Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multi-scale magnetic nanoparticle based optomagnetic bioassay for sensitive DNA and bacteria detection†
Bo
Tian
a,
Teresa
Zardán Gómez de la Torre
b,
Marco
Donolato
c,
Mikkel Fougt
Hansen
c,
Peter
Svedlindh
a and
Mattias
Strömberg
*a
aDepartment of Engineering Sciences, Division of Solid State Physics, Uppsala University, The Ångström Laboratory, Box 534, SE-751 21 Uppsala, Sweden. E-mail: mattias.stromberg@angstrom.uu.se; Tel: +46 18 471 3139
bDepartment of Engineering Sciences, Division of Nanotechnology and Functional Materials, Uppsala University, The Ångström Laboratory, Box 534, SE-751 21 Uppsala, Sweden
cDepartment of Micro- and Nanotechnology, Technical University of Denmark, DTU Nanotech, Building 345 East, DK-2800 Kongens Lyngby, Denmark
First published on 3rd June 2016
Abstract
Benefiting from their rapid readout, highly flexible devices and low-cost portable systems, optomagnetic biosensors have drawn increased attention in recent years as bioassay technologies for small molecules, biomarkers, DNA, and bacteria. Herein, an optomagnetic bioassay strategy suitable for point-of-care diagnostics, utilizing functionalized magnetic nanoparticles (100 nm) with Brownian relaxation behavior is optimized in order to obtain higher detection sensitivity for DNA molecules and bacteria. Presence of target DNA sequences or bacteria changes the dynamic behavior of the magnetic nanoparticles (binding to the target) and thus the optomagnetic response of the sample, which is measured by an optomagnetic setup including a 405 nm laser and a photodetector. The limit of detection is mainly set by the lowest measurable concentration of magnetic nanoparticles. Herein, as new results compared to previous work, we systematically optimize the concentration of 100 nm magnetic nanoparticles to increase the assay sensitivity and lower the limit of detection. To enable biplex detection, we perform this optimization in the presence of larger 250 nm magnetic nanoparticles that do not interact with the target. We show that the optimization and lowering of the 100 nm magnetic nanoparticle concentration result in a limit of detection of 780 fM of DNA coils formed by rolling circle amplification (size of about 1 μm) and 105 CFU per mL Salmonella (for immunoassay). These values are 15 times lower than those reported previously for this readout principle. Finally, we show that the 250 nm magnetic nanoparticles can serve as a second detection label for qualitative biplex detection of DNA coils formed by rolling circle amplification from V. cholerae and E. coli DNA coils using 100 nm and 250 nm magnetic detection nanoparticles, respectively.
1. Introduction
Point-of-care testing (POCT) is defined as simple on-site diagnostics that enables convenient and immediate testing for the patient.1 The development of robust, versatile, and low-cost POCT platforms is expected to have far-reaching implications in many areas such as clinical diagnostics, food safety, drug-abuse control, and lab research.2 The major limitations, hindering current techniques from becoming POCT tools, are the need of sample purification and enrichment of scarce targets.3 Due to the unique advantages of using magnetic nanoparticles (MNPs) for sample pretreatment, and the negligible magnetic background of biological samples, magnetic biosensor methods are considered to be promising tools for POCT.4,5Magnetic readout methods can be categorized mainly into two groups: volumetric and surface-based.5 In contrast to surface-based biosensors, volumetric detection methods are able to detect MNPs bound to biomolecules in the entire sample volume. This makes volumetric biosensors simple and fast3 and particularly suited for the detection of large biological analytes (e.g. proteins and nucleic acid sequences of 20 bases or more), since such molecules have large diffusion times (10–100 μm in the minutes-to-hours timescale).6 Magnetic susceptometers measure the change in the dynamic magnetic properties of MNPs upon probe-target recognition and represent a volumetric detection method that has been utilized to detect various biological targets, such as proteins,7–9 DNA,10,11 and bacteria.12 Compared to miniaturized nuclear magnetic resonance (NMR) systems, which represent an indirect volumetric detection system that detects the signal from H2O instead of MNPs; magnetic susceptometers have simpler readout setups and can achieve multiplex detection using MNPs of different sizes. However, due to the lack of an inherent signal amplification mechanism (while in NMR systems, millions of water molecules can be affected by an individual magnetic particle), magnetic susceptometers are not as sensitive as NMR devices.3,13,14
Optomagnetic sensor systems, which measure a modulation of an optical signal as a consequence of an AC magnetic field induced dynamic response of MNPs (related to Brownian relaxation), provide an alternative method for volumetric detection of MNP-labeled targets.15–17 A 405 nm laser based optomagnetic setup, first reported by Donolato et al.,17 has been utilized to detect ATP,18 biomarkers,19 DNA coils20,21 and bacteria.22 In this biosensor system the sample is subjected to an AC magnetic field and target quantification is accomplished by considering the frequency-dependent second harmonic component of the transmitted laser light intensity, V2 = V′2 + iV′′2 (details can be found in ESI, Section S1†). It should be stressed that the optomagnetic readout can be made compact and at low cost and is suited for use in conjunction with low-cost disposable plastic chips, therefore being suitable for point-of-care diagnostics. However, their reported performance for the detection of DNA coils has so far only been slightly better than that obtained using the magnetic susceptometers discussed above. It should be noted that the limit of detection (LOD) is set by the lowest concentration of MNP labels that can be measured by the setup.23 It is therefore of interest to investigate the impact of lowering of the MNP concentration on the LOD.
