Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Conversion of a non-heme iron-dependent sulfoxide synthase into a thiol dioxygenase by a single point mutation†
Kristina V.
Goncharenko
and
Florian P.
Seebeck
*
Department for Chemistry, University of Basel, St. Johanns-Ring 19, 4056, Basel, Switzerland. E-mail: florian.seebeck@unibas.ch
First published on 14th December 2015
Abstract
EgtB from Mycobacterium thermoresistibile catalyzes O2-dependent sulfur–carbon bond formation between the side chains of Nα-trimethyl histidine and γ-glutamyl cysteine as a central step in ergothioneine biosynthesis. A single point mutation converts this enzyme into a γ-glutamyl cysteine dioxygenase with an efficiency that rivals naturally evolved thiol dioxygenases.
Non-heme iron oxygenases catalyze a broad range of chemically difficult reactions and therefore provide intriguing starting points to develop novel enzymes for industrial applications. Despite remarkable progress in mechanistic enzymology,1–7 only few design studies describing modified non-heme iron enzymes with engineered activities8–12 have explored this potential for technological innovation so far. As a further step into this nascent field of enzyme engineering we present an example of a monooxygenase that was converted into an efficient thiol dioxygenase based on minimal active site redesign.13
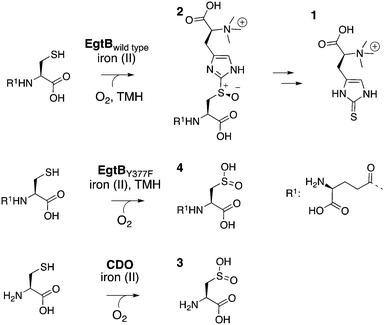
The sulfoxide synthase (EC 1.14.99.50) EgtB catalyzes the central step in ergothioneine biosynthesis (1, Fig. 1).14–16 EgtB mediates carbon–sulfur bond formation between γ-glutamyl cysteine (γGC) and the imidazole ring of Nα-trimethylated histidine (TMH). Concomitant oxidation of the bridging sulfur atom (2, Fig. 1) makes the overall reaction a four-electron oxidation, and classifies EgtB as a monooxygenase.17 The active site of EgtB contains ferrous iron coordinated by a 3-His facial triad, the thiolate side chain of γGC and the imidazole ring of TMH (Fig. 2).16 In this structure the likely O2-binding site is occupied by a water molecule or a hydroxide that also hydrogen bonds to the side chain of Gln55 (2.8 Å), a neighbouring water molecule (3.1 Å) and the side chain a Tyr377 (2.8 Å). In this report we show that substitution of Tyr377 to Phe completely changes the catalytic activity of EgtB.
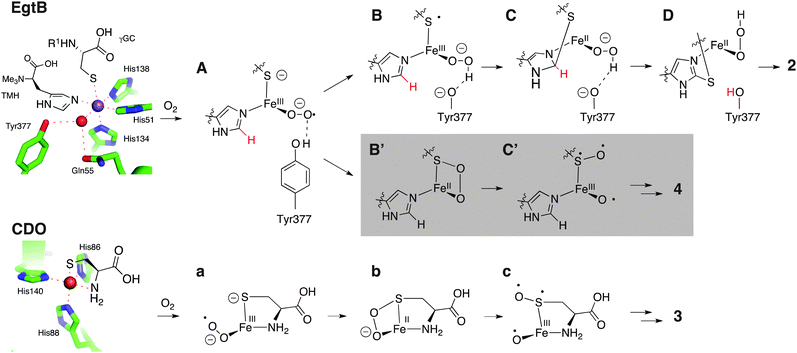 | ||
| Fig. 2 Top: Active site of EgtB from M. thermoresistibile in complex with Mn(II) (magenta sphere), TMH and γGC (PDB code 4X8D).16 One plausible catalytic mechanism has been proposed for EgtBwt: the substrate bound complex reacts with O2 to form an iron(III)–superoxo species (A). Protonation by Tyr377 and reduction by one-electron transfer from γGC leads to the iron(III)–hydroperoxo species (B). C–S bond formation between the γGC radical and TMH (C), deprotonation by Tyr377 and stereospecific sulfoxidation of D to the S-sulfoxide 2 concludes the catalytic cycle. Gray: in the absence of an acidic residue at position 377 species A predominantly reacts to B′ which reacts further to γGC dioxide (4) potentially through a CDO-like mechanism. Bottom: Active site of murine CDO in complex with cysteine (PDB code 4IEW).27 The consensus mechanism of CDO proceeds via a cysteine bond iron(III)–superoxo species (a), followed by irreversible S–O bond formation (b), homolytic O–O bond scission (c), and radical rebound to form cysteine sulfinic acid (3).18–26 | ||
