Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A capillary-based multiplexed isothermal nucleic acid-based test for sexually transmitted diseases in patients†
Gaolian
Xu‡
,
Hang
Zhao‡
,
Jonathan M.
Cooper
and
Julien
Reboud
 *
*
Division of Biomedical Engineering, School of Engineering, University of Glasgow, Oakfield Avenue, Rankine Building, G12 8LT Glasgow, UK. E-mail: Julien.Reboud@glasgow.ac.uk
First published on 8th September 2016
Abstract
We demonstrate a multiplexed loop mediated isothermal amplification (LAMP) assay for infectious disease diagnostics, where the analytical process flow of target pathogens genomic DNA is performed manually by moving magnetic beads through a series of plugs in a capillary. Heat is provided by a water bath and the results are read by the naked eye, enabling applications in low resource settings.
Recently, rapid, high performance nucleic acid testing (NAT) based diagnostic technologies have shown the ability to detect infections early.1 However, the existing technologies for the diagnosis of sexually transmitted diseases, such as chlamydia and gonorrhoea, suffer from limitations in speed, cost, and the significant user expertise required.1 The most widely used technology is based on the polymerase chain reaction (PCR), which attains high performance, but is difficult to carry out in resource limited areas.1 As an alternative, low cost nucleic acid based assays using isothermal amplification have the ability to provide platforms for point-of-care (POC) diagnostics in both low and high resource settings, due to their high sensitivities and specificities.2
Isothermal amplifications, such as the loop-mediated systems (LAMP), also offer the capability of multiplexing,3,4 which in turn can be used to inform treatment in e.g. infectious diseases (where the relative sensitivities to specific drug regimes are often determined genetically). Here we focus on detecting two sexually-transmitted pathogens, Chlamydia trachomatis and Neisseria gonorrhoea, which are prevalent in both high as well as low and middle income countries (LMICs).5 The LAMP reaction generally requires six DNA primers for each target to be detected, limiting the possibilities of simultaneously amplifying multiple targets in single reactions, due to the high level of potential cross-talk between the reactions. This constraint has restricted the development of multiplexing POC devices, since adding independent reactions to a single device in turn increases practical challenges linked to reagent mixing, analysis and temperature control.
Here we show that by exploiting capillary-based platforms for multiplexing and in line sample processing,6–8 through the use of plugs separated by an immiscible phase, we are able to demonstrate a powerless, low-cost, easy-to-use sample-to-answer multiplexed LAMP assay. Our new platform circumvents the requirements for external fluid actuation and electronic thermal control systems, potentially facilitating its deployment as a POC device in resource-limited settings.
The advantages of using capillary-based platforms, such as ease of use and multiplexing DNA amplification reactions of extracted samples, have been realised previously,6 while single target DNA extraction was successfully demonstrated,7 and DNA lysis incorporated only recently within a similar format using a FTA card (Flinders Technology Associates), but requiring multiple pipetting steps.8 In this paper, we now go beyond the current state of the art and show the complete nucleic-acid processing (from crude samples to the answer) in a multiplexed and easy-to-use format.
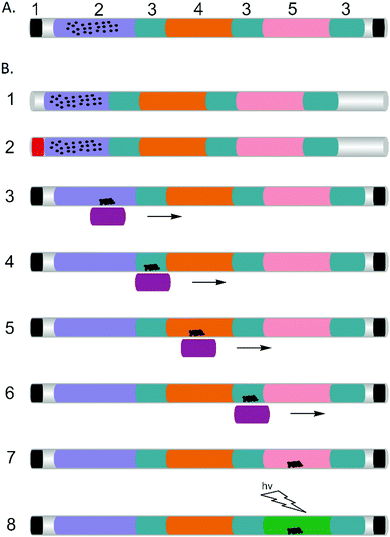
To achieve this, we integrated three different capabilities into the capillary-based system, namely: manual sample manipulation with magnetic beads to extract DNA from complex samples; tolerance to temperature variability (enabling the use of heated water to drive the reaction); and a visual detection of a calcein fluorescent probe on the amplified product, illuminated by a UV flashlight.9
Previously, magnetic beads have been integrated within microfluidic devices for DNA separation and purification,10,11 providing a simple manual method for collection of bound probes or targets without the need for centrifugation or filtration and with no requirement for external power.12 They also advantageously replace previously used FTA cards,8 which are difficult to handle and limit the applicability of previously developed tip-based systems to trained experts. In the assay configuration that we describe, the magnetic beads served as solid carriers for DNA (Fig. 1). By moving a handheld magnet, the DNA passed through a series of discrete process steps, defined by a series of plugs within the capillary, which enabled us to purify and enrich the sample, removing any inhibitors prior to the amplification reaction. The series of plugs simply comprised different aqueous phases separated by mineral oil,13 which also served to prevent evaporation during the amplification step at 60 °C. Multiplexing was enabled by sequentially adding separate plugs containing different reaction mixtures specific to each of the targets.
