Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
13-Helix folding of a β/γ-peptide manifold designed from a “minimal-constraint” blueprint†
Claire M.
Grison
a,
Sylvie
Robin
ab and
David J.
Aitken
*a
aCP3A Organic Synthesis Group, ICMMO, UMR 8182, CNRS, Université Paris-Sud, Université Paris-Saclay, 15 Rue Georges Clemenceau, 91405 Orsay cedex, France. E-mail: david.aitken@u-psud.fr; Fax: +33-(0)169156278; Tel: +33-(0)169153238
bUniversité Paris Descartes, UFR Sciences Pharmaceutiques et Biologiques, 4 Avenue de l'Observatoire, 75270 Paris cedex 06, France
First published on 12th May 2016
Abstract
A bottom-up design rationale was adopted to devise β/γ-peptide foldamer manifolds which would adopt preferred 13-helix conformations, relying on minimal steric imposition brought by the constituent amino acid residues. In this way, a well-defined 13-helix conformer was revealed for short oligomers of trans-2-aminocyclobutanecarboxylic acid and γ4-amino acids in alternation, which gave good topological superposition upon an α-helix motif.
Peptide oligomers which fold regularly around an intramolecular network of non-covalent interactions, principally hydrogen bonds (H-bonds), have been at the forefront of foldamer science since their inception and remain of primary importance today. Through the construction of different sequences of homo- and hetero-oligopeptides containing non-canonical β- or γ-amino acid building blocks, a widely-varied collection of folded conformations has been established.1
The design of particular types of foldamer architecture is a contemporary challenge and one major objective is the preparation of α-helix mimetics. In native proteins, α-helices play a structural role when buried in the hydrophobic core, serving to stabilise the tertiary structure. When completely or partially exposed on the surface of proteins, α-helices play critical roles in the interactions of proteins with other biological molecules.2 In the search for mediators of protein–protein interactions, a number of approaches to develop α-helix mimetics have been considered,3 and some notable successes have been described by employing β-peptide4 or α/β-peptide5 foldamers which adopt helical architectures. For example, Schepartz has elaborated helical β-peptides which can present “hot-spot” side chains along one face, leading to the development of inhibitors of targeted protein–protein interactions,6 inhibitors of HIV fusion,7 and agonists of a particular G protein-coupled receptor.8
The β/γ-hybrid peptide manifold is of special interest because a β/γ-dipeptide has the same number of backbone atoms as a native α-tripeptide. Indeed, in a seminal theoretical study, Hofmann suggested that the 13-helix of a β/γ-peptide might exhibit the same dipole and H-bond orientations as the α-helix.9 Successful isosteric replacements of short α-helical segments by β/γ-peptide fragments in α-peptides have been described.10 Recently, a study of single side-chain β3- and γ4-amino acid insertion into short helical α-peptides revealed that the latter residues were more propitious for helical folding.11 However, with flexible β/γ-peptides a number of stable conformations are accessible, including a 11/13-helix12 and a non-helical 9/8-ribbon.13 Balaram observed a cyclic 13-membered ring (C13) H-bond feature in selected β,γ,β-tripeptides.14 To date, only one description of a 13-helical β/γ-peptide has been reported: Gellman successfully imposed the requisite secondary structure by applying draconian steric constraints upon each constituent residue.15 While this work was a proof-of-principle milestone, the opportunities for the introduction of functional side chains are limited due to the severe steric hindrance in operation.
