Analysis of pre-miR-29b binding conditions to amino acids by using a surface plasmon resonance biosensor
Patrícia
Pereira
a,
Carla
Cruz
a,
João A.
Queiroz
a,
Ana
Figueiras
ab and
Fani
Sousa
*a
aCICS-UBI – Health Sciences Research Centre, University of Beira Interior, Avenida Infante D. Henrique, 6200-506 Covilhã, Portugal. E-mail: fani.sousa@fcsaude.ubi.pt; Fax: +351 275 329 099; Tel: +351 275 329 074
bCNC – Center of Neuroscience and Cell Biology, University of Coimbra, Largo Marquês de Pombal, 3004-517 Coimbra, Portugal
First published on 13th November 2015
Abstract
The aim of this work was to provide binding information between the recombinant pre-miR-29b and L-arginine/L-lysine by surface plasmon resonance (SPR) and circular dichroism (CD). This information brings important insights concerning the characterization of the microRNA binding onto chromatographic supports using these amino acids as specific ligands. Moreover, it is possible to determine some particular conditions enabling the improvement of the binding specificity of the amino acid ligands used to purify microRNA, preserving their integrity. The binding responses were detectable and reproducible, allowing the determination of the equilibrium dissociation constant (KD). Considering the binding affinities, it was verified that the pre-miR-29b binds more strongly to L-arginine (KD between 10−6 and 10−7 M) than to L-lysine (KD between 10−5 and 10−7 M). Remarkably, the results revealed that the ligands possess high affinity to RNA molecules using buffers with low salt concentrations and no binding responses were detected with high salt concentrations, suggesting that electrostatic interactions are mainly responsible for the ligand–analyte interaction. Above all, this study showed the importance of SPR for future screening of other ligands that, like the ones described herein, can be used to design novel microRNA purification platforms which will have a significant impact on biopharmaceutical-based therapeutics.
Introduction
Over the past decade, several hundred small non-coding RNA molecules, known as microRNAs (miRNAs) have emerged as potential biomarkers and drug targets, appearing to be promising for the next generation of diagnostic and therapeutic tools.1–3 These RNA molecules play important roles in modulating various biological and cellular functions and have potential to treat several pathologies such as cancer, cardiovascular and neurodegenerative diseases. This suggests the use of miRNA as a promising approach from a therapeutic point of view.4–6 Human pre-miR-29b was chosen for this work because it belongs to one of the most interesting miRNA families identified in humans, being involved in several regulatory actions, namely in tumor-suppressing and immune-modulating processes, as well as in the regulation of pathways of neurodegenerative diseases.5–7Hence, considering the potential therapeutic application and the mandatory global distribution of these novel therapeutics, it is crucial to develop efficient methods for isolation of pharmaceutical grade miRNAs, with high purity degree and biological activity, envisioning their application in gene therapy. Several strategies are available to isolate and purify miRNA molecules that are chemically synthesized or derived from various biological sources.8,9 However, these methods make use of denaturing conditions and structural modifications in the RNA molecule can occur, by the introduction of tag-sequences. Consequently, these changes can induce RNA conformational alterations which may compromise their activity, and in addition, these procedures are expensive and time-consuming.10,11 More recently, our research group developed an affinity chromatographic method with amino acids as specific ligands, named amino acid-affinity chromatography, applied to efficiently purify different RNA species (total RNA, ribosomal RNA, small RNA, 6S RNA and recombinant pre-miR-29b), on the basis of their biological function or individual chemical structure.12–17 For these reasons, this chromatographic method can be an attractive approach for the miRNA purification, since it can be successfully employed in the purification of a wide range of biomolecules, based on specific and reversible interactions, simulating natural biological processes that are established within the cell. In addition, affinity chromatography is a selective and reliable technique, offering reduced processing time, and capable of preparing therapeutic biomolecules with strict quality assurance.18 However, the problem of finding a suitable ligand in affinity chromatography is not restricted to specificity, but concerns also the binding strength and the kinetics of the ligand–miRNA interaction. This implies that a ligand with an optimized dissociation constant will facilitate the successful operation of affinity chromatography for miRNA purification.
In the past recent years, Cruz and collaborators applied surface plasmon resonance (SPR) for the analysis of the interaction between plasmid DNA and amino acids ligands.19–22 This technique provides both equilibrium and kinetic information about intermolecular interactions and is a powerful tool to study the dynamics of nucleic acid–amino acid interactions and, consequently, to apply these important structural data in the optimization of chromatographic systems.23–26 According to these considerations, a comparative study was performed to characterize and understand the interactions established between pre-miR-29b and L-arginine and L-lysine amino acids. For this purpose, SPR was applied to study the affinity and the binding responses between recombinant pre-miR-29b and L-arginine and L-lysine immobilized on a surface to mimic the affinity support under different experimental conditions, such as the composition and ionic strength of the elution buffer and temperature. This study revealed several interesting characteristics of RNA molecules, including natural affinity interactions that can occur between L-arginine and L-lysine and pre-miRNA, providing important information that can be further applied in unveiling the miRNA chromatographic behavior in these systems. Furthermore, the experimental data allowed us to get binding information to improve pre-miR-29b purification with low sample consumption, using a rapid and automated analysis. Moreover, circular dichroism (CD) was used to evaluate the conformational changes that pre-miR-29b may suffer when exposed to certain experimental conditions. In particular, experimental conditions can influence the interactions established between pre-miRNA and amino acids, as well as the structure of RNA, leading to repulsion or neutralization of the biomolecules, or even inducing stabilization or denaturation of the target RNA.