In previous work published by our group, the optomagnetic biosensor readout was employed for rolling circle amplification (RCA) product detection as well as for a competitive bacterial immunoassay.22,24,25 For DNA detection, DNA coils amplified from target DNA sequences were hybridized to 100 nm MNPs. For the competitive bacterial immunoassay, bacteria were captured by 5 μm magnetic particles (MPs) to form immuno-magnetic aggregates preventing the binding of 100 nm MNPs.
As new results compared to previous work, in the present study, we systematically optimize the concentration of 100 nm MNPs to increase the sensitivity for the detection of DNA coils and bacteria. Measurements at low MNP concentrations require adjustments of the photodetector sensitivity to avoid saturation as well as a careful blocking of the sample container. We show that an equivalent effect is obtained by including larger target-nonspecific MNPs with a diameter of 250 nm. Moreover, we show that the addition of the 250 nm MNPs does not reduce the sensitivity and LOD for the readout based on the 100 nm MNPs. When only the target detected by the 100 nm MNPs is present, we find an LOD for DNA coils formed from a Vibrio cholerae DNA target of 780 fM, which is 15 times lower than previously reported values.24,25 For a Salmonella typhimurium cell immunoassay, we obtain an LOD of 105 CFU per mL, which is also approximately 15 times lower than the value obtained previously.22 When the 100 nm and 250 nm MNPs are functionalized with different detection sequences, we demonstrate qualitative biplex detection of V. cholerae and E. coli DNA coils. It should be emphasized that, for Salmonella detection, although the bacterial immunoassay sensitivity of 105 CFU per mL of our system is close to or slightly higher than that of conventional enzyme-linked immuno-sorbent assay (ELISA)26,27 but lower than the sensitivity of other reported immunoassays (LOD sometimes as low as 103 to 104 CFU per mL),28–31 the proposed technique has the potential to enhance detection speed and accuracy since there is no need to immobilize the target molecule or to remove unbound target molecules.
