NG”, Kubik et al. showed that TALEs have the potential to differentiate 5mC from C at single base resolution.15–17 However, in these studies, the methylation discrimination ability of TALEs was evaluated only by in vitro analysis. In cells, methylated DNA regions are often bound by 5mC binding proteins and tend to form heterochromatin that may inhibit TALEs from binding. It is necessary to evaluate the function of RVD “NG” in living cells. In addition, it has been reported that the methylation-discrimination ability of RVD “NG” is insufficient to completely regulate TALE binding in a methylation-dependent manner.17 Therefore, we searched for RVDs more specific to 5mC within living cells.
To search for non-native RVDs with the desired base preferences, comprehensive analyses of potential RVDs, which covered all 400 possible combinations of amino acid diresidues, were previously performed toward A, C, G, and T.18–20 However, in those studies, the specificities of those RVDs to 5mC were not evaluated. It is possible that there were RVDs that discriminate methylation of cytosine. On the other hand, in some molecular evolution studies of DNA-binding proteins, modification of residues that do not directly interact with DNA led to improved base preferences of the proteins.21–23 Taking into account these points, artificial TALE repeats with ideal 5mC preferences may be generated by modifying both RVD and their neighboring residues. In the current study, a TALE repeat library containing randomized amino acids at RVD and their neighboring residues was screened for 5mC selectivity, and a highly 5mC-selective repeat was successfully identified.
For the screening of new TALE repeats recognizing 5mC, we developed a modified bacterial one-hybrid (B1H) screening that relied on the Dcm methylation system of E. coli, because of the integrity of the Dcm methylation system. Here, the binding of TALEs fused with the omega subunit of a bacterial RNA polymerase to the reporter vector allows the host E. coli to survive.24 The E. coli Dcm methylation system was used to specifically methylate the cytosine base in the target sequence of the reporter vector. Most E. coli strains contain Dcm methylase that methylates the second cytosine in the sequences CCAGG and CCTGG.25 We designed a TALE that targeted the DNA sequence 5′-T![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TATATCCCCC-3′ containing a Dcm methylation site (underlined) and denoted it TALDcm (Fig. 1B). As expected, the reporter vectors extracted from the selection strain were not cleaved by a methylation-sensitive restriction enzyme, PspGI, targeting the sequence CCTGG (Fig. S1A, ESI†). This suggested that the reporter vectors were methylated appropriately. Previous reports indicated that N-terminal repeats were more sensitive to mismatches.26,27 Therefore, to maximize the effect of the repeat corresponding to 5mC, the Dcm methylation site was placed at the 5′-end of the TALDcm target sequence. To select 5mC-specific TALE repeats, a TALDcm library was generated by randomizing four residues (RVD and their neighboring residues) of repeat 2 that corresponded to Dcm methylated cytosine (Fig. 1B). Subsequent B1H screening gave several TALE repeats, but no specific sequence pattern was identified from the obtained mutants (Fig. S2, ESI†). Therefore, the DNA binding preferences of all mutants were evaluated individually by luciferase reporter assays by expressing a TALDcm-based transcription activator in HeLa cells. As reporter plasmids, 3 × TALDcm binding sites were inserted at the promoter of the luciferase gene, creating 3 × TALDcm/pGL3. The plasmids were prepared using Dcm (−) and Dcm (+) E. coli strains, resulting in an unmethylated and methylated status of the second cytosine bases in the 3 × TALDcm binding sites, respectively (Fig. S1B, ESI†).