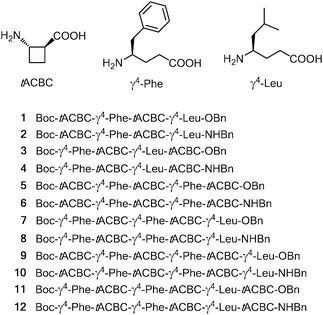
Given the particular attraction of the β/γ-peptide 13-helix as a potential α-helix mimetic, we sought to establish a “minimal constraint” axiom with low steric congestion for this secondary structure. Hofmann's prediction for a stable, right-handed (P) 13-helix implicates a backbone θ torsion angle in the range 90° to 100° for the β-residue component and g+, g+ local conformations for the γ-residues. Following a bottom-up design rationale to accommodate these criteria, we reasoned that a suitable β-amino acid component would be the small, constrained trans-(1S,2S)-2-aminocyclobutanecarboxylic acid (tACBC),16 whose homo-oligomers support a robust 12-helix.17 Unsubstituted γ-aminobutyric acid (GABA) was disfavoured as the γ-component, since it facilitates 9/8-ribbon formation;13 we selected instead a singly substituted (R)-γ4-amino acid. The inclusion of a substituent is in fact pertinent, if the target 13-helix is to provide recognizable side chains on its periphery.
To test this hypothesis we prepared twelve β/γ-peptides, 1–12, for study. The monomer building blocks and the peptide sequences are presented in Fig. 1. The γ-residues, (R)-γ4-Phe and (R)-γ4-Leu, were selected to resemble hydrophobic α-residues which should not interfere with the backbone H-bonding preferences. The features which were varied within this series were the sequence length (tetra-, penta- and hexa-peptides); the N-terminal residue (tACBC or a γ4-amino acid); and the nature of the C-terminal capping group (ester or amide). The twelve peptides were synthesised using standard solution-state peptide coupling techniques (see the ESI†).
All 1H and 13C NMR spectroscopic signals pertinent to conformational analysis were unambiguously assigned to each peptide using standard 1D and 2D pulse sequences in CDCl3. Titration of the CDCl3 solutions with DMSO-d6 revealed a higher titration coefficient for the carbamate and amide NH signals of residues 1 and 2, respectively, than for the other NH signals in each peptide, suggesting that the former NH functions were at least partially solvent exposed and not extensively engaged in hydrogen bonding. This initial observation was qualitatively encouraging, since the folding of the desired 13-helix conformer should not implicate either of the first two NH functions.
Conformational analysis of all twelve peptides 1–12 was carried out using ROESY NMR experiments in CDCl3 solution; interpretation of the data was corroborated by molecular modelling conducted in the form of a hybrid Monte Carlo molecular mechanics (MCMM) conformation search carried out in chloroform medium, using MacroModel and the MMFF force field without restraints. From 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 generated structures the lowest energy conformers (<20 kJ mol−1) were retained and sorted according to their conformer family type (see the ESI†).
000 generated structures the lowest energy conformers (<20 kJ mol−1) were retained and sorted according to their conformer family type (see the ESI†).
In the analysis of tetrapeptides 1–4, short range hydrogen-bonding emerged as the main feature of a rather complex conformational landscape. In each case, ROESY experiments showed short range Hα(i)–NH(i + 1) and Hβ(i)–NH(i + 1) correlations around one or both tACBC residues, as well as Hγ(i)–NH(i + 1) correlations around one or both γ4-amino acid residues. These phenomena are diagnostic of C8 and C9 H-bonding features, which form the basis of a 9/8-ribbon.13 The molecular modelling survey confirmed that the 9/8-ribbon was the most common folded conformer in all four peptides.
Longer distance ROESY Hγ(i)–NH(i + 2) correlations were observed for 2, 3 and 4, as well as Hβ(i)–NH(i + 2) and Hβ(i)–Hα(i + 2) correlations for 3 and 4. These observations were supportive of C13 H-bonds;15 however molecular modelling suggested that the “all-C13” conformers, corresponding to 13-helix structures, were accompanied by mixed conformers featuring combinations of C13 and C8 and/or C9 interactions.
ROESY data for pentapeptides 5–8 featured the propitious long distance correlations Hγ(i)–NH(i + 2), Hβ(i)–NH(i + 2) and Hβ(i)–Hα(i + 2), which are diagnostic for conformers having C13 H-bonding. Furthermore, previously-undescribed Hγ(i)–NH(i + 3) correlations were observed in 5 and 8 (two such correlations for the latter), which were fully consistent with a 13-helix structure. Molecular modelling suggested that 13-helix conformers were dominant for peptides 5 and 8 and made significant contributions to peptides 6 and 7. In all of the low-energy conformers of the latter two peptides, at least two consecutive C13 features were present. The short-range (i)-(i + 1) ROESY correlations shown by these compounds are an integral part of the C13 ROESY signature, rather than being indicative of the presence of alternative C8 or C9 H-bonded conformers.