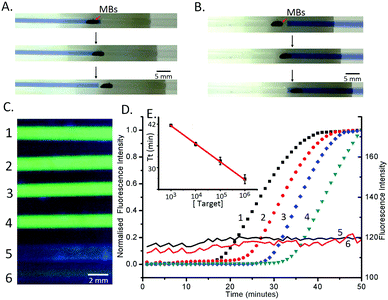
The whole process was completed within 60 min. 1 μl of sample (in this case a model target DNA, at different concentrations in PBS) was manually pipetted into the pre-filled capillary (Fig. 1 and Experimental methods in the ESI†), where it was mixed with lysis buffer, which also contained the magnetic beads. Epoxy was used to seal the ends of the capillary to prevent spillage and the capillary was left at room temperature for 3 minutes to lyse the cells and capture the DNA on the beads (in a low pH and high ionic strength solution). The beads (carrying the DNA) were then subsequently moved through the mineral oil separator into the washing section (Fig. 1B(3–4) and 2A), where they were held for 1 min to remove residues and contaminants from the beads, while retaining the DNA.14 A single wash step was sufficient to purify the DNA to a level that enabled the LAMP reactions to proceed efficiently. The magnet was moved further along the capillary, through an oil separator into the LAMP reaction mixture, where the amplification reaction was performed at 60 °C for 50 minutes, using a water bath as the heat source. This “plug” also served as the elution section, where the DNA was released from the beads and detected using a hand-held UV-flashlight to reveal green fluorescence.15
The results of the LAMP amplification were directly read out visually, and we established a limit of detection of the capillary-based assay by processing ten-fold serially diluted cultured E. coli (Promega), which contained the conserved fragment of C. trachomatis at 2.5 × 103 bacteria per ml (Fig. 2C and D), corresponding to 2.5 copies of the plasmid in 1 μl. This approach was validated using a real-time (RT) amplification approach, where the signal was quantified using fluorescence microscopy4 (Fig. 2D). As the target concentration increased, the exponential phase of signal enhancement started earlier, from ca. 18 minutes for 2.5 × 106 bacteria per ml to 33 minutes for 2.5 × 103 bacteria per ml.
To assess the efficiency of the capillary system, we used the threshold time (Tt) (Fig. 2E), as the reaction time for the fluorescence signal to reach 50% of the maximum,4 which shows a linear decrease with increasing target concentration (on a logarithmic scale). The value of Tt is important as it enables us to quantify the original amount of target DNA present in the reaction after calibration, opening up applications in optimising medical treatment such as those based on viral load quantification.16
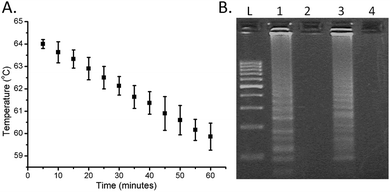
Contrary to more complex DNA amplification techniques, such as PCR, LAMP does not require precise thermal control or cycling of the reaction (previously LAMP has been implemented using pocket warmers6 and thermos containers17). Here we simply poured hot water (ca. 300 ml at 64 °C) into a thermally-insulated polystyrene foam container providing the required stability over 1 h, with the temperature only cooling down to 60 °C over this period (Fig. 3A). The only external energy required for the LAMP was the warming of the water, which can be easily performed even in remote locations. The functionality of the approach was compared with a controlled incubation at 63 °C, using gel electrophoresis analysis of the amplified products (see the ESI† for Experimental details), and showed no appreciable difference (Fig. 3B).
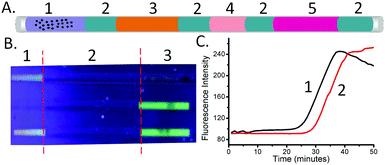
To demonstrate the multiplexed capability of our capillary platform, an additional LAMP reaction mixture plug targeted at the detection of N. gonorrhoeae (see Methods in the ESI† for its composition) was added after the plug targeting C. trachomatis, into a single capillary, separated by mineral oil. Due to the sequential nature of the elution in the two plugs dividing the DNA contents, the volume of the second plug (N. gonorrhoeae) was reduced to 3 μl, compared to that of the first one (C. trachomatis – 5 μl) (Fig. 4A). After amplification, the fluorescence signal in the two sections of each capillary indicated the presence of the targeted sequence of each pathogen simultaneously (Fig. 4B).