2. Experimental section
2.1 Materials
Phi29 polymerase, phi29 buffer, ATP, T4 ligase, MQ water, dNTP mix and bovine serum albumin (BSA) were purchased from Thermo Fisher Scientific. Oligonucleotides used in this study were purchased from Biomers (Germany). All DNA sequences can be found in Table S1.† Ultrapure grade phosphate buffered saline (PBS, 20 ×) was purchased from AMRESCO (Solon, USA). Biotinylated goat anti avidin antibody was purchased from Vector Laboratories Inc. (USA). Biotinylated rabbit anti Salmonella groups was purchased from AbD Serotec. Avidin modified 5 μm MPs (product code 08-18-503, 25 mg mL−1, 3.5 × 108 particles per mL), streptavidin modified 100 nm MNPs (product code 10-19-102, 10 mg mL−1, 6 × 1012 particles per mL), streptavidin modified 250 nm MNPs (product code 09-19-252, 10 mg mL−1, 4.9 × 1011 particles per mL, used in the biplex detection) and protein A modified 250 nm MNPs (product code 09-20-252, 10 mg mL−1, 4.9 × 1011 particles per mL, used to optimize the detection) were purchased from Micromod (Rostock, Germany). Heat-killed Salmonella typhimurium cells (Catalog No. 50-74-01, KPL, Gaithersburg, USA) were used as the positive control sample in the immunoassay.2.2 Conjugation of magnetic particles and magnetic nanoparticles
Conjugations of biotinylated detection oligonucleotide probes to streptavidin modified 100 and 250 nm MNPs were performed according to protocols found in ESI, Section S2.† Conjugations of antibodies to avidin modified 5 μm MPs, streptavidin modified 100 nm MNPs, and protein A modified 250 nm MNPs were performed according to protocols found in ESI, Section S3.† Diluted MP and MNP suspensions were stored at 4 °C for use.2.3 Optomagnetic measurement setup
A detailed description including underlying theory is provided in ESI, Section S1.† The output from one measurement (used for target quantification) is the total intensity of transmitted laser light, V0, the in-phase, V′2, and the out-of-phase, V′′2, components of the complex second harmonic signal of the transmitted light. To compensate for variations in laser light intensity and particle concentration, V′2 was normalized with respect to the simultaneously measured value of V0. Measurements of V′2 and V0 at room temperature were performed in the frequency range 10–104 Hz or 1–104 Hz, using an AC magnetic field amplitude of 2.6 mT applied perpendicular to the propagation direction of the laser light. All measurements were carried out on sample volumes of 65 μL pipetted into a disposable UV-transparent cuvette (REF 67.758.001, SARSTEDT, Nümbrecht, Germany). The optical path through the cuvette was 10 mm.2.4 MNP detection strategies
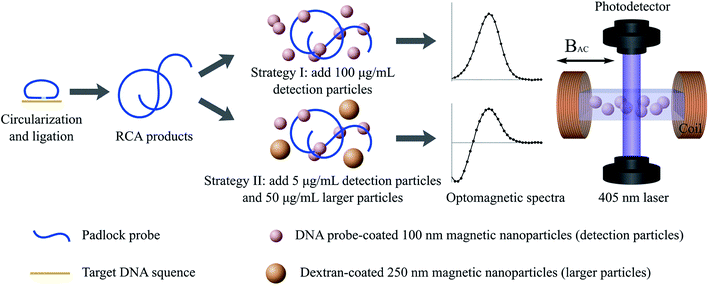
After establishing the detectable concentration ranges for the 100 nm MNPs, we compare two different detection strategies, which are summarized in Table 1.| c 100 nm [μg mL−1] | c 250 nm [μg mL−1] | |
|---|---|---|
| Strategy I | 100 | 0 |
| Strategy II | 5 | 50 |
The first detection strategy corresponds to that employed in previous work.17,20,24,25 The second strategy is that resulting from the optimization of the concentration of 100 nm MNPs. Moreover, it includes 250 nm MNPs with either a target-nonspecific coating (protein A or blocked protein A) or a target-specific coating. Note that the MNP concentrations in Table 1 refer to the values after mixing with the sample volume.
2.5 Padlock probe target recognition, ligation, RCA, and DNA coil detection
Solutions of DNA coils (V. cholerae or E. coli) were synthesized according to protocols found in ESI, Section S4.† Dilution series with DNA coil concentrations of ccoil ranging from 390 fM to 200 pM were prepared by stepwise dilution from the 1 nM DNA coil stock solution with hybridization buffer (0.1 M Tris–HCl, pH 8.0, 0.1 M EDTA, 0.5% Tween-20 and 2.5 M NaCl).DNA detection experiments were carried out by mixing 100 μL DNA coil solution with 100 μL MNP suspension to obtain final MNP concentrations as given in Table 1. The 100 nm MNPs were functionalized with V. cholerae detection probes and the 250 nm MNPs had protein A functional groups on the surface. The sample-MNP suspension was incubated at 55 °C for 20 min after which 65 μL of the suspension was transferred into a cuvette and measured in the optomagnetic setup. The total assay time for DNA detection, including amplification by RCA, was approximately 110 min.
The DNA detection approach is illustrated in Fig. 1; detection strategies I and II are illustrated in the top and bottom rows, respectively.
