Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A de novo self-assembling peptide hydrogel biosensor with covalently immobilised DNA-recognising motifs†
Patrick J. S.
King
a,
Alberto
Saiani
b,
Elena V.
Bichenkova‡
*a and
Aline F.
Miller‡
*b
aManchester School of Pharmacy, the University of Manchester, Stopford Building, Oxford Road, Manchester, M13 9PT, UK. E-mail: Elena.V.Bichenkova@manchester.ac.uk
bSchool of Chemical Engineering and Analytical Science, Manchester Institute of Biotechnology, 131 Princess Street, Manchester, M1 7DN, UK. E-mail: Aline.Miller@manchester.ac.uk
First published on 19th April 2016
Abstract
We report here the first experimental evidence of a self-assembling three-dimensional (3D) peptide hydrogel, with recognition motifs immobilized on the surface of fibres capable of sequence-specific oligonucleotide detection. These systems have the potential to be further developed into diagnostic and prognostic tools in human pathophysiology.
There has been a rapid development of new detection devices and biosensors over the past ten years, in particular, in the biomedical field where identification of single or multiple nucleotide polymorphisms could be crucial in recognising patient susceptibility to certain monogenic or complex diseases.1–6 Modern DNA microarrays typically comprise of tens to thousands of 10–100 μm reaction sites, thus allowing the simultaneous analysis of thousands of nucleic acid sequences in a single experiment.7,8 The majority of these systems rely on the sequence-specific hybridisation of target DNA or RNA sequences, with probe oligonucleotides immobilized to a surface, and typically use optical,9 electrochemical10 or gravimetric detection11 to monitor binding events. There are still major challenges with such technology, which is limiting their successful transition to the commercial market.12–14 These include high fabrication costs, poor sensitivity and accuracy, a lack of reproducibility, and significant divergence in the results of different devices on identical samples.15–17 Moreover, even minor improvements in bioassay performance can very often be accompanied by substantial technological and conceptual complexity, which makes it incompatible with clinical application at point-of-care settings.18 One approach to overcome such issues is to use a 3D polymer network as the solid-phase support. Such materials are relatively inexpensive, amenable to established detection techniques, and can enhance bioassay sensitivity due to a marked increase in analyte storage capacity.19,20 Their functionalization, however, is difficult to control and requires additional synthetic steps. Self-assembling peptide-based hydrogels share the desirable material properties, but in contrast their functionalization can be controlled precisely, on the molecular level using the principles of supramolecular chemistry.21 Although they are currently more expensive to produce synthetically than simple polymers, there has been much recent progress towards reducing their cost significantly through bacterial expression.22 Peptide hydrogels also benefit from providing a more biologically-relevant environment for sensing to occur,23 ‘smart’ responsivity to stimuli such as pH and temperature,24–26 self-healing properties that are amenable to rapid custom array production for personalised medicine, and can be dried for long-term storage, hence protecting oligonucleotides against nuclease degradation.27 These biosensors are envisioned to be used in 96-well plates, or deposited onto chips, with analyte solution introduced onto the surface. Through the use of positively-charged hydrogels (+2 charge per peptide at pH 7, Fig. S1, see the ESI†), we aim to improve the biosensor sensitivity through an increase in the local concentration of negatively-charged target DNA around the sensing elements. The success of this approach has been previously reported in similar systems, such as in mercury detection using acrylamide gels.28
In this pilot study, the proof-of-principle level was achieved by using a simple molecular bio-recognition system which co-assembles with an octapeptide to form hydrogel-based biosensors capable of selectively hybridizing DNA and generating a fluorescence output (Fig. 1). This has never been reported before, and any previous attempts to incorporate DNA aptamers (e.g. via chemical modification during the gelation stage or via non-covalent attachment by mixing DNA aptamers within polymer gels) have not been successful due to the poor homogeneity, lack of reversibility, and undesirable leakage of DNA aptamers.29
The detection of the molecular DNA recognition and binding events was achieved here at the proof-of-principle level through hybridization between a DNA recognition motif CGATTCTGTGTT and a molecular beacon (MB) fluorescent probe F-5′-CGATTCGCCAAACACAGAATCG-3′-D, where F is fluorescein and D is dabcyl. The recognition motif was conjugated to a modified version of the self-assembling peptide and incorporated into the peptide hydrogel through co-assembly with the base peptide (Fig. 1). In isolation, the MB forms an internal six-residue stem region that holds the fluorophore and quencher together to form a FRET pair (Fig. S2, see the ESI†), leading to a complete quenching of fluorescence. The stem region can be displaced by hybridization between the remaining single-stranded residues of the MB and the recognition motif, due to the formation of a more stable duplex comprising 12 Watson–Crick complementary base pairs. This leads to the separation of fluorescein and dabcyl and generates a strong fluorescence signal. The base peptide Val-Lys-Val-Lys-Val-Glu-Val-Lys was used in this pilot study as it is well-known to form flexible fibres comprising antiparallel β-sheets stacked perpendicular to their long axes (where Val is valine, Lys is lysine, and Glu is glutamic acid).30 Above a critical concentration of 10 mM under physiological conditions these entangle to form hydrogels that contain >98% w/v water, with a swelling ratio of 51. The average fibre diameter was found to be 5.27 nm (SD = 0.54 nm, n = 50, Fig. S3, see the ESI†). Moreover, these short peptides have previously been shown to accommodate modified components within their structure, which are displayed and are accessible on the surface of the fibre.31–34 No significant difference in fibre morphology was observed with the addition of analyte oligonucleotides or sensing elements (average diameter 5.02 nm, SD = 0.67 nm, n = 50, Fig. 3).