Finally, we further demonstrated the applicability of the technique in 6 patient samples, obtained as swabs and extracted as part of routine clinical diagnostics at the NHS West of Scotland Specialist Virology Centre. The results (yes/no indication of the presence of the pathogens) were consistent with those obtained using the benchmark assay real-time PCR (Table S1 and ESI†).
Using the ability of the real-time LAMP reaction to quantify the initial DNA concentrations (the amount of DNA present after extraction, before amplification – see Fig. 2), we also explored the influence of the retention time on the quantity of eluted DNA. When the beads were left in the first plug (4 in Fig. 4A), without moving the beads to the second LAMP plug (through the oil plug into 5 in Fig. 4A), the value of Tt was 30.5 ± 1.0 minutes. When the beads were moved through the first plug into the mineral oil, the Tt value for the LAMP mix of the first plug was 35.5 ± 0.5 minute. Using the equation derived from Fig. 2C, we calculated that the recovery of DNA when the beads only pass through the plug of LAMP reaction mix (4 in Fig. 4A) was 15% ± 5% of that when the beads are left in the plug.
In conclusion, in this study, we have reported a capillary-based, magnetically-actuated, multiplexed LAMP assay platform for the rapid diagnosis of C. trachomatis and N. gonorrhoea. Its simplicity, low-cost and low energy requirement (only a tea cup volume of hot water required) holds significant promise for its application in low resource settings.
This work was supported by a College of Science and Engineering Studentship (GX, Glasgow, UK), a Lord Kelvin and Adam Smith Research Fellowship (JR, Glasgow, UK), an EPSRC fellowship (JC, EP/K027611/1), and an ERC Advanced Investigator Award (JC, 340117). All the original data related to this article are within the depository of the University of Glasgow with http://dx.doi.org/10.5525/gla.researchdata.349. Additional data related to this paper may be requested from the authors.
Notes and references
- R. McNerney and P. Daley, Nat. Rev. Microbiol., 2011, 9, 204–213 CrossRef CAS PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- X. Fang, Y. Liu, J. Kong and X. Jiang, Anal. Chem., 2010, 82, 3002–3006 CrossRef CAS PubMed.
- G. Xu, R. N. Gunson, J. M. Cooper and J. Reboud, Chem. Commun., 2015, 51, 2589–2592 RSC.
- World Health Organization, WHO | Report on global sexually transmitted infection surveillance 2013, 2014.
- Y. Zhang, L. Zhang, J. Sun, Y. Liu, X. Ma, S. Cui, L. Ma, J. J. Xi and X. Jiang, Anal. Chem., 2014, 86, 7057–7062 CrossRef CAS PubMed.
- D. Liu, G. Liang, Q. Zhang and B. Chen, Anal. Chem., 2013, 85, 4698–4704 CrossRef CAS PubMed.
- W. Lu, J. Wang, Q. Wu, J. Sun, Y. Chen, L. Zhang, C. Zheng, W. Gao, Y. Liu and X. Jiang, Biosens. Bioelectron., 2016, 75, 28–33 CrossRef CAS PubMed.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Nat. Protoc., 2008, 3, 877–882 CrossRef CAS PubMed.
- B.-I. Haukanes and C. Kvam, Biotechnology, 1993, 11, 60–63 CrossRef CAS PubMed.
- S. J. Reinholt and A. J. Baeumner, Angew. Chem., Int. Ed., 2014, 53, 13988–14001 CrossRef CAS PubMed.
- J. Pipper, M. Inoue, L. F.-P. Ng, P. Neuzil, Y. Zhang and L. Novak, Nat. Med., 2007, 13, 1259–1263 CrossRef CAS PubMed.
- N. M. Adams, A. E. Creecy, C. E. Majors, B. A. Wariso, P. A. Short, D. W. Wright and F. R. Haselton, Biomicrofluidics, 2013, 7, 014104 Search PubMed.
- J. Wen, L. A. Legendre, J. M. Bienvenue and J. P. Landers, Anal. Chem., 2008, 80, 6472–6479 CrossRef CAS PubMed.
- A. Creecy, P. K. Russ, F. Solinas, D. W. Wright and F. R. Haselton, PLoS One, 2015, 10, e0130260 Search PubMed.
- F. Ahmad, G. Seyrig, D. Tourlousse, R. Stedtfeld, J. Tiedje and S. Hashsham, Biomed. Microdevices, 2011, 13, 929–937 CrossRef CAS PubMed.
- P. LaBarre, K. R. Hawkins, J. Gerlach, J. Wilmoth, A. Beddoe, J. Singleton, D. Boyle and B. Weigl, PLoS One, 2011, 6, e19738 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional methods and experimental sections. See DOI: 10.1039/c6cc05679b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |