Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Assessing the potential of photosensitizing flavoproteins as tags for correlative microscopy†
Alberto
Rodríguez-Pulido
a,
Aitziber L.
Cortajarena
ab,
Joaquim
Torra
c,
Rubén
Ruiz-González
c,
Santi
Nonell
 c and
Cristina
Flors
*a
c and
Cristina
Flors
*a
aMadrid Institute for Advanced Studies in Nanoscience (IMDEA Nanoscience) and Nanobiotechnology Unit Associated to the National Center for Biotechnology (CSIC), 28049 Madrid, Spain. E-mail: cristina.flors@imdea.org
bCIC biomaGUNE, Paseo de Miramón 182, E-20009 Donostia-San Sebastian, Spain
cInstitut Químic de Sarrià, Universitat Ramon Llull, 08017 Barcelona, Spain
First published on 7th June 2016
Abstract
Photosensitizing flavoproteins have great potential as tags for correlative light and electron microscopy (CLEM). We examine the photostability of miniSOG mutants and their ability to photo-oxidize diaminobenzidine, both key aspects for CLEM. Our experiments reveal a complex relation between these parameters and the production of different reactive oxygen species.
Correlative (or correlated) light and electron microscopy (CLEM) is a powerful method for dissecting cell and tissue structure and function at high spatial resolution. Each imaging mode provides unique information, and the combination of the two can contribute to a better understanding of the spatiotemporal patterns of protein expression, trafficking, and function.1,2 One of the most popular strategies for CLEM is the use of 3,3′-diaminobenzidine (DAB)-based staining. DAB can be locally oxidized by an enzymatic or a photoinduced reaction to form an insoluble osmiophilic polymer, which provides contrast in EM after staining with osmium tetroxide.3 Critical to these methods is the use of affinity-based or genetically-encoded tags that label specific proteins of interest and that can site-specifically induce DAB polymerization.
We focus here on the CLEM strategy that relies on the photoinduced polymerization of DAB, in which the tag acts as a photosensitizer that generates reactive oxygen species (ROS) that in turn oxidize DAB.4 While this strategy was used decades ago by injecting Lucifer yellow,5 or immunostaining with eosin-conjugated secondary antibodies,6 it has recently regained interest with the development of the genetically-encoded tag miniSOG (for “mini Singlet Oxygen Generator”).7 MiniSOG is a 106 amino acid flavoprotein derived from the LOV domain of phototropin 2 in which the active-site cysteine has been mutated. Upon blue-light irradiation, miniSOG produces enough singlet oxygen to induce DAB photo-oxidation, and has been used successfully in a number of CLEM applications7–11 even if its singlet oxygen generating properties are lower than initially thought.12,13 Improved miniSOG versions would enable a wider applicability of this tag for imaging the localization of low copy number proteins. However, finding better tags for DAB photo-oxidation has been challenging for several reasons. First, the full mechanistic details and the final product of the light-induced DAB polymerization reaction are unknown.14,15 While photosensitized singlet oxygen is thought to be the main responsible for DAB photo-oxidation,6,7 the participation of superoxide (either directly14 or mediated by Mn2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16) as well as the direct photoinduced electron transfer reaction between the photosensitizer and DAB14 has been suggested. The fact that Lucifer yellow, the first dye used for DAB photo-oxidation in CLEM,5 was later shown to produce superoxide and hydrogen peroxide17 also adds some confusion to the literature. In parallel, the variety of ROS interconversion reactions, the difficulty to identify different ROS specifically using indirect chemical probes18 and the complex photophysical behaviour of genetically-encoded photosensitizers12,13,19–22 has precluded a full understanding of the requirements for optimal CLEM tags.
16) as well as the direct photoinduced electron transfer reaction between the photosensitizer and DAB14 has been suggested. The fact that Lucifer yellow, the first dye used for DAB photo-oxidation in CLEM,5 was later shown to produce superoxide and hydrogen peroxide17 also adds some confusion to the literature. In parallel, the variety of ROS interconversion reactions, the difficulty to identify different ROS specifically using indirect chemical probes18 and the complex photophysical behaviour of genetically-encoded photosensitizers12,13,19–22 has precluded a full understanding of the requirements for optimal CLEM tags.
The suitability of photosensitizing dyes as tags for CLEM can be readily tested by monitoring spectroscopically the optical density increase produced upon DAB polymerization,6 since CLEM experiments are very demanding and time-consuming. Herein, we screen the ability of miniSOG mutants to photo-oxidize DAB as well as their fluorescence photobleaching properties in order to predict their performance in CLEM. We compare the DAB photo-oxidation data to direct measurements of singlet oxygen photosensitization efficiency using time-resolved detection of its phosphorescence in the NIR, as well as to their relative generation of other ROS. For this study we have selected three mutants: (i) Q103L,‡ which has been previously shown to have enhanced singlet oxygen photosensitization properties compared to parent miniSOG;19 (ii) Q103V, a novel mutant in which a different hydrophobic residue has been introduced; and (iii) W81F, which is a less efficient singlet oxygen photosensitizer than miniSOG.12
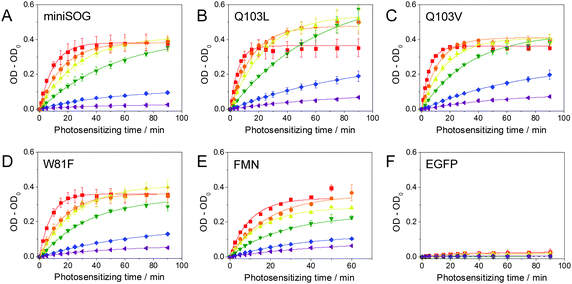
Blue light irradiation of miniSOG and mutants W81F, Q103L and Q103V in the presence of DAB (0.32 mM) induces its photosensitized polymerization, which can be followed spectroscopically as a decrease in the optical density at ∼330 nm and the appearance of a broad band at longer wavelengths with a maximum at ∼450 nm (Fig. 1). The increase in optical density at 450 nm upon increasing irradiation time is compared for all mutants in Fig. 2. The enhanced green fluorescence protein (EGFP), which produces minute quantities of singlet oxygen,23 is used as a negative control. By determining the slope of the initial points of the curve (Fig. S1, ESI†), the relative ability of the mutants to polymerize DAB can be quantified (Fig. 3). Table 1 shows that miniSOG Q103L polymerizes DAB 1.4-fold more efficiently than parent miniSOG. The new variant Q103V also stands out as an efficient photosensitizer for DAB polymerization, with very similar properties to Q103L, confirming that replacement of glutamine by a hydrophobic residue enhances the photosensitizing properties of miniSOG.19 Indeed, the Q103V mutation is even more efficient than Q103L, by a factor of about 1.5, for the purpose of singlet oxygen production (Table 1 and Fig. S2, ESI†), as measured by direct detection of singlet oxygen phosphorescence in the NIR. It is interesting to note that these two variants produce about 4- to 9-fold more singlet oxygen than parent miniSOG, however their ability to photosensitize DAB polymerization in our experimental conditions is only up to about 1.4-fold. We have confirmed that this observation is not due to a saturation effect by performing the experiment at a higher DAB concentration of 0.66 mM, with very similar results (not shown). The prediction from measurements in solution is consistent with transmitted light microscopy experiments that allow monitoring DAB polymerization in the cytosol of bacteria expressing these proteins, in which no dramatic differences can be observed in the performance of miniSOG and mutants Q103L and Q103V in our experimental conditions (Fig. S3, ESI†). As a side note, EGFP induces very low, but observable, DAB polymerization (Table 1), and indeed it has been previously shown that EGFP can be used in CLEM.24,25 The values for FMN have also been included in Fig. 2 and 3, and Table 1 for reference.