The variant enzyme (EgtBY377F) catalyzes dioxygenation of γGC with an efficiency similar to that of naturally evolved cysteine dioxygenases (CDO, EC 1.13.11.20).18,19 CDOs also bind ferrous iron by a 3-His facial triad combined with the thiolate and amine ligands from the substrate (Fig. 2), but the overall structures of EgtB and CDO are unrelated. A wealth of structural, biochemical, spectroscopic and computational investigations suggest that CDO catalysed formation of cysteine sulfinic acid (3) goes though a cysteine bound iron(III)–superoxo species (a, Fig. 2), followed by intermediates b and c.18–26 The structural and functional similarities between the active sites of CDO and EgtB raise the possibility that thiol dioxygenation and sulfoxide synthesis may proceed through at least one common catalytic intermediate.
The following kinetic analysis of EgtBwt and EgtBY377F indicates that an iron(III)–superoxo species (A, a) may be this common intermediate.
To conduct this study we produced EgtBwt and EgtBY377F in Escherichia coli, and we monitored the rate of enzyme catalyzed sulfoxide (2) production using a published HPLC-based assay.16 Michaelis–Menten analysis of this data revealed 103-fold less sulfoxide synthase activity for EgtBY377F than for EgtBwt. This reduction is entirely due to a smaller kcat, since KM,TMH remained unchanged (Table 1, Fig. S1 and S2, ESI†). We also determined the substrate kinetic isotope effect (KIE) using C2-deuterated TMH. Because both enzymes showed a substrate KIE near unity (Fig. S3, ESI†) we concluded that C2–H bond cleavage is not rate limiting in either enzyme and that hydrogen or proton removal from TMH is not an essential function of Tyr377.
| pH | k cat,γGC (s−1) | K M,γGC (μM) | k cat/KM (M−1 s−1) | k cat,TMH (s−1) | K M,TMH (μM) | k cat/KM (M−1 s−1) | |
|---|---|---|---|---|---|---|---|
| a Standard deviation correspond to less than 20% of averaged value. Apparent kcat and kcat/KM in the presence co-substrate in a concentration at least 3-fold higher than the corresponding KM. | |||||||
| Sulfoxide synthase | |||||||
| EgtBwt | 8.0 | 7.5 × 10−1 | 27 | 2.8 × 104 | 8.5 × 10−1 | 10 | 7.4 × 104 |
| EgtBY377F | 8.0 | — | — | — | 0.9 × 10−4 | 10 | 8.5 × 101 |
| EgtBwt | 6.0 | 1.1 | 370 | 2.0 × 103 | 1.2 × 100 | 22 | 5.7 × 104 |
| EgtBY377F | 6.0 | — | — | — | 3.2 × 10−3 | 40 | 8.0 × 101 |
| γGC dioxygenase | |||||||
| EgtBY377F | 8.0 | 1.2 | 110 | 1.1 × 104 | — | — | — |
| EgtBY377F | 6.0 | 0.9 | 320 | 2.7 × 103 | — | — | — |
| CDOmurine19 | 7.5 | 1.8 | 700 | 2.6 × 104 | — | — | — |
When we compared the rates of γGC consumption and sulfoxide production (Fig. S4, ESI†) we found a rather surprising difference between the two enzymes. In EgtBwt catalyzed reactions substrate consumption (kγGC,wt = 0.4 ± 0.02 s−1) and sulfoxide formation (ksulfoxide,wt = 0.3 ± 0.06 s−1) were essentially coupled. By contrast, in EgtBY377F catalysed reactions, γGC consumption was much faster (kγGC,Y377F = 0.6 ± 0.1 s−1) than sulfoxide production (ksulfoxide,Y377F = 0.002 s−1). NMR analysis of the completed reactions revealed that EgtBY377F oxidizes most substrate to γGC dioxide (4) instead (Fig. S5 and S6, ESI†). This new activity still depends on TMH, which indicates that dioxygenation proceeds via the same substrate complex as sulfoxide production. In the absence of TMH both EgtBwt and EgtBY377F catalyzed γGC dioxygenation at a similarly slow rate (kγGC = 0.008 s−1, Fig. S7 and S8, ESI†). Apparently, the single substrate complex is significantly less reactive and its reaction specificity is not influenced by residue 377.