Materials and methods
Materials
L-arginine–Sepharose 4B gel was acquired from Amersham Biosciences (Uppsala, Sweden) and Lysine-Sepharose 4B was obtained from GE Healthcare Biosciences (Uppsala, Sweden). The guanidinium salt and all the chemicals used in the lysis buffer were obtained from Sigma-Aldrich (St Louis, MO, USA). All buffers used for the chromatographic experiments were freshly prepared with sterilized water pre-treated with 0.05% diethyl pyrocarbonate (DEPC; Sigma-Aldrich, St Louis, MO, USA), filtered through a 0.20 μm pore size membrane (Schleicher Schuell, Dassel, Germany) and degassed ultrasonically. The sodium chloride (NaCl) used was purchased from Panreac (Barcelona, Spain), tris(hydroxymethyl) aminomethane (Tris), HEPES sodium salt and L-arginine were from Sigma-Aldrich (St Louis, MO, USA). L-Lysine Hydrochloride was from USB, 3-(N-morpholino)propanesulfonic acid (MOPS) was from Fisher Scientific and, finally, potassium dihydrogen phosphate was from Chem-Lab NV (Zedelgem, Belgium). All the materials used in the experiments were RNase-free.Pre-miR-29b biosynthesis and isolation
The pre-miR-29b used in the experiments was produced by a cell culture of Rhodovulum sulfidophilum DSM 1374 strain (BCCM/LMG, Belgium) modified with the pBHSR1-RM plasmid containing the sequence of pre-miR-29b.27 Growth was carried out at 30 °C under dark-aerobic conditions, using Nutrient Broth medium (1 g L−1 beef extract; 2 g L−1 yeast extract; 5 g L−1 peptone and 30 g L−1 sodium chloride) supplemented with 30 μg mL−1 kanamycin. Cells were recovered by centrifugation and stored at −20 °C. Small RNA was extracted using the acid guanidinium thiocyanate–phenol–chloroform extraction method based on the protocol described by Chomczynski and co-workers.28 Briefly, cells were lysed by adding 5 mL of denaturing cell lysis solution (4 M guanidinium thiocyanate; 25 mM sodium citrate, pH 4.0; 0.5% N-laurosylsarcosine and 0.1 M β-mercaptoethanol). After incubating on ice for 10 min, cellular debris, genomic DNA and proteins were precipitated by gently adding and mixing 5 mL of water–saturated phenol and 0.5 mL of 2 M sodium acetate (pH 4.0). The RNA isolation was achieved by adding 1 mL of chloroform/isoamyl alcohol (49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and by mixing vigorously until two immiscible phases were obtained. The upper aqueous phase, which contained mostly RNA, was recovered and concentrated by the addition of 5 mL of ice-cold isopropanol. Precipitated molecules were recovered by centrifugation at 10
1), and by mixing vigorously until two immiscible phases were obtained. The upper aqueous phase, which contained mostly RNA, was recovered and concentrated by the addition of 5 mL of ice-cold isopropanol. Precipitated molecules were recovered by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 20 min at 4 °C and resuspended in 1.5 mL of lysis solution. It was concentrated again with 1.5 mL of ice-cold isopropanol. After centrifuging for 10 min at 10
000 g for 20 min at 4 °C and resuspended in 1.5 mL of lysis solution. It was concentrated again with 1.5 mL of ice-cold isopropanol. After centrifuging for 10 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g (4 °C), the RNA pellet was washed with 7.5 mL of 75% ethanol and incubated at room temperature for 10 min, followed by a 5 min centrifugation at 10
000 g (4 °C), the RNA pellet was washed with 7.5 mL of 75% ethanol and incubated at room temperature for 10 min, followed by a 5 min centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g (4 °C). The air-dried RNA pellet was solubilized in 1 mL of 0.05% DEPC-treated water. Finally, 260 and 280 nm absorbance of the samples was measured using a Nanodrop spectrophotometer in order to assess RNA quantity and agarose gel electrophoresis was performed to assess purity.
000 g (4 °C). The air-dried RNA pellet was solubilized in 1 mL of 0.05% DEPC-treated water. Finally, 260 and 280 nm absorbance of the samples was measured using a Nanodrop spectrophotometer in order to assess RNA quantity and agarose gel electrophoresis was performed to assess purity.