The peptide–oligonucleotide conjugate was synthesised by covalently linking the oligonucleotide CGATTCTGTGTT recognition sequence to the modified self-assembling peptide Gly-Gly-Val-Lys-Val-Lys-Val-Glu-Val-Lys using thiol-maleimide chemistry (Fig. S4, see the ESI†). The peptide was synthesised using standard Fmoc-based solid phase synthesis procedures and functionalized with N-maleoyl β-alanine at the N-terminus. The oligonucleotide was functionalised with a hexyl-thiol group at the 5′ end. A two-glycine spacer was introduced to provide a flexible linker between the peptide fibres and the DNA recognition motif. The reaction was monitored using reversed-phase HPLC, and the isolated conjugate was characterised using 1H NMR spectroscopy and mass spectrometry (Fig. S5 and S6, see the ESI†).
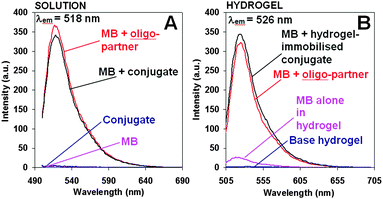
To validate the chosen system, hybridisation between the MB probe and its partner DNA was initially studied in solution (i.e. 100 mM TRIS buffer, pH 7.2, containing 200 mM KCl; full experimental details are given in the ESI†). In isolation, both components (MB and unbound peptide–oligonucleotide conjugate) were entirely fluorescently silent (Fig. 2(A); blue and magenta curves, respectively), but when mixed together in an equimolar ratio in solution, a 100-fold (or greater) increase in MB fluorescence was observed at 518 nm (λex 494 nm, Fig. 2(A); black). This strong fluorescence signal is presumably due to hybridisation of the MB with the oligonucleotide component of the conjugate, causing the MB FRET pair to separate. The intensity of the generated fluorescence band (Fig. 2(A); black) was very similar to that produced by hybridisation of the MB with the free oligonucleotide CGATTCTGTGTT (Fig. 2(A); red), indicating that the binding ability of the oligonucleotide is largely unaffected by its conjugation to the peptide.
 | ||
| Fig. 3 TEM images showing nanofibers that form Val-Lys-Val-Lys-Val-Glu-Val-Lys peptide hydrogels (20 mg ml−1, pH 7), co-assembled with 1 μM peptide–oligonucleotide conjugate and 1 μM partner to form the biosensor (A). Image analysis performed with ImageJ, using intensity profiles taken perpendicular to the fibre long axis (B). No significant difference in the fibre morphology was observed between blank hydrogels (Fig. S3, see the ESI†) and those with incorporated DNA-sensing components. The average fibre widths were 5.27 nm (SD = 0.54 nm, n = 50) and 5.02 nm (SD = 0.67 nm, n = 50), respectively. Full experimental details are given in the Materials and methods section. | ||
To determine whether the hydrogel environment had a detrimental effect on interactions between the MB and its partner, experiments identical to those described previously in TRIS buffer were performed, but within a hydrogel environment (21.5 mM base peptide, pH 7.2). As expected, the hydrogel was fluorescently silent (see Fig. 2(B); blue), while the MB showed some background fluorescence when physically mixed within the hydrogel (Fig. 2(B); magenta), presumably due to some degree of interaction between the MB and the hydrogel. The addition of the partner CGATTCTGTGTT to this system resulted in a 125-fold increase in the fluorescence as compared to the background fluorescence from the base hydrogel (Fig. 2(B); red). This showed a similar level of fluorescence enhancement as compared to that seen in the solution, demonstrating that the hydrogel did not have a detrimental effect on the hybridisation detection process. Interestingly, an 8 nm red-shift of the emission λmax was observed, which presumably suggests that there is an interaction between the DNA and the oppositely-charged peptide fibres.
