Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Broadening substrate specificity of a chain-extending ketosynthase through a single active-site mutation†
Annabel C.
Murphy
*a,
Hui
Hong
a,
Steve
Vance
ab,
R. William
Broadhurst
a and
Peter F.
Leadlay
*a
aDepartment of Biochemistry, University of Cambridge, Sanger Building, 80 Tennis Court Road, Cambridge CB2 1GA, UK. E-mail: acm95@cam.ac.uk; pfl10@cam.ac.uk
bCrescendo Biologics Ltd, Meditrina Building 260, Babraham Research Campus, Cambridge CB22 3AT, UK
First published on 1st June 2016
Abstract
An in vitro model system based on a ketosynthase domain of the erythromycin polyketide synthase was used to probe the apparent substrate tolerance of ketosynthase domains of the mycolactone polyketide synthase. A specific residue change was identified that led to an emphatic increase in turnover of a range of substrates.
The manipulation of biosynthetic pathways provides a useful source of novel analogues of pharmaceutically-important, complex natural products,1 as well as cost-effective and sustainable routes to known compounds.2–4 Modular type I polyketide biosynthesis in particular provides an attractive platform for producing rationally engineered biocatalysts that can generate organic molecules of specified shape and size.5
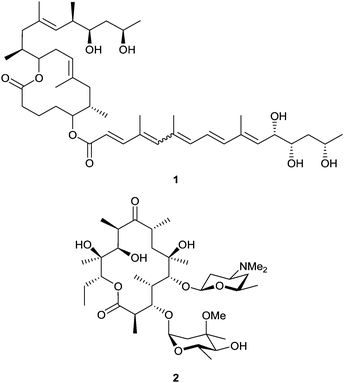
In the biosynthesis of complex reduced polyketides, such as mycolactone (1) and erythromycin A (2) (Fig. 1), by actinomycete bacteria, carbon chains of specific length are produced from small monomers on assembly-line modular polyketide synthase (PKS) multienzymes, each successive module catalysing a different cycle of chain extension.6,7 The key step of carbon–carbon bond formation involves a thioester-templated Claisen condensation reaction, catalyzed by a ketosynthase (KS), between the growing chain tethered to the KS active site, and a chain-extending monomer borne on an acyl carrier protein (ACP) domain. Selection and loading of the chain-extending monomer is carried out by an acyltransferase (AT) domain. The β-ketoacyl-ACP intermediate resulting from condensation may undergo reduction before transfer to the next module, the degree of reduction depending on the presence of ketoreductase, dehydratase and enoyl reductase enzymes in each respective module. Many examples of domain or module replacement, insertion and deletion to give functional chimæric PKSs have been described.8 Unfortunately these hybrid modular PKSs are often much less efficient than the parent native PKS. Early model studies showed that KSs have some intrinsic tolerance for different substrates.9,10 However, the inherent substrate specificity of the KS in the adjacent downstream module may often limit the activity of hybrid modular PKS systems.11–13 The role of KS specificity in assembly-line polyketide biosynthesis has been previously explored in trans-AT PKSs, which lack in-built AT domains in each module. Bio-informatic analysis of their KS sequences has shown that they form distinct clades corresponding to the chemistry of their substrates.14 This structure-selectivity correlation, which has been confirmed by in vitro functional analysis,15–18 implies that native trans-AT KS domains may be generally poor catalysts for extension of non-natural chains. In contrast, the sequences of KS domains from a given cis-AT PKS tend to form a single clade, irrespective of the chemical nature of the substrate at each stage of elongation.14 Although apparently more promising as catalysts operating within hybrid PKS assemblies, the determinants of KS active site specificity in cis-AT systems remain rather poorly understood.8 A better understanding of individual structural features that determine specificity would greatly assist the re-engineering of chimæric systems to improve function.