TATATCCCCC-3′ containing a Dcm methylation site (underlined) and denoted it TALDcm (Fig. 1B). As expected, the reporter vectors extracted from the selection strain were not cleaved by a methylation-sensitive restriction enzyme, PspGI, targeting the sequence CCTGG (Fig. S1A, ESI†). This suggested that the reporter vectors were methylated appropriately. Previous reports indicated that N-terminal repeats were more sensitive to mismatches.26,27 Therefore, to maximize the effect of the repeat corresponding to 5mC, the Dcm methylation site was placed at the 5′-end of the TALDcm target sequence. To select 5mC-specific TALE repeats, a TALDcm library was generated by randomizing four residues (RVD and their neighboring residues) of repeat 2 that corresponded to Dcm methylated cytosine (Fig. 1B). Subsequent B1H screening gave several TALE repeats, but no specific sequence pattern was identified from the obtained mutants (Fig. S2, ESI†). Therefore, the DNA binding preferences of all mutants were evaluated individually by luciferase reporter assays by expressing a TALDcm-based transcription activator in HeLa cells. As reporter plasmids, 3 × TALDcm binding sites were inserted at the promoter of the luciferase gene, creating 3 × TALDcm/pGL3. The plasmids were prepared using Dcm (−) and Dcm (+) E. coli strains, resulting in an unmethylated and methylated status of the second cytosine bases in the 3 × TALDcm binding sites, respectively (Fig. S1B, ESI†).
Initially, we evaluated the methylation discrimination ability of pre-existing RVDs. As expected, TALDcm with a C-specific RVD “HD” at repeat 2 showed significantly higher activity for C than for 5mC. In contrast, comparable activation levels of the C and 5mC reporters were observed when using RVD “NG” at repeat 2 (Fig. S3A, ESI†). These results are in good accordance with the results of an electrophoretic mobility shift assay (EMSA) (Fig. S3B, ESI†), confirming that the discrimination ability of RVD “NG” is not always sufficiently high.
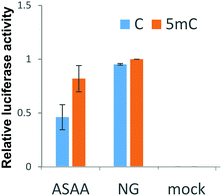
Next, the discrimination ability of all mutants selected from the B1H screening for a methylated cytosine was assessed by luciferase reporter assays. Some mutants showed better discrimination ability than RVD “NG”, but their activation levels were intolerably low (Fig. S4A, ESI†). Intriguingly, the three mutants with “QSAA”, “RNAA”, or “RMAA” repeats, having the consensus sequence “XXAA”, showed relatively high 5mC selectivity. Subsequently, we created an “XXAA” library and screened it by B1H screening. Several of the TALE repeats that were obtained showed a high ability to discriminate 5mC from C. Among them, the “ASAA” repeat showed the highest activity for the 5mC reporter (Fig. 2 and Fig. S4B, ESI†). TALDcm with the “ASAA” repeat activated the luciferase gene in proportion to the methylation percentage of the reporter vectors (Fig. S5, ESI†). This result indicates the methylation-dependent base recognition of the “ASAA” repeat.
The methylation discrimination ability of TALDcm with the “ASAA” repeat in living cells was confirmed by real-time monitoring of luciferase luminescence (Fig. S6, ESI†). At each time point, luciferase activity was always higher in the cells transfected with the methylated compared to the unmethylated reporter. In addition, luciferase activity was greatly reduced for the reporter vector containing mutated TALDcm binding sites, indicating that introduction of the “ASAA” repeat does not impair the overall sequence-specificity of the original TALEs.
EMSAs also supported the methylation-discrimination ability of the “ASAA” repeat (Table 1). Specifically, TALDcm with the “ASAA” repeat showed 1.9-fold stronger binding to 5mC than C, whereas TALDcm with the RVD “NG” repeat showed 1.2-fold stronger binding, although the dissociation constant of TALDcm with the “ASAA” repeat for 5mC was higher than that with the RVD “NG” repeat.
In mammals, cytosine methylation mainly occurs at CpG dinucleotide sites. Therefore, to verify whether the “ASAA” repeat could also recognize 5mC within the CpG context, we designed a TALE that targeted a CpG methylation sequence (Fig. S7A, ESI†). EMSAs showed that the TALE with the “ASAA” repeat had a lower dissociation constant for the 5mC target than for C, while the TALE with the RVD “NG” repeat instead of the “ASAA” repeat had comparable dissociation constants between 5mC and C targets (Table 2). These results indicate that the “ASAA” repeat also preferentially recognizes 5mC within the CpG context, and that the discrimination ability of the “ASAA” repeat is higher than that of the RVD “NG” repeat. Unfortunately, a TALE with two “ASAA” repeats failed to preferentially bind to the target DNA containing two 5mC (Fig. S7B and C, ESI†). This may be because the affinity of the “ASAA” repeat to 5mC is not very strong.