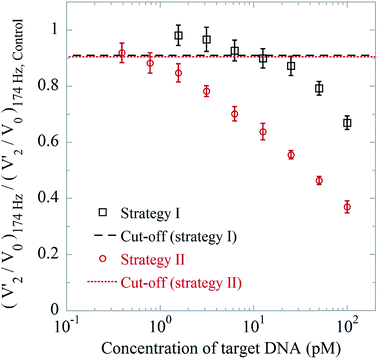
The cut-off value was calculated as the average of V′2/V0 at a frequency of f = 174 Hz for triplicate measurements using a blank control sample (hybridization buffer) minus three times the standard deviation of the triplicate. The frequency of 174 Hz was chosen as it corresponds to the position of the peak from 100 nm MNPs in the V′2/V0 spectra.
2.6 Direct immunoassay for salmonella
Dilution series (from 1.5 × 103 CFU per mL to 108 CFU per mL) containing Salmonella bacteria were prepared by stepwise dilution of a bacteria stock suspension with PBS. In the experiments, the 100 nm MNPs were conjugated with anti Salmonella antibodies and 250 nm protein A (blocked with irrelevant antibodies) MNPs were used. Detection experiments according to strategy I or II were carried out by mixing 2 μL MNP suspension with 198 μL bacteria suspension and incubated at 37 °C for 1 h. The mixture was shaken every 20 min to avoid sedimentation. After incubation, 65 μL of the suspension was transferred into a disposable UV-transparent cuvette for analysis. The total assay time for immunoassay was approximately 70 min.The cut-off value was calculated as the average of V′2/V0 at f = 137 Hz for a triplicate measurement using a blank control sample (PBS) minus three times the standard deviation of the triplicate.
2.7 Biplex detection for E. coli and V. cholerae DNA coils
Four samples (blank sample, 100 pM E. coli DNA coils, 100 pM V. cholerae DNA coils, and a mixture containing 100 pM E. coli and 100 pM V. cholerae DNA coils) were characterized. One hundred microliter of MNP suspension conjugated with V. cholerae (100 nm MNPs) and E. coli (250 nm MNPs) detection probes was added into 100 μL of DNA coil sample. The MNP concentrations were chosen according to detection strategy II. The mixture was incubated at 55 °C for 20 min, and thereafter transferred into a cuvette for analysis.3. Results and discussion
3.1 Determination of the linear range for detection of 100 nm MNPs
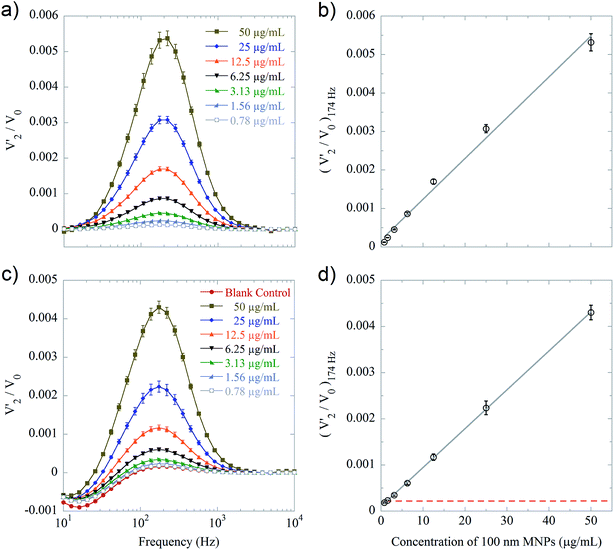
The total intensity of transmitted laser light, V0, the in-phase, V′2, and the out-of-phase, V′′2, components of the complex second harmonic signal of the transmitted light, were recorded outputs from the analysis and used for target quantification. The frequency dependence of V′2/V0 is closely related to the Brownian relaxation dynamics of the used MNPs (details can be found in ESI, Section S1†). For free 100 nm MNPs, a characteristic peak in the V′2/V0 spectrum is located at approximately 170 Hz. MNPs bound to their matching target have a much larger hydrodynamic size and thus their signal is shifted to frequencies below the lowest frequency used in the measurements. Thus, the binding of MNPs is observed as a decrease of the amplitude of the peak from free MNPs in the V′2/V0 spectra. This reduction can be used to quantify the target.As mentioned above, the sensitivity of the system is strongly correlated to the concentration of detection MNPs used in the assay. For lower concentrations of detection MNPs, a larger fraction of the MNPs will be affected by binding to a certain target concentration. Thus, the sensitivity is expected to be improved by lowering the concentration of detection MNPs until the noise dominates. In previous work, concentrations of around 100 μg mL−1 of 100 nm detection MNPs have been employed for bioassays utilizing the present setup.17,20,24,25 To investigate the linear detection range for a single species of MNPs, suspensions of 100 nm detection MNPs with MNP concentrations c100 nm between 100 μg mL−1 to 0.78 μg mL−1 were measured in triplicate and the average value of (V′2/V0)174 Hz was calculated. For fixed settings of the setup, we first found a linear signal for MNPs from c100 nm = 6.25 to 100 μg mL−1 (R2 > 0.997) (Fig. S3†) and that lower concentrations of MNPs were essentially undetectable. Further studies established that this observation resulted from:
(1) Saturation of the transimpedance amplifier of the photodetector due to the higher transmissivity of the suspension for lower c100 nm (Fig. S4†). This corresponded to values of V0 of about 10 V and could be mitigated by either reducing the gain of the transimpedance amplifier or by introducing an absorbing material in the light path (Fig. S5a†).