A valuable framework for detailed analysis of KS specificity has been provided by X-ray crystal structures determined for several KS domains.16,19–22 Structure-based sequence alignment of both cis-AT and trans-AT KS domains has revealed three variable regions within KS domains; a “clasping loop”, a “dimer interface loop” and an “active site cap” (Fig. S1, ESI†).21 In an earlier comparison of the erythromycin PKS (6-deoxyerythronolide B synthase, DEBS) KS5AT5 and KS3AT3 structures,20 the difference in conformation adopted by the dimer interface loop was similarly suggested to contribute to the different substrate specificity of KS3 and KS5. Roles in determining substrate specificity in trans-AT KS domains have been plausibly assigned to the amino acid residue immediately N-terminal of the essential active site Cys,15,17,18 but there have been no equivalent successes for the KS domains of cis-AT PKSs. Also, at the outset of our work there was no evidence that engineered sequence changes in cis-AT KSs might improve tolerance for non-natural acyl-ACP substrates.
A new approach to this question is suggested by the existence of a natural cis-AT PKS showing an exceptionally high level of sequence identity (>97%) between KS domains in different extension modules, even though the KSs act on substrates that vary significantly in chain length and chemical functionality. This is the PKS responsible for generating mycolactone (1) in Mycobacterium ulcerans, the causative agent of Buruli ulcer.23–25 This multienzyme appears to offer a unique natural example of broad KS substrate tolerance. In principle, mycolactone modules might serve as universal building blocks in combinatorial polyketide biosynthesis. Unfortunately, the slow-growing mycolactone producer is intractable for genetic manipulation,26 and the PKS is not active upon heterologous expression.27 Insertion of a myc KS domain into a heterologous PKS assembly line to replace the resident KS is not expected to be effective, due to deleterious effects on KS:AT and KS:ACP protein:protein interfaces.
To learn from the remarkable tolerance of myc KS domains, we instead analysed in silico the active site differences between modelled mycolactone KS domains and the experimentally-determined crystal structure of the DEBS KS3AT3 didomain (EryKS3AT3).20 Guided by this comparison, we then replaced specific amino acids in the EryKS3 active site by their mycolactone KS counterparts, and determined the condensation activity of each mutant KS3 enzyme towards a panel of surrogate thioester substrates in vitro. We report here that although most of the mutants conserved the catalytic properties of the parent enzyme, the replacement of a specific alanine residue by tryptophan markedly improved both catalytic turnover and the ability of the enzyme to act on non-natural substrates, encouraging the view that the performance of chimæric cis-AT PKS multienzymes may be improved by active-site engineering.
In silico comparison between the KS domains of the mycolactone PKS and the EryKS3AT3 crystal structure: an in silico model of the KS domain from MlsA2 was generated using Phyre229 and compared with the EryKS3AT3 didomain crystal structure,20 to identify amino acid residues likely to define the extended substrate binding pocket of the KS domain. The catalytically essential residues Cys202, His337 and His377 (EryKS3 domain numbering) occupy the same positions as their MlsA2 counterparts. However, seven EryKS3 residues (Ala154, Lys155, Phe156, Val173, Ala230, Phe263 and Phe265) are replaced by other amino acids in MlsA2 KS (Fig. S2, ESI†). Except for Ala154 and Phe265, the substitutions are the same in all 16 mycolactone KS domains (Lys155Ala, Phe156Glu, Val173Met, Ala230Thr, and Phe263Thr). Ala154 is replaced either by Gly or Trp, and Phe265 is either conserved or replaced by Trp. These seven residues were therefore selected for mutagenesis of the EryKS3 domain, to determine their potential role in promoting broader substrate specificity.
To gain insight into specificity determinants in cis-AT KS domains, we used a sequence alignment of 199 domains to compute the sequence variability at a given position of a cis-AT KS domain, expressed as the percentage of sequences where the consensus residue is present (Fig. S3 and S8, ESI†). This showed that for cis-AT KS domains the “dimer-interface loop” is the most prominently variable. Residues Ala154, Lys155, Phe156 chosen for mutagenesis in this study reside within this loop.