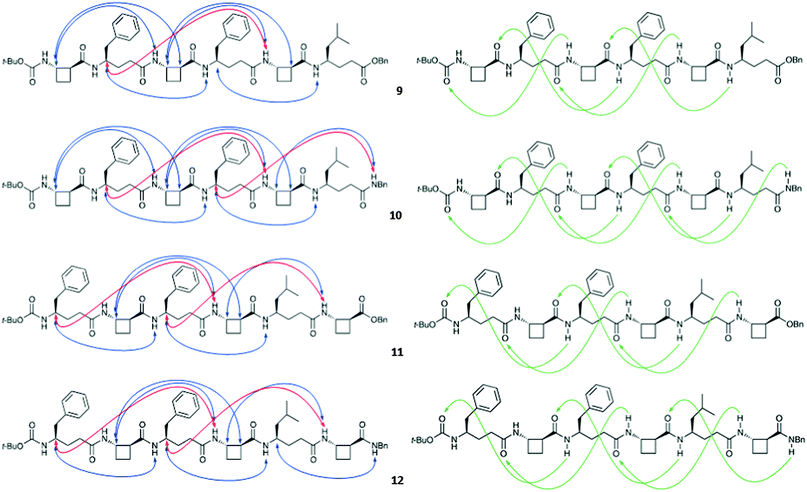
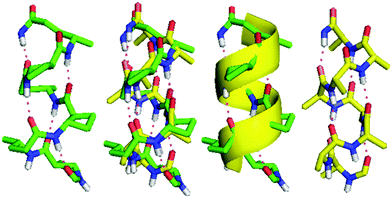
The four hexapeptides 9–12 provided extensive ROESY correlation data which were entirely consistent with a dominant 13-helix conformer in each case. For hexapeptide ester 9, the presence of a second conformer was suspected, in which the C-terminal of the 13-helix had given way to the C8 feature around tACBC-5. Molecular modelling suggested that in fact, for all four peptides, minor contributions might arise from N- or C-terminal “tightening” of the H-bonding pattern; however the 13-helix essentially remained the central feature of the conformer landscape. Fig. 2 shows the ROESY correlations observed for peptides 9–12 along with the 13-helix H-bonding networks deduced from these data.
The solution-state IR absorption spectra of peptides 1–12 in chloroform (see the ESI†) confirmed the general trends for the folding predilection described above. In all cases, free carbamate (residue 1) and amide (residue 2) NH vibrations appeared in the region of 3430–3450 cm−1. For peptides 1–4, low frequency (H-bonded) amide NH bands were discernible at around 3260 cm−1 for C8 motifs and at around 3350 cm−1 for C9 motifs. For pentapeptides 5–8, these features were less well defined and the curves evolved towards increased absorption in the zone between these two frequencies, concomitant with significant C13 contributions. For peptides 9–12 the most intense absorption was in the region at around 3310 cm−1, in agreement with the predominance of 13-helix conformers. It is noteworthy that the 12-helix formed by homo-oligomers of tACBC has a C12 H-bonded amide NH absorption centred at 3300 cm−1 in the same solvent.17
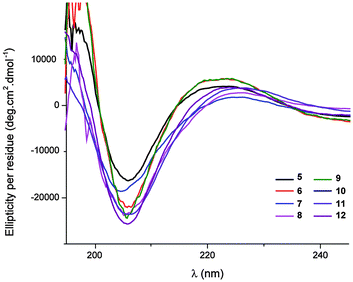
The peptides considered here were insoluble in water. The far-UV circular dichroism (CD) spectra were recorded in MeOH (0.20 mM) for the pentapeptides and hexapeptides 5–12, which showed a considerable or substantial population of 13-helix conformers. The common features show a small positive Cotton effect at around 225 nm and a stronger negative Cotton effect at 206 nm, with a mean-residue ellipticity [Θ] reaching a significant value of 25 × 103 deg cm2 dmol−1 for the latter (Fig. 3). While the CD spectrum of a β/γ-peptide 13-helix has been neither calculated nor reported, these data are very close to the CD signature of a right-handed β-peptide 12-helix in the same solvent.17,18 This observation is entirely consistent with the prevalence of the β/γ-peptide 13-helix conformer in polar protic solution.