The performance of a self-assembling DNA hydrogel biosensor was directly compared with the analogous detection system in solution using identical concentrations of the MB and conjugate. To form the hydrogel-based biosensor, non-functionalised peptide (21.5 mM, pH 7) was co-assembled with the peptide–oligonucleotide conjugate (1 μM; giving a doping level of 4.65 × 10−3% within the hydrogel, Fig. 1). As such the oligonucleotide sequence should be exposed on the surface of the fibres, ready to hybridize with the complementary MB. As expected, the functionalised hydrogel remained fluorescently silent over 24 hours of pre-incubation at 20 °C. Gratifyingly, a strong fluorescence was immediately observed upon the addition of the MB (Fig. 2(B); black), with a similar intensity to that observed previously in homogeneous solutions (i.e. in TRIS buffer). The fluorescence emission of this fully-assembled model system was also red-shifted by 8 nm showing a fluorescence emission at λmax at 526 nm. The rate of signal development was found to be approximately halved in a hydrogel environment compared with that in solution, as may be expected due to a slower rate of diffusion. The half-maximal signal was observed at 7.9 and 4.3 minutes after preparation, respectively (Fig. S7, see the ESI†).
To estimate the lowest limit of the conjugate doping level at which a detectable signal could still be confidently measured, an array of the hybridisation experiments were carried out (Fig. S8, see the ESI†), where the concentration of the peptide–oligonucleotide ranged from 200 to 0.02 nM. The base peptide concentration remained the same throughout the experiments at 16.1 mM to form weaker hydrogels, thus improving response times. The resulting doping level of the peptide–oligonucleotide conjugate relative to the non-functionalised peptide was varied, therefore, from 1.24 × 10−3 to 1.24 × 10−7%. To achieve this, 250 μl of the hydrogel biosensor containing different doping levels of the peptide–oligonucleotide conjugate was deposited into individual wells in a 96-well plate, and incubated for 1 h at 20 °C. The MB (50 μl aliquot) was introduced on top of hydrogel at molar concentrations identical to those of the DNA recognition components of the conjugate, and fluorescence spectra were recorded after 24 h of the incubation at 20 °C. As can be seen from Fig. S8B (see the ESI†), the lowest concentration of the conjugate in the hydrogel that allowed detection with a reproducible signal (with the signal-to-noise ratio of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was 50 pM. This corresponds to a conjugate doping level of only 3.11 × 10−7%. The lowest concentration at which an immediate response can be observed was 2 nM (1.24 × 10−5% doping, Fig. S8A, see the ESI†).
1) was 50 pM. This corresponds to a conjugate doping level of only 3.11 × 10−7%. The lowest concentration at which an immediate response can be observed was 2 nM (1.24 × 10−5% doping, Fig. S8A, see the ESI†).
These experiments allowed us to explore the possibility of reducing the overall cost of bio-assay systems by considerably lowering the doping levels of the functionalised peptides within the hydrogel. Taking this further, we evaluated the analytical performance and capability of this novel biosensor by estimating the limit of detection (LoD) for this model assay system. This corresponded to the lowest quantity of the MB that can be reliably distinguished from the ‘blank’ sample, with a 95% confidence interval. The detection limit was estimated from the mean value of the ‘blank’ samples (n = 132, Meanblank), the standard deviation of the ‘blank’ (SDblank) and the standard deviation of the ‘low concentration sample’ test replicates (n = 88, SDlcs) of a sample containing 50 pM analyte, using the published protocol.35 Firstly, the limit of ‘blank’ (LoB) was estimated using eqn (1):
| LoB = Meanblank + 1.645(SDblank) | (1) |
| LoD = LoB + 1.645(SDlcs) | (2) |
In conclusion, we have demonstrated that de novo designed peptide-based hydrogels can be used as a 3D solid support on which oligonucleotides can be immobilized and be used to detect complementary sequences, with a detection limit of 22 pM. The lowest doping level of the conjugate in the hydrogel that allowed the detection of a reproducible fluorescence signal (with the signal-to-noise ratio of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was found to be 3.11 × 10−7%. This has not been accomplished before, and compares favourably with established 2D systems that use similar detection techniques. The unique properties of peptide hydrogels also enable the biomolecule detection under biologically-relevant conditions, minimize unwanted probe–probe interactions, and have the potential to hugely increase the analyte storage capacity of these devices. Here we have described a proof-of-concept system in which recognition elements are covalently attached to the hydrogel, and an MB sequence was used as a detector to signify the hybridisation event. Future work will focus on reversing the system, attaching the MB fluorescent probe to the peptide fibres to enable detection of unmodified, biologically-relevant sequences. Once accomplished, this technology has the potential to be applied towards the detection of other biomolecules, and even small molecules through the use of aptamers, which is of pressing need in areas such as drug identification, biomedical diagnostics, and pollutants in environmental monitoring.