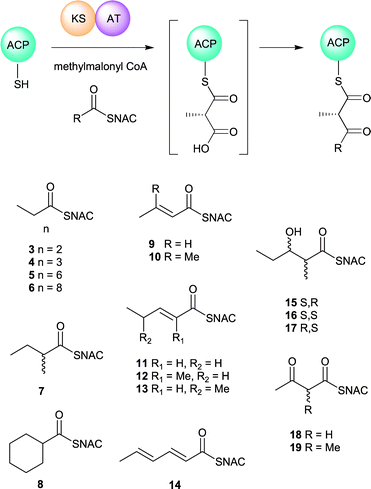
Mass spectrometric assay for in vitro ketosynthase activity: the catalytic competence of recombinant EryKS3AT3 has previously been monitored using a radio-TLC based assay involving radiolabelled N-acetylcysteamine (NAC) thioester 15.30 More recent studies on trans-AT KS domains have successfully used mass spectrometry to assay both self-acylation of the KS15,16 and subsequent Claisen condensation.17 We also chose a mass spectrometric method to directly monitor the formation of ketide-ACP condensation products, starting from an acyl-NAC thioester, recombinant EryACP3 and either methylmalonyl- or malonyl-CoA. Acyl-NAC thioesters are convenient surrogate substrates for EryKS3AT3 even though where Km values have been reported they are 2–3 orders of magnitude higher than those of the corresponding acyl-ACP substrates.10 First, we confirmed that when diketide NAC-thioester 15 was incubated with methylmalonyl-EryACP3 in the presence of EryKS3AT3, as described in the ESI,† a new acyl-EryACP3 species was formed whose mass corresponded to that of the expected Claisen condensation product. A panel of acyl-NAC thioesters was then synthesised (Fig. 2) and assayed as substrates for Claisen condensation by KS3. This panel included acyl-thioesters of varying chain length; acyl-thioesters containing each of the functional groups routinely encountered during polyketide biosynthesis (β-keto-, β-hydroxy- and 2-enoyl-thioesters); and acyl-thioesters bearing an alkyl branch. For each substrate, the percentage of ACP bearing the respective ketide condensation product after 1 h of incubation was measured (Table 1). Acyl transfer from SNAC thioesters to holo-EryACP3 was an observable background reaction, but this was not accelerated by the presence of EryKS3AT3, and is presumably the result of direct chemical thioester–thiol exchange. Substrates featuring α,β-unsubstituted thioesters were also susceptible to side reactions involving 1,4-conjugate addition of phosphopantetheine. Of the substrates tested, as well as the diketide thioester 15, the alkanoyl thioesters 3 and 4, the C-2 branched thioesters 7 and 8, and the 2,3-unsaturated thioester 10 yielded modest amounts (2–6%) of β-ketoacyl-ACP species.