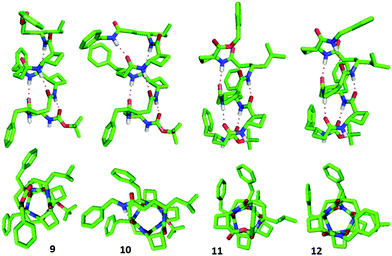
The 13-helix conformers generated by the MCMM conformational search for each of the four hexapeptides 9–12 were subjected to ab initio geometry optimization by DFT using GAUSSIAN 09 and the B3LYP/6-311G(d,p) basis set in chloroform medium (Fig. 4) (for details, see the ESI†).
Four (for peptides 9 and 11) or five (for peptides 10 and 12) standard hydrogen bonds form the basis of the right-handed (P) 13-helical structures, displaying C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N distances within the range of 1.78 Å to 2.01 Å and O⋯H–N angles within the range of 169.7° to 178.9°. In all peptides, the γ4-amino acid residues each adopted the anticipated g+, g+ conformations, showing θ and ζ torsion angles falling in the ranges of 44° to 62° and 56° to 63°, respectively, for those residues which were involved in the H-bonding network. For the tACBC residues the backbone θ torsion angle values fell within the narrow range of 100° to 106°, which was close to the θ value ranges previously observed for tACBC in an 8-helix foldamer (101° to 104°)19 or a 9/8-ribbon (99° to 102°),13 although perceptibly greater than the range observed for tACBC in its homo-oligomeric 12-helix (96° to 100°).17 However, when the φ and ψ torsion angles (averaging around −102° and −106°, respectively) are taken into account, tACBC approaches convincingly the idealized topology and pitch rise for the β-amino acid component in a β/γ-peptide 13-helix: the theoretical values for φ, θ and ψ average around −96°, 94° and −115°, respectively, in Hofmann's idealized unconstrained model.9
O⋯H–N distances within the range of 1.78 Å to 2.01 Å and O⋯H–N angles within the range of 169.7° to 178.9°. In all peptides, the γ4-amino acid residues each adopted the anticipated g+, g+ conformations, showing θ and ζ torsion angles falling in the ranges of 44° to 62° and 56° to 63°, respectively, for those residues which were involved in the H-bonding network. For the tACBC residues the backbone θ torsion angle values fell within the narrow range of 100° to 106°, which was close to the θ value ranges previously observed for tACBC in an 8-helix foldamer (101° to 104°)19 or a 9/8-ribbon (99° to 102°),13 although perceptibly greater than the range observed for tACBC in its homo-oligomeric 12-helix (96° to 100°).17 However, when the φ and ψ torsion angles (averaging around −102° and −106°, respectively) are taken into account, tACBC approaches convincingly the idealized topology and pitch rise for the β-amino acid component in a β/γ-peptide 13-helix: the theoretical values for φ, θ and ψ average around −96°, 94° and −115°, respectively, in Hofmann's idealized unconstrained model.9
The topology of the β/γ-peptide 13-helical backbone was compared with a model of the native α-helical backbone based on oligo-Ala (Fig. 5). The 13-helix of peptide 10 and the α-helix model pleasingly exhibited the same orientation of the helix dipole and close similarities in helix pitch and diameter.
In conclusion, the judicious combination of the small cyclic β-amino acid tACBC and a singly-substituted γ4-amino acid provides sufficient steric imposition to induce regular folding of a β/γ-peptide manifold into a well-defined 13-helix conformer. This secondary structure overlays very well with a representative α-helical peptide; since its backbone is relatively unhindered, it may constitute a useful manifold for the design of mimetics of natural α-helical fragments.