(2) Nonspecific binding of the detection MNPs to the cuvette wall. This nonspecific binding affected a significant fraction of the MNPs for lower c100 nm. We found that it could be mitigated by adding 0.1% BSA to the reaction buffer. The comparison of the results obtained with and without blocking can be seen in Fig. S3 and S5b.†
Using optimum conditions (photodetector gain reduction and 0.1% BSA blocking), we obtained the spectra shown in Fig. 2a with the values of (V′2/V0)174 Hzvs. c100 nm shown in Fig. 2b. The results correspond to a linear detection range from c100 nm = 0.78 to 50 μg mL−1 (R2 > 0.992).
Fig. 2c and d show the results of a study of (V′2/V0)174 Hzvs. c100 nm where 250 nm protein A MNPs have been added to a final concentration of 50 μg mL−1. Due to the higher absorption of the mixed MNP suspension, these measurements were not affected by saturation of the photodetector (Fig. S4†). Moreover, the larger MNPs seemed to preferentially bind to and block the cuvette surface such that results for the 100 nm MNPs with and without BSA blocking were essentially the same. The results correspond to essentially the same linear detection range of c100 nm from 0.78 to 50 μg mL−1 (R2 > 0.999). We also tested the effect of using larger MNPs in other concentrations (25 μg mL−1 and 75 μg mL−1, data not shown), and the results were similar to the test using 50 μg mL−1. The spectra from the two MNP types are well separated such that the signal from the 250 nm MNPs has very little overlap with the peak due to 100 nm MNPs. Also, the V′2 peak has positive and negative sign for 100 nm and 250 nm MNPs, respectively (as mentioned in ref. 17, the sign of the peak is determined by the MNP size, susceptibility and laser wavelength). The appearance of well separated V′2 spectra is likely the reason that the same range of 100 nm MNP concentrations can be resolved in the two cases. Thus, we have found that the same sensitivity to 100 nm MNPs can be obtained for a single population of MNPs under optimal conditions (BSA blocking, reduced photodetector gain) as for a mixed population of MNPs without blocking.
Below, we compare the results obtained using only 100 nm MNPs with a final concentration of 100 μg mL−1 corresponding to previously published results (detection strategy I) with those obtained using 100 nm MNPs with a final concentration of 5 μg mL−1 in the presence of 50 μg mL−1 of 250 nm protein A MNPs (detection strategy II). This comparison reveals the improvement obtained by reducing the concentration of the 100 nm MNPs and also demonstrates that this improvement can be obtained even in the presence of a population of larger MNPs. Finally, we demonstrate that the two populations of MNPs can be used for biplex detection.