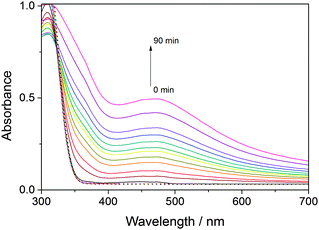 | ||
| Fig. 1 Solid lines correspond to irradiation times of 0, 5, 10, 15, 20, 25, 30, 40, 50, 60, 75 and 90 minutes. | ||
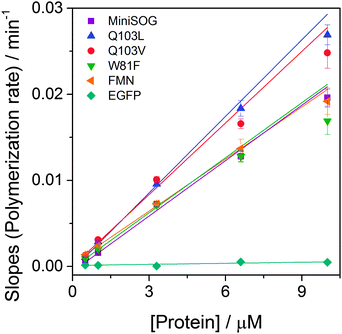 | ||
| Fig. 3 Slopes calculated from the initial points of the curves in Fig. 2, which reflect the rate of DAB polymerization, as a function of protein concentration. | ||
| Relative ability to polymerize DAB | Φ Δ | Relative production of other ROS | |
|---|---|---|---|
| a Ref. 12. b Ref. 19. c This work. d In dPBS, ref. 19. e In dPBS, this work. f Ref. 27. g Ref. 23. h n.d. = Not determined. | |||
| miniSOG | 1 | 0.03a, 0.05b | 1 |
| miniSOG W81F | 1.0 ± 0.1 | 0.01a | 4.3 ± 1.0 |
| miniSOG Q103L | 1.4 ± 0.2 | 0.19b,c, 0.25d,e | 1.5b, 2.2 ± 0.5c |
| miniSOG Q103V | 1.3 ± 0.2 | 0.29c, 0.39e | 2.4 ± 0.4 |
| FMN | 0.9 ± 0.1 | 0.51f, 0.57e | 1.0 ± 0.3 |
| EGFP | 0.02 ± 0.01 | <0.004g | n.d.h |
In order to gain more information about the mechanism for DAB polymerization, PBS was replaced by a deuterated buffer (dPBS), as D2O greatly increases the lifetime of singlet oxygen and thus its ability to encounter a reaction partner.26 Unexpectedly, no substantial difference was found between both solvents (Fig. S4, ESI†). It is worth noting that the interaction between DAB and singlet oxygen is complex, as it leads to DAB photo-oxidation as well as to physical quenching of singlet oxygen.14 The lack of an observable isotope effect on the photosensitized DAB polymerization suggests that the lifetime of singlet oxygen is not a limiting factor in this interaction.
The production of ROS other than singlet oxygen was also tested using the fluorescent probe dihydroethidium19,28 (Table 1 and Fig. S5, ESI†). It was found that W81F is the mutant that produces highest amounts of ROS. However, W81F polymerizes DAB less efficiently than the other mutants, which does not allow extracting a clear correlation between the generation of ROS other than singlet oxygen and the ability to polymerize DAB. Moreover, it was previously shown that KillerRed,29 a genetically-encoded photosensitizer that produces other ROS than singlet oxygen,20 does not polymerize DAB and is therefore not suitable for CLEM.7
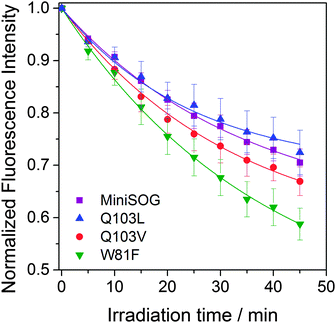
Since photostability is a crucial aspect when assessing fluorophore performance in fluorescence microscopy, we monitored the rates of fluorescence photobleaching of miniSOG mutants. Fig. 4 shows that W81F is the least and Q103L the most photostable of the mutants tested, although the latter very close to miniSOG. Clearly, the trend in fluorescence photobleaching does not correlate with the self-sensitization of singlet oxygen, i.e. Q103L produces 19 times more singlet oxygen than W81F but it is the most photostable. On the other hand, W81F is the least photostable and the most efficient producing other ROS, suggesting that these two properties are related. In parallel, oxygen-independent pathways may contribute to photobleaching of these proteins. Indeed, miniSOG has similar photostability to other flavin binding fluorescent proteins (FbFPs) that have not been reported to photosensitize ROS.30 We have confirmed that photobleaching shown in Fig. 4 is irreversible and does not correspond to reversible semiquinone formation,31,32 as already reported for miniSOG in solution.30 While the detailed mechanisms for FbFPs photobleaching are not known and merit further investigation, it has been shown that constraining the FMN chromophore improves photostability in these proteins.33
In conclusion, simple screening of DAB polymerization and fluorescence photobleaching is useful to predict the performance of flavoproteins as novel photosensitizers for CLEM. This is a valuable test since the ability to polymerize DAB does not clearly correlate with the efficiency of the photosensitizers to generate singlet oxygen or other ROS. MiniSOG mutants with a hydrophobic residue in position 103 induce somewhat more efficient DAB polymerization, and we report the novel mutant Q103V has a 1.5 higher efficiency of singlet oxygen production than the previously reported leucine mutant.19 Moreover, our fluorescence photobleaching experiments show that photooxidation by self-sensitized singlet oxygen is not a major photobleaching pathway in miniSOG variants in these conditions, and that the photosensitization of other ROS may play a more significant role. These observations are important for the development of improved flavoproteins for CLEM and other microscopy applications.34–38
This work has been funded by the Spanish Ministerio de Economía y Competitividad (RyC2011-07637, MAT2012-34487, BIO2012-34835, CTQ2013-48767-C3-1-R, CTQ2015-71896-REDT, MAT2015-66605-P) and the European Commission Marie Curie Actions (FP7-PEOPLE-2010-IRG-246688, FP7-PEOPLE-2011-CIG-303620, FP7-PEOPLE-2011-COFUND-291803). We thank the Regan Lab at Yale University for the plasmid encoding EGFP.39
Notes and references
- P. de Boer, J. P. Hoogenboom and B. N. Giepmans, Nat. Methods, 2015, 12, 503–513 CrossRef CAS PubMed.