We also determined the Michaelis–Menten parameters for EgtBY377F catalyzed γGC dioxygenation (Table 1). Considering that this protein may never have evolved to catalyze this alternative reaction it is particularly striking that its catalytic efficiency closely matches that of naturally evolved CDOs (Table 1).
The kinetic parameters for EgtBY377F catalyzed dioxygenation are also remarkably similar to those for EgtBwt catalyzed sulfoxide synthesis (Table 1). Given the similar apparent KM in both enzymes for both substrates we have no indication that the substitution of residue 377 affected binding of TMH or γGC. The fact that both enzymes oxidize γGC at similar rates further suggest that the efficiency of O2 binding and activation have not changed either. However, the resulting ternary complex (A, Fig. 2) does behave quite differently in the wild-type and variant protein.
In the presence of ascorbate EgtBwt and EgtBY377F catalyze many hundreds of turnovers, without any sign of inactivation. Without ascorbate EgtBwt oxidizes to the inactive iron(III) form after approximately 100 turnovers,16 corresponding to an autoxidation rate of kautoxidation = 0.01 s−1 (Fig. S9, ESI†). This inactivation is reversible by addition of ascorbate,28,29 and is best explained by unproductive decay of the iron(III)–superoxo species A to superoxide and ferric EgtB.30–33 EgtBY377F inactivates 10-fold faster (kautoxidation = 0.1 s−1, Fig. S10, ESI†), indicating that the initial iron coordinated oxygen species may be destabilized by the Tyr377 to Phe substitution (Fig. 2).
This substitution also influences the solvent KIE on EgtB. The sulfoxide synthase activity of EgtBwt and the γGC dioxygenase activity of EgtBY377F are both characterized by a solvent KIE near unity (1.2 ± 0.2 and 0.9 ± 0.1, Fig. S11 and S12, ESI†). In contrast, the sulfoxide synthase activity of EgtBY377F exhibited a solvent KIE of 1.9 ± 0.1 (Fig. S11, ESI†), indicating that one or multiple protons or hydrogen atoms are being transferred in the rate limiting step. Because the dioxygenase activity is not affected by solvent deuteration we conclude that this transfer occurs exclusively on sulfoxide synthase pathway. In the context of the proposed mechanisms for EgtB and CDO (Fig. 2) the most likely candidate for this solvent isotope sensitive step would be protonation of the iron(III)–superoxo intermediate A. Protonation of this oxygen species may be important to increase the thiyl radical character of the γGC ligand (B), which in turn could attack the imidazole ring of TMH (C). In EgtBY377F the iron coordinated superoxide is not protonated an may instead attack the electron deficient sulfur atom on γGC (A to B′). The analogous S–O bond forming step (a to b) has been found as the first irreversible step in CDO catalyzed cysteine dioxygenation (Fig. 2).18–26
According to this model, species A can either react via irreversible proton transfer to intermediate B, or via irreversible S–O bond formation leading to intermediate B′ simply depending on the availability of an acidic proton in the active site. We did indeed observe a 3.5-fold increase in kcat for EgtBY377F catalyzed sulfoxide synthesis when the reaction pH was lowered from 8.0 to 6.0 (Table 1). The observed sulfoxide synthase activity is a hyperbolic function of proton concentration with a half-saturation point (KM,proton) near pH 7 (Fig. S13, ESI†). This dependence is consistent with a general acid mechanism in which the phosphate buffer (pKa,monoanion = 7.2), or an alternative protein residue with a similar pKa can replace Tyr377 as an indirect proton source. The kcat of γGC dioxygenase activity of the same protein proved nearly constant in the same pH range (Table 1) which is in agreement with the proposition that acid catalysis is less important in the first irreversible step of thiol dioxygenation (A to B′ or a to b′).