3.2 Optimized bioassay for DNA detection
We employ and compare the result of detection strategy II with those obtained using detection strategy I for the readout of DNA coil concentrations. The average values of (V′2/V0)174 Hz, normalized with respect to the peak value of a blank control (hybridization buffer), are shown in Fig. 3. In detection strategy I (black squares in Fig. 3), the LOD was 12.5 pM, which corroborated previous results (LODs of approximately a few pM in the measurement volume).17,20,24,25 From the results of detection strategy II (red circles in Fig. 3), we found an LOD of 780 fM, which is about 15 times lower. The V′2/V0 spectra of the DNA coil measurements are shown in Fig. S6.†3.3 Optimized bioassay for immuno-detection
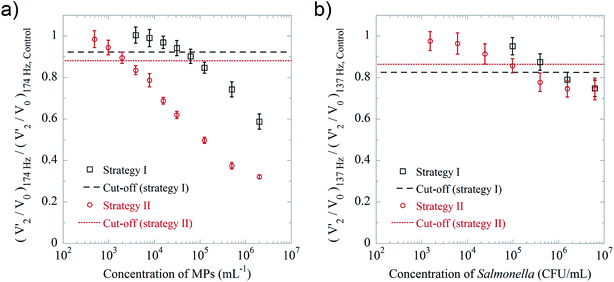
In the previous work we have used biotinylated goat anti avidin antibody conjugated 5 μm MPs as a bacteria model since they benefit from a large number of binding sites and strong binding reaction between streptavidin and biotin.22 To compare the two presented detection strategies, we again used this bacteria model and streptavidin coated 100 nm MNPs as labels (detection particles). The 250 nm MNPs used in detection strategy II were blocked with irrelevant antibodies. Fig. 4a shows the average values of (V′2/V0)174 Hz, normalized to those obtained for the blank control, vs. the MP concentration. We found LODs of 6 × 104 and 4 × 103 particles per mL for detection strategies I and II, respectively. This means that, in an ideal situation, e.g., abundant binding sites and strong reactions, the use of detection strategy II can improve the sensitivity of the optomagnetic immunoassay by a factor of 15 compared to detection strategy I. This is due to the lower concentration of detection MNPs in detection strategy II. The V′2/V0 spectra of the MP measurements are shown in Fig. S7.†To further evaluate the applicability of the optomagnetic immunoassay, a Salmonella cell detection was performed using the two strategies. The protocol was similar to that used for the “bacteria model” detection, except that the 100 nm MNPs were coated with biotinylated rabbit anti Salmonella antibodies, and the peak frequency of antibody coated 100 nm MNPs was about 137 Hz. From the dose–response curve shown in Fig. 4b, we found LODs of 1.6 × 106 and 105 CFU per mL for detection strategies I and II, respectively. Thus, also in this case, the lower concentration of 100 nm detection MNPs in detection strategy II results in a factor of 15 improvement compared to detection strategy I. The V′2/V0 spectra of the Salmonella immunoassay are shown in Fig. S8.† The reduction of the normalized values of (V′2/V0)137 Hz for the Salmonella detection was not as large as for the MP detection. This can be attributed to three major reasons: (1) the affinity of the antibody–antigen pair (dissociation constant, Kd, ranges from 10−11 to 10−8 M) is at least 104 times weaker than the streptavidin-biotin pair (Kd = 10−15 M); (2) once the detection MNPs bind to small bacterial fragments in the suspension, their hydrodynamic size only slightly changes, thus the peak reduction is not significant; (3) the total intensity of the transmitted laser light, V0, is strongly influenced by the amount of bacteria suspended in the sample, especially for bacterial concentrations higher than 106 CFU per mL, leading to an increase of (V′2/V0)137 Hz.
3.4 Biplex E. coli and V. cholerae target DNA sequence detection
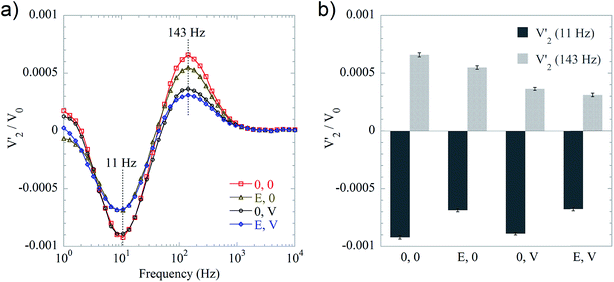
A biplex DNA detection format was evaluated to demonstrate the usefulness of including 250 nm MNPs as detection labels. In the biplex bioassay strategy, we employed 100 and 250 nm MNPs as detection labels that were able to bind to DNA coils originating from V. cholerae and E. coli, respectively. The V′2 spectrum of non-bound probe conjugated 250 nm MNPs was characterized by a valley located at 11 Hz, while the V′2 spectrum of non-bound probe conjugated 100 nm MNPs was characterized by a peak at 143 Hz. Four representative samples were measured, namely a blank sample (hybridization buffer), a sample containing 100 pM E. coli target sequence sample, a sample containing 100 pM