- H. D. Ou, T. J. Deerinck, E. Bushong, M. H. Ellisman and C. C. O'Shea, Methods, 2015, 90, 39–48 CrossRef CAS PubMed.
- R. C. Graham, Jr. and M. J. Karnovsky, J. Histochem. Cytochem., 1966, 14, 291–302 CrossRef.
- C. Meisslitzer-Ruppitsch, C. Rohrl, J. Neumuller, M. Pavelka and A. Ellinger, J. Microsc., 2009, 235, 322–335 CrossRef CAS PubMed.
- A. R. Maranto, Science, 1982, 217, 953–955 CAS.
- T. J. Deerinck, M. E. Martone, V. Lev-Ram, D. P. Green, R. Y. Tsien, D. L. Spector, S. Huang and M. H. Ellisman, J. Cell Biol., 1994, 126, 901–910 CrossRef CAS PubMed.
- X. Shu, V. Lev-Ram, T. J. Deerinck, Y. Qi, E. B. Ramko, M. W. Davidson, Y. Jin, M. H. Ellisman and R. Y. Tsien, PLoS Biol., 2011, 9, e1001041 CAS.
- D. Boassa, M. L. Berlanga, M. A. Yang, M. Terada, J. Hu, E. A. Bushong, M. Hwang, E. Masliah, J. M. George and M. H. Ellisman, J. Neurosci., 2013, 33, 2605–2615 CrossRef CAS PubMed.
- D. Boassa, P. Nguyen, J. Hu, M. H. Ellisman and G. E. Sosinsky, Front. Cell. Neurosci., 2015, 8, 468 Search PubMed.
- C. Cleyrat, A. Darehshouri, M. P. Steinkamp, M. Vilaine, D. Boassa, M. H. Ellisman, S. Hermouet and B. S. Wilson, Traffic, Copenhagen, Denmark, 2014, vol. 15, pp. 961–982 Search PubMed.
- A. Ludwig, G. Howard, C. Mendoza-Topaz, T. Deerinck, M. Mackey, S. Sandin, M. H. Ellisman and B. J. Nichols, PLoS Biol., 2013, 11, e1001640 CAS.
- R. Ruiz-González, A. L. Cortajarena, S. H. Mejias, M. Agut, S. Nonell and C. Flors, J. Am. Chem. Soc., 2013, 135, 9564–9567 CrossRef PubMed.
- F. M. Pimenta, R. L. Jensen, T. Breitenbach, M. Etzerodt and P. R. Ogilby, Photochem. Photobiol., 2013, 89, 1116–1126 CrossRef CAS PubMed.
- J. Natera, W. Massad, F. Amat-Guerri and N. García, J. Photochem. Photobiol., A, 2011, 220, 25–30 CrossRef CAS.
- J. A. Litwin, Folia Histochem. Cytochem., 1979, 17, 3–28 CAS.
- M. J. Steinbeck, A. U. Khan, W. H. Appel, Jr. and M. J. Karnovsky, J. Histochem. Cytochem., 1993, 41, 1659–1667 CrossRef CAS PubMed.
- J. P. Martin and N. Logsdon, Photochem. Photobiol., 1987, 46, 45–53 CrossRef CAS PubMed.
- N. Macia and B. Heyne, J. Photochem. Photobiol., A, 2015, 306, 1–12 CrossRef CAS.
- M. Westberg, L. Holmegaard, F. M. Pimenta, M. Etzerodt and P. R. Ogilby, J. Am. Chem. Soc., 2015, 137, 1632–1642 CrossRef CAS PubMed.
- R. B. Vegh, K. M. Solntsev, M. K. Kuimova, S. Cho, Y. Liang, B. L. Loo, L. M. Tolbert and A. S. Bommarius, Chem. Commun., 2011, 47, 4887–4889 RSC.