We are grateful to Mr J.-P. Baltaze (ICMMO) for help with NMR experiments. The award of a French MESR doctoral research scholarship (to C. M. G.) is acknowledged.
Notes and references
- (a) Foldamers: Structure, Properties, and Applications, ed. S. Hecht and I. Huc, Wiley-VCH Verlag GmbH & Co, Weinheim, 2007 Search PubMed; (b) G. Guichard and I. Huc, Chem. Commun., 2011, 47, 5933 RSC; (c) D. J. Hill, M. J. Mio, R. B. Prince, T. S. Hughes and J. S. Moore, Chem. Rev., 2001, 101, 3893 CrossRef CAS PubMed; (d) T. A. Martinek and F. Fülöp, Chem. Soc. Rev., 2012, 41, 687 RSC; (e) P. G. Vasudev, S. Chatterjee, N. Shamala and P. Balaram, Chem. Rev., 2011, 111, 657 CrossRef CAS PubMed; (f) A. Roy, P. Prabhakaran, P. K. Baruah and G. J. Sanjayan, Chem. Commun., 2011, 47, 11593 RSC; (g) W. S. Horne and S. H. Gellman, Acc. Chem. Res., 2008, 41, 1399 CrossRef CAS PubMed; (h) D. Seebach, D. F. Hook and A. Glättli, Biopolymers, 2006, 84, 23 CrossRef CAS PubMed; (i) R. P. Cheng, S. H. Gellman and W. F. DeGrado, Chem. Rev., 2001, 101, 3219 CrossRef CAS PubMed.
- (a) D. J. Barlow and J. M. Thornton, J. Mol. Biol., 1988, 201, 601 CrossRef CAS PubMed; (b) T. E. Creighton, Proteins: Structures and Molecular Properties, W.H. Freeman & Co, New York, 2nd edn, 1992 Search PubMed; (c) G. T. Kilosanidze, A. S. Kutsenko, N. G. Esipova and V. G. Tumanyan, Protein Sci., 2004, 13, 351 CrossRef CAS PubMed.
- (a) M. Pelay-Gimeno, A. Glas, O. Koch and T. N. Grossmann, Angew. Chem., Int. Ed., 2015, 54, 8896 CrossRef CAS PubMed; (b) V. Azzarito, K. Long, N. S. Murphy and A. J. Wilson, Nat. Chem., 2013, 5, 161 CrossRef CAS PubMed; (c) B. N. Bullock, A. L. Jochim and P. S. Arora, J. Am. Chem. Soc., 2011, 133, 14220 CrossRef CAS PubMed; (d) J. Gardner and M. M. Harding, Org. Biomol. Chem., 2007, 5, 3577 RSC.
- M. Hintersteiner, T. Kimmerlin, G. Garavel, T. Schindler, R. Bauer, N.-C. Meisner, J.-M. Seifert, V. Uhl and M. Auer, ChemBioChem, 2009, 10, 994 CrossRef CAS PubMed.
- (a) R. W. Cheloha, A. Maeda, T. Dean, T. J. Gardella and S. H. Gellman, Nat. Biotechnol., 2014, 32, 653 CrossRef CAS PubMed; (b) L. M. Johnson, S. Barrick, M. V. Hager, A. McFedries, E. A. Homan, M. E. Rabaglia, M. P. Keller, A. D. Attie, A. Saghatelian, A. Bisello and S. H. Gellman, J. Am. Chem. Soc., 2014, 136, 12848 CrossRef CAS PubMed; (c) B. J. Smith, E. F. Lee, J. W. Checco, M. Evangelista, S. H. Gellman and W. D. Fairlie, ChemBioChem, 2013, 14, 1564 CrossRef CAS PubMed; (d) M. D. Boersma, H. S. Haase, K. J. Peterson-Kaufman, E. F. Lee, O. B. Clarke, P. M. Colman, B. J. Smith, W. S. Horne, W. D. Fairlie and S. H. Gellman, J. Am. Chem. Soc., 2012, 134, 315 CrossRef CAS PubMed; (e) W. S. Horne, M. D. Boersma, M. A. Windsor and S. H. Gellman, Angew. Chem., Int. Ed., 2008, 47, 2853 CrossRef CAS PubMed.