In conclusion, we identified Tyr377 as a catalytic residue in EgtB from M. thermoresistibile. Mutation of this residue to Phe did not measurably affect substrate binding or O2 activation, but instead changed the dominant activity of this enzyme. The remaining sulfoxide synthase activity of EgtBY377F is characterized by an increased solvent KIE and significant dependence on buffer pH. These observations are best explained with a mechanistic model suggesting that (i) the sulfoxide synthase and the thiol dioxygenase reaction pathways share a common intermediate, (ii) that this intermediate is the iron(III)–superoxo species A, and (iii) that protonation by Tyr377 is essential to move this species towards sulfoxide synthesis, and away from γGC dioxygenation. This report the first example of a non-heme iron enzyme which could be engineered to efficiently catalyze a completely different reaction type than the parent enzyme.
CDOs and EgtB belong to entirely unrelated protein families and evolved along different selective pressures. The fact that EgtBY377F catalyzes thiol dioxygenation with similar efficiency as CDOs makes the two enzymes a stunning example of accidental convergent evolution. We anticipate that detailed comparison of the two catalyst will prove a fruitful avenue to advance our current understanding of both reaction types.18–26
K. V. G. is a recipient of a Swiss Government Fellowship for Excellence; F. P. S. is supported by the “Professur für Molekulare Bionik” and by an ERC starting grant.
Notes and references
- J. E. Baldwin and M. Bradley, Chem. Rev., 1990, 90, 1079–1088 CrossRef CAS.
- J. C. Price, E. W. Barr, T. E. Glass, C. Krebs and J. M. Bollinger Jr., J. Am. Chem. Soc., 2003, 125, 13008–13009 CrossRef CAS PubMed.
- M. Costas, M. P. Mehn, M. P. Jensen and L. Que Jr., Chem. Rev., 2004, 104, 939–986 CrossRef CAS PubMed.
- E. G. Kovaleva and J. D. Lipscomb, Nat. Chem. Biol., 2008, 4, 186–193 CrossRef CAS PubMed.
- W. A. van der Donk, C. Krebs and J. M. Bollinger Jr., Curr. Opin. Stuct. Biol., 2010, 20, 1–11 CrossRef PubMed.
- M. M. Mbughuni, M. Chakrabarti, J. A. Hayden, E. L. Bominaar, M. P. Hendrich, E. Münck and J. D. Lipscomb, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16788–16793 CrossRef CAS PubMed.
- K. Ray, F. F. Pfaff, B. Wang and W. Nam, J. Am. Chem. Soc., 2014, 136, 13942–13958 CrossRef CAS PubMed.
- S. L. Groce and J. D. Lipscomb, J. Am. Chem. Soc., 2003, 125, 11780–11781 CrossRef CAS PubMed.
- J. Schlosrich, K. L. Eley, P. J. Crowley and T. D. H. Bugg, ChemBioChem, 2006, 7, 1899–1908 CrossRef CAS PubMed.
- S. M. Pratter, C. Konstantinovices, C. M. L. Giuro, E. Leitner, D. Kumar, S. P. de Visser, G. Grogan and G. D. Straganz, Angew. Chem., Int. Ed., 2013, 52, 9677–9681 CrossRef CAS PubMed.
- H. M. O'Hare, F. Huang, A. Holding, O. W. Choroba and J. B. Spencer, FEBS Lett., 2006, 580, 3445–3450 CrossRef PubMed.