V. cholerae target sequence sample, and a sample containing a mixture of both of these two target sequences. As shown in Fig. 5a, each sample has its own signature in the V′2 spectrum in terms of valleys/peaks. For samples containing the E. coli target sequence (yellow triangles and blue diamonds), the absolute values of the valleys are smaller than the blank sample (red squares). For the spectra of V. cholerae target sequence containing samples, i.e., black circles and blue diamonds, the peak at 143 Hz is lower than for the blank sample. Valley/peak values of the four representative samples are presented in the bar graph Fig. 5b. From the biplex detection results, we conclude that the setup with detection strategy II can be further applied for qualitative multiplex bioassays by using MNPs of different hydrodynamic sizes.4. Conclusions
In this work, as new results compared to our previous studies, we aimed to study the impact of lowering the concentration of 100 nm magnetic nanoparticle detection labels in an optomagnetic bioassay. We found that the concentration of 100 nm MNPs can be significantly reduced, but also as a consequence that care should be taken to avoid saturation of the photodetector and nonspecific binding of the MNPs for low concentrations. Under optimized conditions, we found a concentration range of MNPs giving rise to a linear signal of 0.78 to 50 μg mL−1. We have systematically investigated the impact of using an MNP concentration of 5 μg mL−1 and compared the results to those obtained for a concentration of 100 μg mL−1, which has been used in previous studies in the literature. Further, to prepare for biplex detection, we have added a population of 250 nm MNPs to study the impact on the signal for the 100 nm MNPs. We found that the larger MNPs tended to reduce the transmitted light intensity such that saturation effects of the photodetector were avoided. Further, protein A functionalized particles seemed to preferentially block the sample cuvette such that the nonspecific binding of the 100 nm MNPs was reduced. In our studies, we found that the same concentration range of 100 nm MNPs could be resolved with and without the presence of the larger MNPs. The main reason for this is that the spectra from the two MNP sizes only overlap very little. We compared the performance of assays detecting DNA coils and E. coli cells using 5 μg mL−1 of 100 nm MNPs with that using 100 μg mL−1 of 100 nm MNPs and found in both cases a factor of 15 improvement. Finally, we demonstrated qualitative biplex detection of DNA coils formed from V. cholerae and E. coli using the two MNP populations.Acknowledgements
This work was financially supported by Swedish Research Council Formas (Project no. 221-2012-444, 221-2014-574 and 2011-1692) and The Swedish Foundation for Strategic Research.References
- P. B. Luppa, C. Muller, A. Schlichtiger and H. Schlebusch, TrAC, Trends Anal. Chem., 2011, 30, 887–898 CrossRef CAS.
- V. Gubala, L. F. Harris, A. J. Ricco, M. X. Tan and D. E. Williams, Anal. Chem., 2012, 84, 487–515 CrossRef CAS PubMed.
- H. Lee, T. H. Shin, J. Cheon and R. Weissleder, Chem. Rev., 2015, 115, 10690–10724 CrossRef CAS PubMed.
- H. C. Tekin and M. A. Gijs, Lab Chip, 2013, 13, 4711–4739 RSC.
- D. Issadore, Y. I. Park, H. Shao, C. Min, K. Lee, M. Liong, R. Weissleder and H. Lee, Lab Chip, 2014, 14, 2385–2397 RSC.
- S. O. Kelley, C. A. Mirkin, D. R. Walt, R. F. Ismagilov, M. Toner and E. H. Sargent, Nat. Nanotechnol., 2014, 9, 969–980 CrossRef CAS PubMed.
- A. P. Astalan, F. Ahrentorp, C. Johansson, K. Larsson and A. Krozer, Biosens. Bioelectron., 2004, 19, 945–951 CrossRef CAS PubMed.
- C. Y. Hong, C. C. Wu, Y. C. Chiu, S. Y. Yang, H. E. Horng and H. C. Yang, Appl. Phys. Lett., 2006, 88, 212512 CrossRef.
- K. Enpuku, Y. Tamai, T. Mitake, M. Matsuo, A. Tsukamoto, T. Mizoguchi and A. Kandori, Appl. Phys. Express, 2009, 2, 037001 CrossRef.
- M. Strömberg, J. Göransson, K. Gunnarsson, M. Nilsson, P. Svedlindh and M. Strømme, Nano Lett., 2008, 8, 816–821 CrossRef PubMed.
- T. Zardán Gómez de la Torre, A. Mezger, D. Herthnek, C. Johansson, P. Svedlindh, M. Nilsson and M. Strømme, Biosens. Bioelectron., 2011, 29, 195–199 CrossRef PubMed.