Pre-miR-29b purification
Chromatographic experiments were performed in an ÄKTA Avant system with UNICORN 6.1 software (GE Healthcare). A 10 mm diameter × 20 mm long (about 2 mL) column was packed with commercial L-arginine–Sepharose 4B gel. This support is characterized by the manufacturer as a cross-linked 4% beaded agarose matrix with a 12-atom spacer and an extent of labeling between 14 and 20 μmoL/mL. The column was first equilibrated with 0.28 M of NaCl in 10 mM Tris–HCl buffer (pH 8.0). Small RNA samples (30 μg) were applied onto the column using a 100 μL loop at a flow-rate of 1 mL min−1. After the elution of unbound species with 0.28 M of NaCl in 10 mM Tris–HCl buffer (pH 8.0), the ionic strength of the buffer was increased to 0.36 M NaCl in order to elute the pre-miR-29b. The most retained species were finally eluted with 0.5 M NaCl in 10 mM Tris–HCl buffer (pH 8.0), as previously described.14 The absorbance of the eluate was continuously monitored at 260 nm. All chromatographic runs were performed at 20 °C, by using a specific column containing a water jacket tube connected to a circulating water bath to maintain the temperature. Pre-miR-29b fractions were pooled according to the chromatograms obtained, and were concentrated and desalted with Vivaspin concentrators. Posteriorly, the integrity and purity of pre-miR-29b samples isolated from R. sulfidophilum were also analyzed by vertical electrophoresis using an Amersham Biosciences system (GE Healthcare) with 10% polyacrylamide gel supplemented with 8 M urea. Electrophoresis was performed at a running voltage of 125 V for 90 min in TBE buffer, pH 8.3, and samples were previously denatured with 97.5% formamide. Gels were visualized using a UV transilluminator (UVItec, Cambridge) after staining with GreenSafe Premium (NZYTech, Lisbon, Portugal).SPR measurements
All SPR experiments were performed using a Biacore T200 Biosensor software v 1.0 (Biacore, GE Healthcare, Sweden) with carboxymethylated dextran-coated sensor chips (CM5 research grade). L-Arginine and L-lysine (0.2 M in 0.1 M borate buffer, pH 8.5) were immobilized on the CM5 sensor chip surface, to mimic immobilized ligands in affinity chromatographic supports, based on the protocol described by Cruz and co-workers.19 Briefly, the immobilization of L-arginine and L-lysine was performed through amine coupling chemistry using HBS and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) as a running buffer (7 min, 5 μL min−1) in the flow cells 2 and 3, respectively. Flow cell 1 was used as the reference control for all experiments, which was treated as the other cells, without amino acids. After immobilization, the surfaces of all flow cells were blocked with 1 M ethanolamine–HCl (pH 8.5), followed by HBS-EP injection to stabilize the baseline. The immobilized density averaging for L-arginine was 245.6 RU and for L-lysine was 242.2 RU. Human pre-miR-29b samples used to collect affinity binding data were prepared by serial dilutions to obtain the desired concentration range (2.5 to 0.0195 μM) with adequate buffers, for each experiment. Duplicate injections of each pre-miR-29b sample were analyzed in a random order and a buffer blank was flowed over the L-arginine and L-lysine surface, as well as over the reference surface, at a flow rate of 2 μL min−1 for 7 min. No regeneration solution was required since all pre-miR-29b solutions were removed from the surface chip during the dissociation time. Thus, equilibrium binding experiments were performed under similar conditions to those used in the affinity chromatography assays (temperature, pH, ionic strength of the buffer and competitive ligand), in order to understand the biomolecular interactions occurring between ligands and pre-miR-29b. Several buffers were studied to determine which are suitable to be used with RNA allowing the establishment of interactions involved in miRNA purification, namely (i) 10 mM Tris–HCl, pH 8.0; (ii) 10 mM and 100 mM HEPES buffer, pH 7.4; (iii) 10 mM and 50 mM phosphate buffer, pH 8.0; (iv) 10 mM MOPS, pH 8.0; (v) 280 mM and 500 mM NaCl in 10 mM Tris–HCl, pH 8.0; (vi) 200 mM NaCl supplemented with 8 mM L-arginine in 10 mM Tris–HCl, pH 8.0 and, finally, (vii) 200 mM NaCl supplemented with 10 mM L-lysine in 10 mM Tris–HCl, pH 8.0. As previously mentioned, two different temperatures, 4 and 20 °C, were also investigated to address their influence on the interactions. To correct the bulk refractive index background, the signal obtained for the reference surface was subtracted from sensorgrams for the amino acid immobilized on cell