1) was found to be 3.11 × 10−7%. This has not been accomplished before, and compares favourably with established 2D systems that use similar detection techniques. The unique properties of peptide hydrogels also enable the biomolecule detection under biologically-relevant conditions, minimize unwanted probe–probe interactions, and have the potential to hugely increase the analyte storage capacity of these devices. Here we have described a proof-of-concept system in which recognition elements are covalently attached to the hydrogel, and an MB sequence was used as a detector to signify the hybridisation event. Future work will focus on reversing the system, attaching the MB fluorescent probe to the peptide fibres to enable detection of unmodified, biologically-relevant sequences. Once accomplished, this technology has the potential to be applied towards the detection of other biomolecules, and even small molecules through the use of aptamers, which is of pressing need in areas such as drug identification, biomedical diagnostics, and pollutants in environmental monitoring.
The authors would like to thank the EPSRC National Mass Spectrometry Centre in Swansea for MS analysis of the peptidyl-oligonucleotide conjugate samples. PK also thanks the EPSRC (EP/H04986X/1) for financial support. AS thanks the EPSRC for financial support (EP/K016210/1). This work was also supported by the Wellcome Trust ‘Research Consortia’ Scheme [105610/Z/14/Z]. All research data supporting this publication are directly available within this publication.
Notes and references
- F. Payne, J. D. Cooper, N. M. Walker, A. C. Lam, L. J. Smink, S. Nutland, H. E. Stevens, J. Hutchings and J. A. Todd, J. Leukocyte Biol., 2007, 81, 581–583 CrossRef CAS PubMed.
- A. P. Chiang, J. S. Beck, H. J. Yen, M. K. Tayeh and T. E. Scheetz, et al. , Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 6287–6292 CrossRef CAS PubMed.
- R. Kuwano, A. Miyashita, H. Arai, T. Asada and M. Imagawa, Hum. Mol. Genet., 2006, 15, 2170–2182 CrossRef CAS PubMed.
- Y. Takata, D. Hamada, K. Miyatake, S. Nakano, F. Shinomiya, C. R. Scafe, V. M. Reeve, D. Osabe, M. Moritani and K. Kunika, Arthritis Rheum., 2007, 56, 30–42 CrossRef CAS PubMed.
- M. A. Noureddine, X. J. Qin, S. A. Oliveira, T. J. Skelly and J. van der Walt, Hum. Genet., 2005, 117, 27–33 CrossRef CAS PubMed.
- E. Henke, J. Perk, J. Vider, P. Candia, Y. Chin, D. B. Solit, V. Ponomarev, L. Cartegni, K. Manova, N. Rosen and R. Benezra, Nat. Biotechnol., 2008, 26, 91–100 CrossRef CAS PubMed.
- J. Lamartine, J. Mater. Sci. Eng., 2006, 26, 354 CAS.
- F. Bertucci, R. Houlgatte, C. Nguyen, P. Viens, R. R. Jordan and D. Birnbaum, Lancet Oncol., 2001, 2, 674 CrossRef CAS PubMed.
- A. Charrier, N. Candoni, N. Liachenko and F. Thibaudan, Biosens. Bioelectron., 2007, 22, 1881–1886 CrossRef CAS PubMed.
- X. Yao, X. Li, F. Toledo, C. Zurita-Lopez, M. Gutova, J. Momand and F. Zhou, Anal. Biochem., 2006, 354, 220–228 CrossRef CAS PubMed.