| Repeat 3 | K d (μM) | Relative Kd (C/5mC) | |
|---|---|---|---|
| C | 5mC | ||
| a Determined by EMSA. | |||
| ASAA | 2.3 ± 0.2 | 1.5 ± 0.2 | 1.6 |
| NG | 0.9 ± 0.1 | 0.8 ± 0.1 | 1.1 |
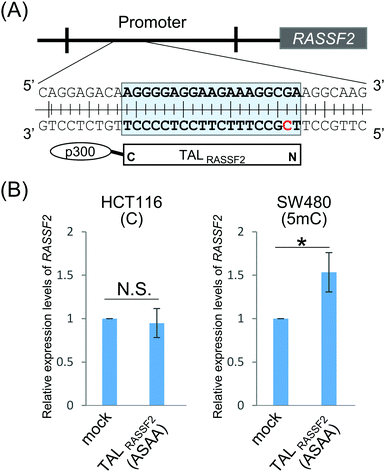
Finally, we explored the ability of the “ASAA” repeat to regulate endogenous gene expression dependent on the methylation status of genomic DNA. A TALE targeting an endogenous gene, Ras association domain-containing protein 2 (RASSF2), was designed and denoted TALRASSF2. RASSF2 works as a tumor suppressor gene and the RASSF2 protein induces apoptosis in tumor cells.28,29 In many colorectal tumor cell lines, the promoter region of RASSF2 is highly methylated and thus RASSF2 hypermethylation is a potential marker for cancer diagnosis.30 SW480 and HCT116 cells were reported to have different RASSF2 methylation statuses.28 Bisulfite sequencing showed that RASSF2 was highly methylated in SW480 but not in HCT116 cells (Fig. S8, ESI†). TALRASSF2, which targets the RASSF2 promoter region including CpG dinucleotides, was fused to a transcription activator, p300 histone acetyltransferase, to activate RASSF2 by binding TALRASSF2 to the target genomic region (Fig. 3A). TALRASSF2-p300 was expressed in SW480 and HCT116 cells, and the expression levels of RASSF2 mRNA were evaluated by RT-qPCR. When using the “ASAA” repeat, significant gene activation was induced only in SW480 cells that had a highly methylated RASSF2 promoter (Fig. 3B). This result was not due to differences of cell types or chromatin states because TALRASSF2 with the RVD “NG” repeat instead of the “ASAA” repeat activated the gene in both cell types (Fig. S9, ESI†). Therefore, the result suggested that the “ASAA” repeat contributed to the selective binding to 5mC.
In conclusion, we successfully created a highly 5mC-selective TALE repeat by Dcm methylation-dependent B1H screening of the TALE library. The new repeat showed high 5mC discrimination ability even for genomic DNA. One reason for the successful screening targeting 5mC may be the expanded randomization strategy. Although further improvement of the affinity is desired, the new repeat enabled us to use TALEs as an easily designable tool to detect 5mC at user-defined sites in living cells. For example, fluorescently labeled TALE has been used to visualize endogenous sequences to study chromatin dynamics.31,32 Using 5mC-selective TALEs, time-lapse observations of the methylation status at specific sites can be realized. Furthermore, TALEs have been used as artificial transcriptional regulators, nucleases, and epigenetic modulators.33–35 Thus, methylation status-dependent gene regulation is possible using the 5mC-selective TALE repeat in contrast to the existing 5mC identification methods. In addition, there are many kinds of modified nuclear bases besides 5mC.36,37 Our strategy to obtain new functional TALE repeats is applicable to other modified nuclear bases. This study should provide new ways of exploring the biological functions of 5mC and other modified nuclear bases.
We thank Feng Zhang for the plasmids used to construct the TALEs, Warner Greene for plasmids, Scot Wolfe for plasmids and cells for B1H screening, and Hiromu Suzuki for the SW480 and HCT116 cells. This work was supported in part by JSPS KAKENHI 16H03281 (M. I.) and 15J09770 (S. T.), JST CREST and the Naito Foundation.
Notes and references
- J. A. Law and S. E. Jacobsen, Nat. Rev. Genet., 2010, 11, 204 CrossRef CAS PubMed.