surfaces. In addition, subtraction of the blank injection responses (running buffer without pre-miR-29b) was performed prior to each collection of binding data following exactly the same time course as the runs with pre-miR-29b. The equilibrium dissociation constants (KD) were determined by averaging of the resonance unit values (RU) in the plateau region of the sensorgrams (300 to 400 s) to an affinity model, [Req] = Rmax − (1/(1 + KD/[A])), where Req represents the amount of analyte complexed with the ligand on the surface, [A] is the analyte concentration, and Rmax is the maximum binding capacity of the surface. Rmax was also determined by the previous mathematical model. All data processing and analyses were performed by using BIAevaluation software v.4.1.Structural analysis of pre-miR-29b samples by circular dichroism (CD)
CD was used to monitor the structural behavior of pre-miR-29b (50 μg mL−1) when subjected to different buffer conditions. CD spectra were obtained using a quartz rectangular cell with an optical path length of 0.1 cm at a constant temperature of 25 °C, in a Jasco J-1850 Spectrophotometer (Jasco, Easton, MD, USA). Spectra were recorded from 320 to 215 nm at a scan speed of 10 nm min−1 and a spectral bandwidth of 1 nm. All measurements were conducted under a constant nitrogen gas flow, to purge the ozone generated by the light source of the instrument. The data were collected in triplicate and the average spectra are presented for each sample after subtraction of the buffer contribution. The CD signal was converted to molar ellipticity. Noise in the data was smoothed using Jasco J-1850 software, including the fast Fourier transform algorithm, which allows enhancement of most noisy spectra without distorting their peak shape.Results and discussion
MiRNAs represent an important target of a wide analysis, particularly relevant, in the diagnostic of several disorders, as well as in basic and applied science research. For this reason, the preparation of highly purified miRNAs in large quantity arises as one of the most important challenges in the development of therapeutic strategies based on the use of miRNAs.29 In this study, pre-miR-29b biosynthesis was obtained from a cell culture of recombinant Rhodovulum sulfidophilum DSM 1374 strain in a standard bacterial culture.15 However, the RNAs obtained from this culture need to be further purified in order to isolate the target pre-miR-29b from other host small RNAs. Thus, pre-miR-29b isolation was achieved using an increasing sodium chloride gradient in L-arginine-affinity chromatography, as described by Pereira and co-workers.14 The quality of the pure pre-miR-29b stock solution was examined by polyacrylamide gel electrophoresis, as shown in Fig. 1. In affinity chromatography, the elution strategy can be performed either specifically, using a competitive agent, or non-specifically, by changing the pH, ionic strength or polarity of the buffer.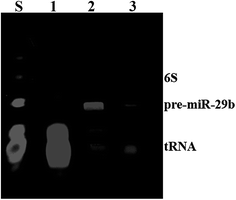 | ||
| Fig. 1 Polyacrylamide gel electrophoresis analysis of pure pre-miR-29b samples. Lane S, Initial sample; Lanes 1, transfer RNAs; Lane 2, pre-miR-29b; Lane 3, pre-miR-29b plus transfer RNAs. | ||
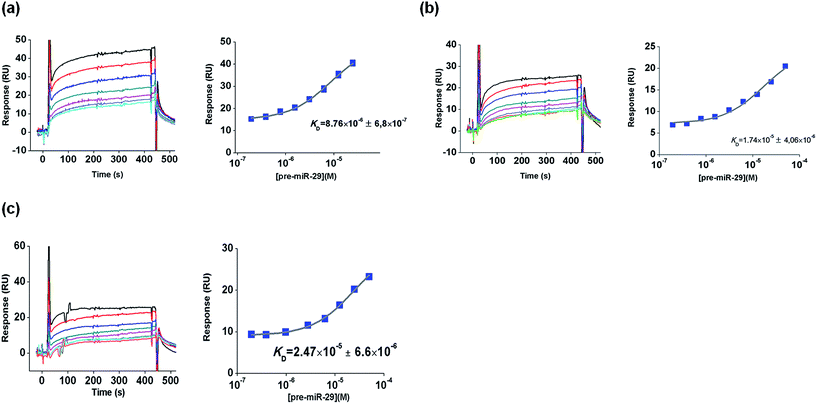
Hence, this SPR study was developed in order to characterize the interactions between pre-miR-29b and L-arginine and L-lysine as biospecific ligands. For this purpose, changes in the elution buffer composition, ionic strength or temperature were performed in order to observe their effects on biomolecular interactions. The amino acids L-arginine and L-lysine were chosen because they are conserved amino acids in the active center of the Argonaute protein, which is part of the RNA-induced silencing complex (RISC), suggesting that they have an important role in the cellular recognition of the 3′ overhang of the pre-miRNA.30,31 So, it is expected that this biorecognition can be exploited to implement a chromatographic purification methodology for pre-miR-29b. Initially, L-arginine and L-lysine were immobilized on a CM5 sensor chip via amine coupling chemistry at pH 8.5 (below their isoelectric point), as described by Cruz and collaborators.19 This protocol of immobilization ensures that the guanidine functional groups (ε-amino group) of L-arginine and L-lysine are positively charged and the α-NH2 group is attached to the reactive ester groups of the chip. All SPR equilibrium data were fitted to a steady state bimolecular interaction model. The SPR binding profile (square-shaped) was identical for all complexes and some are exemplified in Fig. 2.