- H. L. Grossman, W. R. Myers, V. J. Vreeland, R. Bruehl, M. D. Alper, C. R. Bertozzi and J. Clarke, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 129–134 CrossRef CAS PubMed.
- J. B. Haun, T. J. Yoon, H. Lee and R. Weissleder, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 291–304 CrossRef CAS PubMed.
- E. Harel, L. Schroder and S. J. Xu, Annu. Rev. Anal. Chem., 2008, 1, 133–163 CrossRef CAS PubMed.
- K. Aurich, S. Nagel, G. Glockl and W. Weitschies, Anal. Chem., 2007, 79, 580–586 CrossRef CAS PubMed.
- S. H. Chung, M. Grimsditch, A. Hoffmann, S. D. Bader, J. Xie, S. Peng and S. Sun, J. Magn. Magn. Mater., 2008, 320, 91–95 CrossRef CAS.
- M. Donolato, P. Antunes, R. S. Bejhed, T. Zardán Gómez de la Torre, F. W. Østerberg, M. Strömberg, M. Nilsson, M. Strømme, P. Svedlindh, M. F. Hansen and P. Vavassori, Anal. Chem., 2015, 87, 1622–1629 CrossRef CAS PubMed.
- J. Yang, M. Donolato, A. Pinto, F. G. Bosco, E. T. Hwu, C. H. Chen, T. S. Alstrom, G. H. Lee, T. Schafer, P. Vavassori, A. Boisen, Q. Lin and M. F. Hansen, Biosens. Bioelectron., 2016, 75, 396–403 CrossRef CAS PubMed.
- P. Antunes, D. Watterson, M. Parmvi, R. Burger, A. Boisen, P. Young, M. A. Cooper, M. F. Hansen, A. Ranzoni and M. Donolato, Sci. Rep., 2015, 5, 16145 CrossRef CAS PubMed.
- M. Donolato, P. Antunes, T. Zardán Gómez de la Torre, E. T. Hwu, C. H. Chen, R. Burger, G. Rizzi, F. G. Bosco, M. Strømme, A. Boisen and M. F. Hansen, Biosens. Bioelectron., 2015, 67, 649–655 CrossRef CAS PubMed.
- A. Mezger, J. Fock, P. Antunes, F. W. Østerberg, A. Boisen, M. Nilsson, M. F. Hansen, A. Ahlford and M. Donolato, ACS Nano, 2015, 9, 7374–7382 CrossRef CAS PubMed.
- B. Tian, R. S. Bejhed, P. Svedlindh and M. Strömberg, Biosens. Bioelectron., 2015, 77, 32–39 CrossRef PubMed.
- Y. R. Chemla, H. L. Crossman, Y. Poon, R. McDermott, R. Stevens, M. D. Alper and J. Clarke, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 14268–14272 CrossRef CAS PubMed.
- R. S. Bejhed, T. Zardán Gómez de la Torre, M. Donolato, M. F. Hansen, P. Svedlindh and M. Strömberg, Biosens. Bioelectron., 2015, 66, 405–411 CrossRef CAS PubMed.
- R. S. Bejhed, T. Zardán Gómez de la Torre, P. Svedlindh and M. Strömberg, Biotechnol. J., 2015, 10, 469–472 CrossRef CAS PubMed.
- L. P. Mansfield and S. J. Forsythe, Lett. Appl. Microbiol., 2000, 31, 279–283 CrossRef CAS PubMed.
- O. Lazcka, F. J. Del Campo and F. X. Munoz, Biosens. Bioelectron., 2007, 22, 1205–1217 CrossRef CAS PubMed.
- Y. Y. Wong, S. P. Ng, M. H. Ng, S. H. Si, S. Z. Yao and Y. S. Fung, Biosens. Bioelectron., 2002, 17, 676–684 CrossRef CAS PubMed.
- L. J. Yang and Y. B. Li, Analyst, 2006, 131, 394–401 RSC.
- S. H. Ko and S. A. Grant, Biosens. Bioelectron., 2006, 21, 1283–1290 CrossRef CAS PubMed.
- W. Chunglok, D. K. Wuragil, S. Oaew, M. Somasundrum and W. Surareungchai, Biosens. Bioelectron., 2011, 26, 3584–3589 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ay00721j |
| This journal is © The Royal Society of Chemistry 2016 |