- J. Pan, Biochem. Eng. J., 2007, 35, 183 CrossRef CAS.
- A. Mateescu, Y. Wang, J. Dostalek and U. Jonas, Membranes, 2012, 2, 40–69 CrossRef CAS PubMed.
- J. Liu, Soft Matter, 2011, 7, 6757–6767 RSC.
- A. Sassolas, B. D. Leca-Bouvier and L. J. Blum, Chem. Rev., 2008, 108, 109–139 CrossRef CAS PubMed.
- S. Draghici, P. Khatri, A. C. Eklund and Z. Szallasi, Trends Genet., 2006, 22, 101–109 CrossRef CAS PubMed.
- A. Abdullah-Sayani, J. M. Bueno-de-Mesquita and M. J. Van de Vijver, Nat. Clin. Pract. Oncol., 2006, 3, 501–516 CrossRef CAS PubMed.
- P. K. Tan, T. J. Downey, E. L. Spitznagel, P. Xu, D. Fu, D. S. Dimitrov, R. A. Lempicki, B. M. Raaka and M. C. Cam, Nucleic Acids Res., 2003, 31, 5676–5684 CrossRef CAS PubMed.
- E. V. Bichenkova, Z. Lang, X. Yu, C. Rogert and K. T. Douglas, Biochim. Biophys. Acta, Gene Regul. Mech., 2011, 1809(1), 1–23 CrossRef CAS PubMed; Y. Wang, A. Brunsen, U. Jonas, J. Dostalek and W. Knoll, Anal. Chem., 2009, 81, 9625–9632 CrossRef PubMed.
- Y. Wang, A. Brunsen, U. Jonas, J. Dostalek and W. Knoll, Anal. Chem., 2009, 81, 9625–9632 CrossRef CAS PubMed.
- Y. Helwa, N. Dave, R. Froidevaux, A. Samadi and J. Liu, ACS Appl. Mater. Interfaces, 2012, 4(4), 2228–2233 CAS.
- H. Hosseinkhani, P. Hong and D. Yu, Chem. Rev., 2013, 113, 4837–4861 CrossRef CAS PubMed.
- C. Sonmez, K. J. Nagy and J. P. Schneider, Biomaterials, 2015, 37, 62–72 CrossRef CAS PubMed.
- S. H. Gerhke, L. H. Uhden and J. F. McBride, J. Controlled Release, 1998, 55, 21–33 CrossRef.
- A. L. Boyle and D. N. Woolfson, Chem. Soc. Rev., 2011, 40, 4295–4306 RSC.
- D. Chow, M. L. Nunalee, D. W. Lim, A. J. Simnick and A. Chilkoti, Mater. Sci. Eng., 2008, 62, 125–155 Search PubMed.
- X. P. Yang, X. H. Pan, J. Blyth and C. R. Lowe, Biosens. Bioelectron., 2008, 23, 899–905 CrossRef CAS PubMed.
- P. Dubruel, L. Dekie, B. Christiaens, B. Vanloo, M. Rosseneu, J. Vandekerckhove, M. Mannisto, A. Urtti and E. Schacht, Biomacromolecules, 2003, 4, 1177–1183 CrossRef CAS PubMed.
- N. Dave, M. Y. Chan, P. J. Huang, B. D. Smith and J. Liu, J. Am. Chem. Soc., 2010, 132, 12668–12673 CrossRef CAS PubMed.
- M. Guvendiren, H. D. Lu and J. A. Burdick, Soft Matter, 2012, 8, 260–272 RSC.
- D. Roberts, C. Rochas, A. Saiani and A. F. Miller, Langmuir, 2012, 28, 16196–16206 CrossRef CAS PubMed.
- C. Hickling, H. S. Toogood, A. Saiani, N. Scrutton and A. F. Miller, Macromol. Rapid Commun., 2014, 35, 868–874 CrossRef CAS PubMed.
- S. Piluso, H. C. Cassell, J. L. Gibbons, T. E. Waller, N. J. Plant, A. F. Miller and G. Cavalli, Soft Matter, 2013, 9, 6752–6756 RSC.
- A. Maslovskis, N. Tirelli, A. Saiani and A. F. Miller, Soft Matter, 2011, 7, 6025–6033 RSC.
- S. Boothroyd, A. Saiani and A. F. Miller, Macromol. Symp., 2008, 273, 139–145 CrossRef CAS.
- D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29(1), S49–S52 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6cc01433j |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2016 |