- M. Gunsior, J. Ravel, G. L. Challis and C. A. Townsend, Biochemistry, 2004, 43, 663–674 CrossRef CAS PubMed.
- M. D. Toscano, K. J. Woycechowsky and D. Hilvert, Angew. Chem., Int. Ed., 2007, 46, 3212–3236 CrossRef CAS PubMed.
- Y. Ishikawa, S. E. Israel and D. B. Melville, J. Biol. Chem., 1974, 249, 4420–4427 CAS.
- F. P. Seebeck, J. Am. Chem. Soc., 2010, 132, 6632–6633 CrossRef CAS PubMed.
- K. V. Goncharenko, A. Vit, W. Blankenfeldt and F. P. Seebeck, Angew. Chem., Int. Ed., 2015, 54, 2821–2824 CrossRef CAS PubMed.
- A. Vit, G. T. Mashabela, W. Blankenfeldt and F. P. Seebeck, ChemBioChem, 2015, 16, 1490–1496 CrossRef CAS PubMed.
- B. S. Pierce, J. D. Gardner, L. J. Bailey, T. C. Brunold and B. G. Fox, Biochemistry, 2007, 46, 8569–8578 CrossRef CAS PubMed.
- W. Li and B. S. Pierce, Arch. Biochem. Biophys., 2015, 565, 49–56 CrossRef CAS PubMed.
- S. Aluri and S. P. de Visser, J. Am. Chem. Soc., 2007, 129, 14846–14847 CrossRef CAS PubMed.
- C. R. Simmons, K. Krishnamoorthy, S. L. Granett, D. J. Schuller, J. E. Dominy, T. P. Begley, M. H. Stipanuk and P. A. Karplus, Biochemistry, 2008, 47, 11390–11392 CrossRef CAS PubMed.
- D. Kumar, W. Thiel and S. P. de Visser, J. Am. Chem. Soc., 2011, 133, 3869–3882 CrossRef CAS PubMed.
- J. A. Crawford, W. Li and B. S. Pierce, Biochemistry, 2011, 50, 10241–10253 CrossRef CAS PubMed.
- E. Blaesi, J. D. Gardner, B. G. Fox and T. C. Brunold, Biochemistry, 2013, 52, 6040–6051 CrossRef CAS PubMed.
- R. J. Souness, T. Kleffmann, E. P. Tchesnokov, S. M. Wilbanks, G. B. Jameson and G. N. Jameson, Biochemistry, 2013, 52, 7606–7617 CrossRef CAS PubMed.
- E. J. Blaesi, B. G. Fox and T. C. Brunold, Biochemistry, 2014, 63, 5759–5770 CrossRef PubMed.
- C. M. Driggers, R. B. Cooley, B. Sankaran, L. L. Hirschberger, M. H. Stipanuk and P. A. Karplus, J. Mol. Biol., 2013, 425, 3121–3136 CrossRef CAS PubMed.
- G. T. Mashabela and F. P. Seebeck, Chem. Commun., 2013, 49, 7714–7716 RSC.
- H. Song, M. Leninger, N. Lee and P. Liu, Org. Lett., 2013, 15, 4854–4857 CrossRef CAS PubMed.
- R. E. Brantley, S. J. Smerdon, A. J. Wilkinson, E. W. Singelton and J. S. Olson, J. Biol. Chem., 1993, 268, 6993–7010 Search PubMed.
- S. C. Trewick, T. F. Henshaw, R. P. Hausinger, T. Lindahl and B. Sedgwick, Nature, 2002, 419, 174–178 CrossRef CAS PubMed.
- J. K. Crowell, W. Li and B. S. Pierce, Biochemistry, 2014, 53, 7541–7548 CrossRef CAS PubMed.
- I. G. Denisov and S. G. Sligar, in Cytochrome P450, ed. P. R. Ortiz de Montellano, Springer International Publishing, Switzerland, 2015 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S13, and detailed experimental procedures. See DOI: 10.1039/c5cc07772a |
| This journal is © The Royal Society of Chemistry 2016 |