- Z. D. Smith and A. Meissner, Nat. Rev. Genet., 2013, 14, 204 CrossRef CAS PubMed.
- S. E. Brown, M. F. Fraga, I. C. G. Weaver, M. Berdasco and M. Szyf, Epigenetics, 2007, 2, 54 CrossRef PubMed.
- G. Egger, G. Liang, A. Aparicio and P. A. Jones, Nature, 2004, 429, 457 CrossRef CAS PubMed.
- S. J. Cokus, S. Feng, X. Zhang, Z. Chen, B. Merriman, C. D. Haudenschild, S. Pradhan, S. F. Nelson, M. Pellegrini and S. E. Jacobsen, Nature, 2008, 452, 215 CrossRef CAS PubMed.
- F. Santos, B. Hendrich, W. Reik and W. Dean, Dev. Biol., 2002, 241, 172 CrossRef CAS PubMed.
- S. Kobayakawa, K. Miike, M. Nakao and K. Abe, Genes Cells, 2007, 12, 447 CrossRef CAS PubMed.
- C. Desjobert, M. E. Maï, T. Gérard-Hirne, D. Guianvarc'h, A. Carrier, C. Pottier, P. B. Arimondo and J. Riond, Epigenetics, 2015, 10, 82 CrossRef PubMed.
- S. Çelik-Uzuner, Y. Li, L. Peters and C. O'Neill, In Vitro Cell. Dev. Biol.: Anim. DOI:10.1007/s11626-016-0075-4.
- J. Boch, H. Scholze, S. Schornack, A. Landgraf, S. Hahn, S. Kay, T. Lahaye, A. Nickstadt and U. Bonas, Science, 2009, 326, 1509 CrossRef CAS PubMed.
- A. J. Bogdanove and D. F. Voytas, Science, 2011, 333, 1843 CrossRef CAS PubMed.
- T. Gaj, C. A. Gersbach and C. F. Barbas, Trends Biotechnol., 2013, 31, 397 CrossRef CAS PubMed.
- D. Deng, P. Yin, C. Yan, X. Pan, X. Gong, S. Qi, T. Xie, M. Mahfouz, J. K. Zhu, N. Yan and Y. Shi, Cell Res., 2012, 22, 1502 CrossRef CAS PubMed.
- J. Valton, A. Dupuy, F. Daboussi, S. Thomas, A. Marechal, R. Macmaster, K. Melliand, A. Juillerat and P. Duchateau, J. Biol. Chem., 2012, 287, 38427 CrossRef CAS PubMed.
- G. Kubik, M. J. Schmidt, J. E. Penner and D. Summerer, Angew. Chem., Int. Ed., 2014, 53, 6002 CrossRef CAS PubMed.
- G. Kubik and D. Summerer, ChemBioChem, 2015, 16, 228 CrossRef CAS PubMed.
- G. Kubik, S. Batke and D. Summerer, J. Am. Chem. Soc., 2015, 137, 2 CrossRef CAS PubMed.
- J. Yang, Y. Zhang, P. Yuan, Y. Zhou, C. Cai, Q. Ren, D. Wen, C. Chu, H. Qi and W. Wei, Cell Res., 2014, 24, 628 CrossRef CAS PubMed.
- A. Juillerat, C. Pessereau, G. Dubois, V. Guyot, A. Marechal, J. Valton, F. Daboussi, L. Poirot, A. Duclert and P. Duchateau, Sci. Rep., 2015, 5, 8150 CrossRef CAS PubMed.
- J. C. Miller, L. Zhang, D. F. Xia, J. J. Campo, I. V. Ankoudinova, D. Y. Guschin, J. E. Babiarz, X. Meng, S. J. Hinkley, S. C. Lam, D. E. Paschon, A. I. Vincent, G. P. Dulay, K. A. Barlow, D. A. Shivak, E. Leung, J. D. Kim, R. Amora, F. D. Urnov, P. D. Gregory and E. J. Rebar, Nat. Methods, 2015, 12, 465 CrossRef CAS PubMed.
- S. Tsuji, S. Futaki and M. Imanishi, Biochem. Biophys. Res. Commun., 2013, 441, 26 CrossRef PubMed.