Biomolecular interactions between pre-miR-29b and L-arginine
Recently, our research group has demonstrated the potential of the arginine amino acid in the purification of RNA, suggesting that the binding mechanism involves multiple non-covalent interactions with RNA, due to its ability to interact in different conformational arrangements, the length of its side chain, the guanidine functional group and its ability to produce hydrogen bond geometries.12,17,32 Thereby, since RNA is negatively charged due to the phosphate groups in the backbone, it is easy to predict favorable electrostatic interactions between RNA phosphate groups and L-arginine.Interestingly, the SPR sensorgrams obtained with the L-arginine ligand present response profiles in the shape of a square, indicating rapid dissociation (450 s), i.e., the binding assays suggest that pre-miR-29b biomolecules interact with the immobilized L-arginine, and that these interactions are reversible. The KD values are listed in Table 1. At 20 °C, in the range of concentrations tested, SPR responses for L-arginine-pre-miR-29b were obtained in Tris–HCl, NaCl and NaCl supplemented with L-lysine buffers. The equilibrium dissociation constants demonstrate that Tris–HCl (10 mM) promotes lower ligand–analyte affinity (KD = 5.11 × 10−6 ± 4.4 × 10−7 M) when compared with the values obtained for NaCl (KD = 4.06 × 10−7 ± 3.5 × 10−8 M). In fact, in chromatography, the Tris–HCl buffer can be used to promote the retention of total small RNAs onto the L-arginine support, but the application of NaCl in the binding buffer is responsible for the higher selectivity towards the target miRNA. On the other hand, negligible (≤5 RU) non-specific binding was detected when using the following buffers: phosphate (10 and 50 mM), HEPES (10 and 100 mM) and MOPS (10 mM), regardless of their concentration, indicating that L-arginine does not interact with pre-miR-29b under these conditions (data not shown).
| Buffers | |||||||
|---|---|---|---|---|---|---|---|
| Tris–HCl (10 mM) | MOPS (10 mM) | HEPES (10 mM) | NaCl (280 mM) | L-Arginine (8 mM) | L-Lysine (10 mM) | ||
| L-Arginine | 20 °C | 5.11 × 10−6 ± 4.4 × 10−7 | — | — | 4.06 × 10−7 ± 3.5 × 10−8 | — | 3.53 × 10−6 ± 1.9 × 10−7 |
| 4 °C | 1.34 × 10−6 ± 2.3 × 10−7 | — | — | — | — | — | |
| L-Lysine | 20 °C | 3.24 × 10−7 ± 4.6 × 10−8 | 1.74 × 10−5 ± 4.1 × 10−6 | 2.47 × 10−5 ± 6.6 × 10−6 | 9.96 × 10−7 ± 3.5 × 10−8 | 2.08 × 10−6 ± 4.4 × 10−7 | 2.28 × 10−6 ± 6.3 × 10−7 |
| 4 °C | 8.76 × 10−6 ± 6.8 × 10−7 | 2.31 × 10−5 ± 3.1 × 10−6 | 3.95 × 10−6 ± 1.5 × 10−7 | — | — | — | |
In contrast, the ionic strength of buffer (10 mM Tris–HCl supplemented with NaCl) has a significant effect on the binding of pre-miR-29b. The SPR response to the pre-miR-29b solution in 500 mM NaCl was very low (<5 RU), implying that the chromatographic binding and elution of pre-miR-29b may be manipulated with this parameter. In the sensorgrams was notorious the dissociation in the first 100 s using 500 mM NaCl (data not shown), indicating that the high content of NaCl results in the loss of pre-miR-29b binding. Our results also revealed that when a moderate salt concentration (280 mM NaCl) is used, high binding responses were obtained (KD = 4.06 × 10−7 ± 3.5 × 10−8 M). In general, the affinity of the pre-miR-29b with the L-arginine surface was high (KD ≥ 10−6 M) at 20 °C. These results may be correlated with the chromatographic experiments, where the total retention of pre-miR-29b could be achieved using a low concentration of NaCl (280 mM) and high concentrations (500 mM NaCl) lead to its elution.14 Besides, the salt concentration can also have a positive effect on RNA folding as well as on the stabilization of non-covalent RNA–amino acid interactions during the purification, once the salt allows the single-stranded nucleic acid molecules to form stable hydrogen bonds between their complementary bases. In this case, the chromatographic interaction was promoted by using a low ionic strength buffer and the selective elution of the bound species was achieved by increasing the NaCl concentration.14 Moreover, it was verified that the pre-miR-29b was eluted at a higher ionic strength, indicating a stronger interaction than other contaminants. We also evaluated the influence of a competing agent (L-arginine and L-lysine) in the buffer, as a way to conclude about the use of these amino acids to promote binding or elution of the pre-miR-29b to the L-arginine ligand. The experiments were carried out using different concentrations, from 8 to 19 mM of L-arginine and 10 to 16 mM of L-lysine in the 200 mM NaCl buffer. However, for the highest concentrations, the binding of pre-miR-29b to the L-arginine ligand was not verified (results not shown). In these experiments, a binding response for the condition of 10 mM of L-lysine in 200 mM NaCl was only obtained (KD = 3.53 × 10−6 ± 1.9 × 10−7 M), while 8 mM of L-arginine resulted in a negligible (≤5 RU) sensor response to the pre-miR-29b solution. This result is in accordance with some evidence obtained in L-arginine-affinity chromatography, once this condition can be used for the pre-miR-29b elution, as it functions as a competitive agent. This behavior can be due to the positive character of these amino acids that promotes the binding of pre-miR-29b to the free amino acid present in the buffer by electrostatic interactions, inducing the elution of bound pre-miR-29b. Moreover, the results suggest that the elution of bound pre-miR-29b can be more effective when using L-lysine in the buffer. As demonstrated in Table 1, the KD value of 10 mM L-lysine is higher than the value obtained at 280 mM NaCl, indicating that the affinity of pre-miR-29b to the L-arginine ligand is higher for the 280 mM NaCl condition, and lysine is expected to weaken the interaction.