| EryKS3AT3 | A154G | A154W | K155A | F156Q | V173M | A230T | F263T | F265W | |
|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 4 | 46 | 3 | T | 6 | 4 | T | |
| 4 | 3 | 6 | 83 | 6 | 7 | 4 | 2 | 23 | |
| 5 | 77 | T | 3 | T | |||||
| 6 | 13 | ||||||||
| 7 | 1 | 5 | 20 | 2 | T | T | T | T | |
| 8 | 2 | 2 | 72 | T | 5 | 2 | 1 | 4 | |
| 9 | 2 | ||||||||
| 10 | 6 | 17 | 91 | 16 | 18 | 11 | 8 | 10 | |
| 11 | 1 | 5 | |||||||
| 12 | 2 | ||||||||
| 13 | 24 | ||||||||
| 14 | 33 | ||||||||
| 15 | 2 | 3 | 7 | 5 | 2 | 2 | T | 4 | |
| 16 | |||||||||
| 17 | T | T | T | ||||||
| 18 | |||||||||
| 19 |
Different mycolactone KS domains efficiently carry out condensation with either malonyl- or methylmalonyl-ACP as extender units25 so we also sought to evaluate the effect of the chosen active site mutations on the ability of EryKS3AT3 to catalyze condensation with malonyl-ACP. In fact, even native EryKS3AT3 has been reported to catalyse condensation with malonyl-ACP as the extender unit in vitro, albeit less efficiently than with the natural methylmalonyl-ACP substrate.30 The malonyl-CoA-specific AT from MLS module 9 was cloned and expressed, and used to prepare malonyl-ACP3 (see Fig. S7, ESI†) in situ from holo-EryACP3. EryKS3AT3 and a NAC thioester substrate were added to initiate condensation. Wild type KS3AT3 accepted the same NAC thioester substrates and gave similar yields of condensation product from both malonyl- and methylmalonyl-ACP (see Table S4, ESI†). These results show that the KS3 does not discriminate between these two ACP bound extender units, consistent with previous studies.31
The point mutation Ala154Trp dramatically improves in vitro ketosynthase condensing activity against several substrates: having established the reactivity of wild type EryKS3AT3 with this range of NAC thioester substrates and with two alternative extender units, the mutant EryKS3AT3 enzymes were tested using the same conditions (Table 1 and Table S4, Fig. S5, ESI†). Most mutations were found to exert little effect on either substrate (SNAc or extender unit) specificity or the overall yield of the reaction, with two notable exceptions. First, the condensation activity was completely abolished in the EryKS3AT3 mutant Phe265Trp, for all substrates tested, although its acyl transferase activity remained intact (data not shown). The residue is located (Fig. S2, ESI†) close in space to the catalytic triad required for KS-catalyzed condensation, and although Trp is tolerated in this position in certain mycolactone KS domains, it may be that here the increased steric bulk of the tryptophan sidechain interferes with either initial acylation of the KS or the condensation reaction itself. In contrast, mutant Ala154Trp showed a markedly increased substrate tolerance, giving β-ketoacyl-ACP product from nearly all SNAC thioesters tested, with the exception of β-ketoacyl SNAC thioesters 18 and 19. This mutant also showed a significant increase in turnover of substrates compared to the wild type enzyme. To confirm these preliminary results, side-by-side comparison of Ala154Trp with the wild type was repeated with the inclusion of an internal standard to confirm the stability of the ACP-bound Claisen condensation products.18 Side reactions proved to be less problematic in these experiments and improved turnover was observed for both wild type and mutant EryKS3AT3. However, Ala154Trp continued to be both significantly more promiscuous and a more effective catalyst. Detectable levels of condensation product could be observed for substrate 5 even for the wild type, but only the mutant gave condensation products from substrates 6, 9 and 11–14 (examples shown in Fig. S5, ESI†). Comparative time courses for the wild type and mutant were carried out using substrate 4, and initial rates were extracted by fitting the data to an equation that allows derivation of initial rates from reaction progress curves32 (Fig. S6, ESI†). This confirmed that the initial rate with Ala154Trp is 4.5-fold greater than with the wild type.
The Ala154Gly mutant behaved similarly to the wild type enzyme, even though this substitution is found in certain mycolactone domains, so the observed effect is not simply related to the size of the side-chain at this position. The observed increase in both substrate tolerance and catalytic efficiency for Ala154Trp suggests an important role for this region of the active site. Analysis of the EryKS3AT3 crystal structure using Pymol (http://www.pymol.org) showed that at least local rearrangement of the active site would be necessary to accommodate a Trp side-chain in place of Ala154. This residue is located at the start of the “dimer-interface loop”,21 a region that shows a distinctive lack of sequence conservation in cis-AT KS domains (Fig. S3, ESI†), indicating a possible role in substrate selection. This region is only partly structured in the crystal structure of EryKS3AT3,20 and it may be that in the Ala154Trp mutant this loop is significantly re-ordered. Not only does residue Ala154 lie within the active site close to the dimer interface, but it is also between two regions that cryo-EM studies have recently implicated in docking interactions between the KS and both its ‘upstream’ and intramodular ACP28 (Fig. S1, ESI†). The exact mechanism by which this mutation modulates KS specificity and KS:ACP binding interactions remains to be defined by structural studies, which are in progress. Meanwhile, this present finding mirrors an analogous finding of improved substrate tolerance made for a single active site residue substitution in an AT domain in the erythromycin PKS.33 It provides important encouragement for further exploration of targeted mutagenesis of residues predicted to shape, or control access to, KS active sites, with a view to expanding the range of acyl-ACPs accepted.