- B. M. Lamb, A. C. Mercer and C. F. Barbas, Nucleic Acids Res., 2013, 41, 9779 CrossRef CAS PubMed.
- B. P. Hubbard, A. H. Badran, J. A. Zuris, J. P. Guilinger, K. M. Davis, L. Chen, S. Q. Tsai, J. D. Sander, J. K. Joung and D. R. Liu, Nat. Methods, 2015, 12, 939 CrossRef CAS PubMed.
- M. B. Noyes, X. Meng, A. Wakabayashi, S. Sinha, M. H. Brodsky and S. A. Wolfe, Nucleic Acids Res., 2008, 36, 2547 CrossRef CAS PubMed.
- B. R. Palmer and M. G. Marinus, Gene, 1994, 143, 1 CrossRef CAS PubMed.
- J. F. Meckler, M. S. Bhakta, M.-S. Kim, R. Ovadia, C. H. Habrian, A. Zykovich, A. Yu, S. H. Lockwood, R. Morbitzer, J. Elsaesser, T. Lahaye, D. J. Segal and E. P. Baldwin, Nucleic Acids Res., 2013, 41, 4118 CrossRef CAS PubMed.
- A. Juillerat, G. Dubois, J. Valton, S. Thomas, S. Stella, A. Marechal, S. Langevin, N. Benomari, C. Bertonati, G. H. Silva, F. Daboussi, J. C. Epinat, G. Montoya, A. Duclert and P. Duchateau, Nucleic Acids Res., 2014, 42, 5390 CrossRef CAS PubMed.
- K. Akino, M. Toyota, H. Suzuki, H. Mita, Y. Sasaki, M. Ohe-Toyota, J. P. J. Issa, Y. Hinoda, K. Imai and T. Tokino, Gastroenterology, 2005, 129, 156 CrossRef CAS.
- W. N. Cooper, L. B. Hesson, D. Matallanas, A. Dallol, A. von Kriegsheim, R. Ward, W. Kolch and F. Latif, Oncogene, 2009, 28, 2988 CrossRef CAS PubMed.
- J. Shi, G. Zhang, D. Yao, W. Liu, N. Wang, M. Ji, N. He, B. Shi and P. Hou, Am. J. Cancer Res., 2012, 2, 116 CAS.
- Y. Miyanari, C. Z. Birling and M. E. T. Padilla, Nat. Struct. Mol. Biol., 2013, 20, 1321 CAS.
- H. Ma, P. Reyes-Gutierrez and T. Pederson, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 21048 CrossRef CAS PubMed.
- J. Hu, Y. Lei, W.-K. Wong, S. Liu, K.-C. Lee, X. He, W. You, R. Zhou, J.-T. Guo, X. Chen, X. Peng, H. Sun, H. Huang, H. Zhao and B. Feng, Nucleic Acids Res., 2014, 42, 4375 CrossRef CAS PubMed.
- V. M. Bedell, Y. Wang, J. M. Campbell, T. L. Poshusta, C. G. Starker, R. G. Krug, W. Tan, S. G. Penheiter, A. C. Ma, A. Y. H. Leung, S. C. Fahrenkrug, D. F. Carlson, D. F. Voytas, K. J. Clark, J. J. Essner and S. C. Ekker, Nature, 2012, 491, 114 CrossRef CAS PubMed.
- M. L. Maeder, J. F. Angstman, M. E. Richardson, S. J. Linder, V. M. Cascio, S. Q. Tsai, Q. H. Ho, J. D. Sander, D. Reyon, B. E. Bernstein, J. F. Costello, M. F. Wilkinson and J. K. Joung, Nat. Biotechnol., 2013, 31, 1137 CrossRef CAS PubMed.
- M. Tahiliani, K. P. Koh, Y. Shen, W. A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L. M. Iyer, D. R. Liu, L. Aravind and A. Rao, Science, 2009, 324, 930 CrossRef CAS PubMed.
- S. Ito, L. Shen, Q. Dai, S. C. Wu, L. B. Collins, J. A. Swenberg, C. He and Y. Zhang, Science, 2011, 333, 1300 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6cc06824c |
| This journal is © The Royal Society of Chemistry 2016 |