Considering that RNA is an unstable molecule and has a very short half-life once extracted, the effect of temperature on the miRNA–arginine binding is a critical parameter in RNA purification.33 In contrast to the results achieved for the temperature of 20 °C, at 4 °C the SPR response of L-arginine and pre-miR-29b using 10 mM Tris–HCl buffer was only found. The dissociation constants of Tris–HCl are of the same order of magnitude at 20 and 4 °C (KD = 1.34 × 10−6 ± 2.3 × 10−7 M), but a slightly higher affinity was found for 4 °C, suggesting stronger interactions at lower temperatures. Non-specific binding (≤5 RU) was detected when using the other buffers, regardless of their concentration, indicating that no significant interactions occur between pre-miR-29b and L-arginine ligand under these conditions (data not shown). The KD values obtained are consistent with the chromatographic results previously reported by Pereira and collaborators,14 where the pre-miR-29b retention in the L-arginine–agarose support was higher for the lowest temperature studied (4 °C), suggesting that the temperature is a critical factor in the binding pre-miR-29b.
Summarizing, the affinity order of pre-miR-29b to L-arginine was 280 mM NaCl > 10 mM Tris–HCl ≈ 10 mM L-lysine in 200 mM NaCl, with NaCl promoting the highest affinity for the pre-miR-29b to the L-arginine surface. Hence, this comparative study reinforces the idea that the binding mechanism inherent to the biorecognition between the L-arginine amino acid and pre-miR-29b molecules can result from the combination of several phenomenological interactions such as electrostatic interactions, multiple hydrogen bond and van der Waals forces.17 Thus, it is reasonable to suggest that the retention of RNA in L-arginine is due to its side chain, which can promote multicontacts with RNA backbone or bases, according to RNA folding.32,34,35 Additionally, recent studies with saturation transfer difference-nuclear magnetic resonance spectroscopy and SPR reported that guanine polynucleotides interact preferably with L-arginine through the sugar-phosphate backbone.18,19 Thus, another explanation for these findings is related to pre-miR-29 structural features that seem to be relevant on its distinct retention behavior in the arginine ligand. Pre-miR-29b is a small RNA molecule with a stem-loop shape consisting of two long irregular double-stranded stem regions, which are interrupted by a largely single-stranded internal loop. In addition, along the bulge of pre-miR-29b and through the continuous stem sequences there are several guanines, which were described to interact preferably with arginine. Hence, the nucleotide base composition and exposure can also favor other biomolecular interactions, responsible for the specificity and biorecognition found in the L-arginine ligand.