- (a) E. A. Harker and A. Schepartz, ChemBioChem, 2009, 10, 990 CrossRef CAS PubMed; (b) E. A. Harker, D. S. Daniels, D. A. Guarracino and A. Schepartz, Bioorg. Med. Chem., 2009, 17, 2038 CrossRef CAS PubMed.
- O. M. Stephens, S. Kim, B. D. Welch, M. E. Hodsdon, M. S. Kay and A. Schepartz, J. Am. Chem. Soc., 2005, 127, 13126 CrossRef CAS PubMed.
- E. V. Denton, C. J. Craig, R. L. Pongratz, J. S. Appelbaum, A. E. Doerner, A. Narayanan, G. I. Schulman, G. W. Cline and A. Schepartz, Org. Lett., 2013, 15, 5318 CrossRef CAS PubMed.
- C. Baldauf, R. Günther and H.-J. Hofmann, J. Org. Chem., 2006, 71, 1200 CrossRef CAS PubMed.
- (a) R. R. Araghi, C. Jäckel, H. Cölfen, M. Salwiczek, A. Völkel, S. C. Wagner, S. Wieczorek, C. Baldauf and B. Koksch, ChemBioChem, 2010, 11, 335 CrossRef PubMed; (b) I. L. Karle, A. Pramanik, A. Bannerjee, S. Bhattacharjya and P. Balaram, J. Am. Chem. Soc., 1997, 119, 9087 CrossRef CAS.
- Y.-H. Shin, D. E. Mortenson, K. A. Satyshur, K. T. Forest and S. H. Gellman, J. Am. Chem. Soc., 2013, 135, 8149 CrossRef CAS PubMed.
- G. V. M. Sharma, V. B. Jadhav, K. V. S. Ramakrishna, P. Jayaprakash, K. Narsimulu, V. Subash and A. C. Kunwar, J. Am. Chem. Soc., 2006, 128, 14657 CrossRef CAS PubMed.
- C. M. Grison, S. Robin and D. J. Aitken, Chem. Commun., 2015, 51, 16233 RSC.
- P. G. Vasudev, K. Ananda, S. Chatterjee, S. Aravinda, N. Shamala and P. Balaram, J. Am. Chem. Soc., 2007, 129, 4039 CrossRef CAS PubMed.
- L. Guo, A. M. Almeida, W. Zhang, A. G. Reidenbach, S. H. Choi, I. A. Guzei and S. H. Gellman, J. Am. Chem. Soc., 2010, 132, 7868 CrossRef CAS PubMed.
- (a) V. Declerck and D. J. Aitken, Amino Acids, 2011, 41, 587 CrossRef CAS PubMed; (b) C. Fernandes, E. Pereira, S. Faure and D. J. Aitken, J. Org. Chem., 2009, 74, 3217 CrossRef CAS PubMed; (c) C. Fernandes, C. Gauzy, Y. Yang, O. Roy, E. Pereira, S. Faure and D. J. Aitken, Synthesis, 2007, 2222 CAS.
- C. Fernandes, S. Faure, E. Pereira, V. Théry, V. Declerck, R. Guillot and D. J. Aitken, Org. Lett., 2010, 12, 3606 CrossRef CAS PubMed.
- (a) H.-S. Lee, F. A. Syud, X. Wang and S. H. Gellman, J. Am. Chem. Soc., 2001, 123, 7721 CrossRef CAS PubMed; (b) X. Wang, J. F. Espinosa and S. H. Gellman, J. Am. Chem. Soc., 2000, 122, 4821 CrossRef CAS.
- A. Altmayer-Henzien, V. Declerck, J. Farjon, D. Merlet, R. Guillot and D. J. Aitken, Angew. Chem., Int. Ed., 2015, 54, 10807 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures, CD, IR and NMR analyses, and molecular modelling data. See DOI: 10.1039/c6cc02142e |
| This journal is © The Royal Society of Chemistry 2016 |