This work was supported by project grants from the Biotechnology and Biological Sciences Research Council (BBSRC) U. K. to P. F. L. (BB/J007250/1), from the Wellcome Trust to R. W. B. (094252/Z/10/Z), and a Herchel Smith Postdoctoral Fellowship from the University of Cambridge to A. C. M.
References
- H. Sun, Z. Liu, H. Zhao and E. L. Ang, Drug Des., Dev. Ther., 2015, 9, 823 Search PubMed.
- E. Kim, B. S. Moore and Y. J. Yoon, Nat. Chem. Biol., 2015, 11, 649 CrossRef CAS PubMed.
- A. C. Murphy, Nat. Prod. Rep., 2011, 28, 1406 RSC.
- C. J. Paddon, et al. , Nature, 2013, 496, 528 CrossRef CAS PubMed.
- A. Hagan, S. Poust, T. de Rond, J. L. Fortman, L. Katz, C. J. Petzold and J. D. Keasling, ACS Synth. Biol., 2016, 5, 21 CrossRef PubMed.
- C. Hertweck, Angew. Chem., Int. Ed. Engl., 2009, 48, 4688 CrossRef CAS PubMed.
- J. Staunton and K. J. Weissman, Nat. Prod. Rep., 2001, 18, 380 RSC.
- K. J. Weissman, Nat. Prod. Rep., 2016, 33, 203 RSC.
- J. A. Chuck, M. McPherson, H. Huang, J. R. Jacobsen, C. Khosla and D. E. Cane, Chem. Biol., 1997, 4, 757 CrossRef CAS PubMed.
- N. Wu, S. Y. Tsuji, D. E. Cane and C. Khosla, J. Am. Chem. Soc., 2001, 123, 6465 CrossRef CAS PubMed.
- C. J. Rowe, I. U. Böhm, I. P. Thomas, B. Wilkinson, B. A. Rudd, G. Foster, A. P. Blackaby, P. J. Sidebottom, Y. Roddis, A. D. Buss, J. Staunton and P. F. Leadlay, Chem. Biol., 2001, 8, 475 CrossRef CAS PubMed.
- K. Watanabe, C. C. C. Wang, C. N. Boddy, D. E. Cane and C. Khosla, J. Biol. Chem., 2003, 278, 42020 CrossRef CAS PubMed.
- B. Busch, N. Ueberschaar, S. Behnken, Y. Sugimoto, M. Werneburg, N. Traitcheva, J. He and C. Hertweck, Angew. Chem., Int. Ed. Engl., 2013, 52, 5285 CrossRef CAS PubMed.
- T. Nguyen, K. Ishida, H. Jenke-Kodama, E. Dittmann, C. Gurgui, T. Hochmuth, S. Taudien, M. Platzer, C. Hertweck and J. Piel, Nat. Biotechnol., 2008, 26, 225 CrossRef CAS PubMed.
- M. Jenner, S. Frank, A. Kampa, C. Kohlhaas, P. Pöplau, G. S. Briggs, J. Piel and N. J. Oldham, Angew. Chem., Int. Ed. Engl., 2013, 52, 1143 CrossRef CAS PubMed.
- D. C. Gay, G. Gay, A. J. Axelrod, M. Jenner, C. Kohlhaas, A. Kampa, N. J. Oldham, J. Piel and A. T. Keatinge-Clay, Structure, 2014, 22, 444 CrossRef CAS PubMed.