Biomolecular interactions between pre-miR-29b and L-lysine
Previous studies have shown that the main interactions occurring between nucleic acids and the L-lysine amino acid are electrostatic, because of its amine groups.36–38 Moreover, L-lysine has also some characteristics, such as the lateral side chain with significant length and ability to interact with several RNA classes even in different conformational rearrangements, which may produce hydrogen bond geometries, reinforcing the establishment of multiple non-covalent interactions involved in the specific recognition mechanism of nucleic acids.38 Indeed, besides the multiple interactions (electrostatic interactions, van de Waals forces and hydrogen bonds) that L-lysine can establish with the RNA backbone or with the more exposed bases, several small RNAs can also interact and be recognized by the presence of hydrophobic interactions with the aliphatic portion of a L-lysine side chain.15 Considering this complex biorecognition mechanism, it is important to study the influence of experimental conditions on the interaction occurring between pre-miR-29b and L-lysine amino acid, resulting in different binding affinities. It was verified that pre-miR-29b binds more strongly to L-lysine in 10 mM Tris–HCl and 280 mM NaCl at 20 °C (KD = 3.24 × 10−7 ± 4.6 × 10−8 M and KD = 9.96 × 10−7 ± 3.5 × 10−8 M, respectively) than in 10 mM HEPES (KD = 2.47 × 10−5 ± 6.6 × 10−6 M) or 10 mM MOPS buffers (KD = 1.74 × 10−5 ± 4.1 × 10−6 M). In fact, these findings are in agreement with the results reported in the literature, where Tris–HCl buffer is commonly used to promote the binding of RNA to L-lysine. Negligible binding (≤5 RU) was detected when the following buffers were used: phosphate (10 mM and 50 mM) and HEPES (100 mM), indicating that pre-miR-29b has no affinity to L-lysine under these conditions. Similarly to the results obtained to L-arginine, the ionic strength of buffer (10 mM Tris–HCl supplemented with NaCl or with amino acids) has an effect on the binding of pre-miR-29b to L-lysine. Our findings demonstrate that when high concentrations of salt (500 mM NaCl) or amino acids are used, no binding responses of pre-miR-29b are observed and, consequently, these conditions can be used for the elution of the target biomolecule in chromatography. The addition of 10 mM of L-lysine in 200 mM NaCl resulted in a binding response characterized by KD = 2.28 × 10−6 ± 6.3 × 10−7 M and the supplementation with 8 mM of L-arginine in 200 mM NaCl resulted in a KD = 2.08 × 10−6 ± 4.4 × 10−7 M. Although these concentrations of competing agents induce binding of pre-miR-29b, the interaction is weaker when compared with Tris–HCl. Briefly, the affinity order of pre-miR-29b binding to L-lysine was 10 mM Tris–HCl ≈ 280 mM NaCl > 10 mM L-lysine ≈ 8 mM L-arginine in 200 mM NaCl > 10 mM HEPES ≈ 10 mM MOPS, thus Tris–HCl and NaCl buffers induced greater affinity to the L-lysine surface, followed by HEPES and MOPS (Table 1).In this study the effect of decreasing the temperature to 4 °C on the binding of pre-miR-29b to the L-lysine surface was also evaluated, comparing with the binding at 20 °C. The binding responses are similar to those obtained at 20 °C, illustrating that the interactions between L-lysine and pre-miR-29b also occur under these conditions. However, some differences were found in KD values determined at 4 °C (3.24 × 10−7 M to 8.76 × 10−6 M for 10 mM Tris–HCl; 2.47 × 10−5 M to 3.95 × 10−6 M for 10 mM HEPES and 1.74 × 10−5 to 2.31 × 10−5 M for 10 mM MOPS, at 20 and 4 °C, respectively). In general, the KD values obtained are consistent with the chromatographic results, in which the lowest temperature studied induced higher pre-miR-29b retention onto the L-lysine–agarose support (data not shown). Globally, these results can also be related to the pre-miRNA structure, which presents continuous stem sequences mostly containing guanines that are described to preferentially interact with L-lysine due to hydrogen bonds that are established with one of the donor atoms.39,40
Briefly, to compare the salt influence on the binding of pre-miR-29b to the L-arginine and L-lysine surfaces, several buffers were analyzed. In general, the maximum RU signal was higher for the L-lysine than for the L-arginine surface (Fig. 3). The experiments showed a low resonance response level with both surfaces (normalized signal). It was found that the maximum binding between pre-miR-29b and immobilized L-arginine and L-lysine was observed in 280 mM NaCl, followed by 10 mM Tris–HCl, whereas the lowest binding was observed in 200 mM NaCl supplemented with 8 mM L-arginine or 10 mM L-lysine, 10 mM HEPES and 10 mM MOPS (Fig. 3). Thus, our affinity data show that Tris–HCl and NaCl are optimal buffers for interaction assays and distinct buffer environments such as MOPS or HEPES did not enhance the binding to L-arginine and L-lysine. Overall, these results provide suitable conditions to induce pre-miRNA binding, thereby increasing the selectivity and improving efficient microRNA separation. It was also proved that under these conditions (ionic strength and buffer composition) the electrostatic interactions are favored between the amino positive groups of the L-arginine and L-lysine and the negative phosphate groups of the pre-miR-29b, mainly with guanosine. These results demonstrated that SPR allows making a screening of ligands that, like the ones described herein, can be used to design and implement novel miRNA purification methods.