- J. Torra, A. Burgos-Caminal, S. Endres, M. Wingen, T. Drepper, T. Gensch, R. Ruiz-González and S. Nonell, Photochem. Photobiol. Sci., 2015, 14, 280–287 CAS.
- R. B. Vegh, K. B. Bravaya, D. A. Bloch, A. S. Bommarius, L. M. Tolbert, M. Verkhovsky, A. I. Krylov and K. M. Solntsev, J. Phys. Chem. B, 2014, 118, 4527–4534 CrossRef CAS PubMed.
- A. Jimenez-Banzo, S. Nonell, J. Hofkens and C. Flors, Biophys. J., 2008, 94, 168–172 CrossRef CAS PubMed.
- M. Grabenbauer, W. J. C. Geerts, J. Fernandez-Rodriguez, A. Hoenger, A. J. Koster and T. Nilsson, Nat. Methods, 2005, 2, 857–862 CrossRef CAS PubMed.
- M. Grabenbauer, Methods Cell Biol., 2012, 111, 117–138 CAS.
- P. R. Ogilby, Chem. Soc. Rev., 2010, 39, 3181–3209 RSC.
- J. Baier, T. Maisch, M. Maier, E. Engel, M. Landthaler and W. Baumler, Biophys. J., 2006, 91, 1452–1459 CrossRef CAS PubMed.
- K. Takemoto, T. Matsuda, N. Sakai, D. Fu, M. Noda, S. Uchiyama, I. Kotera, Y. Arai, M. Horiuchi, K. Fukui, T. Ayabe, F. Inagaki, H. Suzuki and T. Nagai, Sci. Rep., 2013, 3, 2629 Search PubMed.
- M. E. Bulina, D. M. Chudakov, O. V. Britanova, Y. G. Yanushevich, D. B. Staroverov, T. V. Chepurnykh, E. M. Merzlyak, M. A. Shkrob, S. Lukyanov and K. A. Lukyanov, Nat. Biotechnol., 2006, 24, 95–99 CrossRef CAS PubMed.
- M. Wingen, J. Potzkei, S. Endres, G. Casini, C. Rupprecht, C. Fahlke, U. Krauss, K. E. Jaeger, T. Drepper and T. Gensch, Photochem. Photobiol. Sci., 2014, 13, 875–883 CAS.
- E. F. Yee, R. P. Diensthuber, A. T. Vaidya, P. P. Borbat, C. Engelhard, J. H. Freed, R. Bittl, A. Moglich and B. R. Crane, Nat. Commun., 2015, 6, 10079 CrossRef CAS PubMed.
- S. Chapman, C. Faulkner, E. Kaiserli, C. Garcia-Mata, E. I. Savenkov, A. G. Roberts, K. J. Oparka and J. M. Christie, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20038–20043 CrossRef CAS PubMed.
- J. M. Christie, K. Hitomi, A. S. Arvai, K. A. Hartfield, M. Mettlen, A. J. Pratt, J. A. Tainer and E. D. Getzoff, J. Biol. Chem., 2012, 287, 22295–22304 CrossRef CAS PubMed.
- T. L. To, M. J. Fadul and X. Shu, Nat. Commun., 2014, 5, 4072 CAS.
- A. M. Buckley, J. Petersen, A. J. Roe, G. R. Douce and J. M. Christie, Curr. Opin. Chem. Biol., 2015, 27, 39–45 CrossRef CAS PubMed.
- A. Mukherjee and C. M. Schroeder, Curr. Opin. Biotechnol., 2015, 31, 16–23 CrossRef CAS PubMed.
- T. Drepper, T. Gensch and M. Pohl, Photochem. Photobiol. Sci., 2013, 12, 1125–1134 CAS.
- A. S. Mishin, V. V. Belousov, K. M. Solntsev and K. A. Lukyanov, Curr. Opin. Chem. Biol., 2015, 27, 1–9 CrossRef CAS PubMed.
- R. P. Ilagan, E. Rhoades, D. F. Gruber, H. T. Kao, V. A. Pieribone and L. Regan, FEBS J., 2010, 277, 1967–1978 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Materials and methods, Fig. S1 and S2. See DOI: 10.1039/c6cc03119f |
| ‡ We follow the same residue numbering criteria as the original miniSOG paper.7 In ref. 19 this residue was referred to as 102. |
| This journal is © The Royal Society of Chemistry 2016 |