- C. Kohlhaas, M. Jenner, A. Kampa, G. S. Briggs, J. P. Afonso, J. Piel and N. J. Oldham, Chem. Sci., 2013, 4, 3212 RSC.
- M. Jenner, J. P. Afonso, H. R. Bailey, S. Frank, A. Kampa, J. Piel and N. J. Oldham, Angew. Chem., Int. Ed. Engl., 2015, 54, 1817 CrossRef CAS PubMed.
- Y. Tang, C.-Y. Kim, I. I. Mathews, D. E. Cane and C. Khosla, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11124 CrossRef CAS PubMed.
- Y. Tang, A. Y. Chen, C.-Y. Kim, D. E. Cane and C. Khosla, Chem. Biol., 2007, 14, 931 CrossRef CAS PubMed.
- J. R. Lohman, M. Ma, J. Osipiuk, B. Nocek, Y. Kim, C. Chang, M. Cuff, J. Mack, L. Bigelow, H. Li, M. Endres, G. Babnigg, A. Jochimiak, G. N. Phillips Jr. and B. Shen, Proc. Natl. Acad. Sci. U. S. A., 2015, 54, 6842 Search PubMed.
- T. Bretschneider, J. B. Heim, D. Heine, R. Winkler, B. Busch, B. Kusebauch, T. Stehle, G. Zocher and C. Hertweck, Nature, 2013, 502, 124 CrossRef CAS PubMed.
- T. P. Stinear, A. Mve-Obiang, P. L. Small, W. Frigui, M. J. Pryor, R. Brosch, G. A. Jenkin, P. D. Johnson, J. K. Davies, R. E. Lee, S. Adusumilli, T. Garnier, S. F. Haydock, P. F. Leadlay and S. T. Cole, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1345 CrossRef CAS PubMed.
- H. Hong, J. B. Spencer, J. L. Porter, P. F. Leadlay and T. P. Stinear, ChemBioChem, 2005, 6, 643 CrossRef CAS PubMed.
- H. Hong, C. Demangel, S. J. Pidot, P. F. Leadlay and T. P. Stinear, Nat. Prod. Rep., 2008, 25, 447 RSC.
- J. L. Porter, N. J. Tobias, S. J. Pidot, S. Falgner, K. L. Tuck, A. Vettiger, H. Hong, P. F. Leadlay and T. P. Stinear, PLoS One, 2013, 8, e70520 CAS.
- J. L. Porter, N. J. Tobias, H. Hong, K. L. Tuck, G. A. Jenkin and T. P. Stinear, Microbiology, 2009, 155, 1923 CrossRef CAS PubMed.
- S. Dutta, J. R. Whicher, D. A. Hansen, W. A. Hale, J. A. Chemler, G. R. Congdon, A. R. H. Narayan, K. Håkansson, D. H. Sherman, J. L. Smith and G. Skiniotis, Nature, 2014, 510, 512 CrossRef CAS PubMed.
- L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass and M. J. E. Sternberg, Nat. Protoc., 2015, 10, 845 CrossRef CAS PubMed.
- A. Y. Chen, N. A. Schnarr, C.-Y. Kim, D. E. Cane and C. Khosla, J. Am. Chem. Soc., 2006, 128, 3067 CrossRef CAS PubMed.
- I. Koryakina, J. B. McArthur, M. M. Draelos and G. J. Williams, Org. Biomol. Chem., 2013, 11, 4449 CAS.
- W. Cao and E. M. De La Cruz, Sci. Rep., 2013, 3, 2658 Search PubMed.
- K. Bravo-Rodriguez, S. Klopries, K. R. M. Koopmans, U. Sundermann, S. Yahiaoui, J. Arens, S. Kushnir, F. Schulz and E. Sanchez-Garcia, Chem. Biol., 2015, 22, 1425 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6cc03501a |
| This journal is © The Royal Society of Chemistry 2016 |