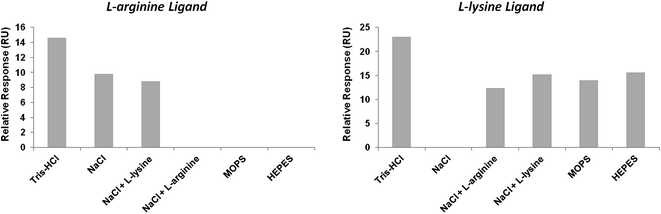 | ||
| Fig. 3 Relative resonance response (RU) with different running buffers at 450 s after injection of pre-miR-29b at a concentration of 1.25 μM, at 20 °C, in L-arginine and L-lysine surfaces. | ||
Pre-miR-29b structural analysis
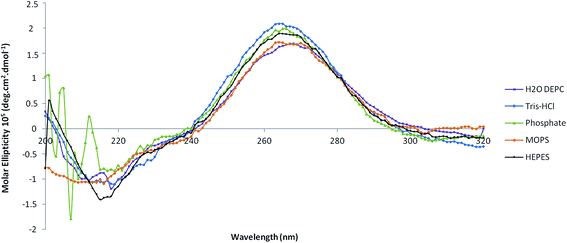
Both ligands promoted the biorecognition of pre-miR-29b as well as an effective interaction under most conditions studied. To select the buffer that is able to enhance the pre-miR-29b binding to the amino acid ligands and to evaluate the effect of the different conditions in the pre-miR-29b structural stability, four buffers were analyzed (Tris–HCl, HEPES buffer, MOPS, phosphate buffer) by circular dichroism (CD) spectroscopy. Thus, structural characterization of the pre-miR-29b under different experimental conditions gives additional information about the interactions with the ligands and also about the suitable conditions that must be used in chromatographic procedures to purify the target biomolecules while maintaining its stability and biological activity. CD technique offers good resolution, provides sensitive spectra and it has been widely used in structural determination of biomolecules (proteins, nucleic acids, etc.). Quantitative analysis of CD spectra is possible, but, as with any assay, the correct and proper interpretation of the results depends on an accurate understanding of the procedure. The spectra in Fig. 4 are given in terms of molar ellipticity thus making it possible to directly compare the intensity of the bands obtained for different samples. CD spectra of the pre-miR-29b show characteristic peaks around 210 nm (negative maximum signal) and 265 nm (positive maximum signal). The comparison with the CD spectrum of established nucleic acid structures confirms that the pre-miRNA possesses a stem-loop structure because the CD spectral pattern typically indicates the B-form RNA duplex.15,41 Although, CD is not sensible to detect the contamination of other cytosol miRNAs, it is sensible to detect the contamination of other biomolecules. However, the pre-miR-29 subjected to this analysis has a purity of 98%, and it is unlikely that the sample is contaminated, and changes in the CD spectra resulting from contamination are not expected.In Fig. 4 are shown the CD spectra resulting from the evaluation of pre-miR-29b in four buffers, after an initial 15 min incubation in Tris–HCl buffer (10 mM) and HEPES (10 mM). The CD spectra of the pre-miRNA sample in Tris–HCl and HEPES show typical CD spectrum of RNA, with a positive band at 265 nm and a negative band at 210 nm, similar to that obtained with DEPC water. However, a significant difference in the band magnitude of the pre-miR-29b spectra was found (Fig. 4). These results are consistent with the responses obtained with SPR, suggesting that Tris–HCl is a valuable choice for promoting pre-miRNA-L-arginine/lysine binding interactions in future applications. In the presence of HEPES or MOPS, the electrostatic interaction between the L-arginine and the L-lysine surface and the pre-miR-29b backbone could be more favored, since these buffers at neutral pH are a zwitterionic molecule with a positive charge on one of the protonated nitrogens. This can reduce the repulsive electrostatic interaction between charged phosphate groups of pre-miR-29b, stabilizing the pre-miRNA molecule and favoring the interaction with the amino acids. By analyzing the CD spectra of pre-miR-29b in MOPS (10 mM), a decrease of intensity of the positive band as well as the negative band is visible. By the CD spectral analysis of Fig. 4, a strong modification in the RNA structure of the phosphate buffer is evident. These data can also justify why pre-miR-29b molecules do not interact with the L-arginine or L-lysine supports in the presence of phosphate buffer due to the increase of the repulsions with the pre-miR-29b backbone. In contrast, in the presence of salt (280 mM NaCl), phosphate groups of the pre-miR-29b backbone are effectively screened by the salt counterions resulting in a more compact and less regular structure. The data obtained from SPR can play an important role in the optimization of chromatographic-based purification processes since it allows the determination of buffers which favor interactions of the target biomolecule with the chromatographic support. Moreover, the conjugation with the CD technique allows the confirmation of which buffers are able to maintain the stability of the miRNA.
Conclusions
The binding affinity quantification between pre-miR-29b and L-arginine and L-lysine surfaces was examined by using a SPR technique, under different conditions, in order to mimic the immobilized ligand on an affinity chromatographic support, and to verify the effect of these conditions on the interactions and stability of the molecule. Therefore, several experimental conditions for amino acid ligand–pre-miR-29b binding were characterized, and an optimal set of buffers as well as temperature conditions were defined. Distinct buffers affected the interaction strength with an increased response in the presence of Tris–HCl and NaCl. These results provide suitable conditions to induce pre-miRNA binding, thereby increasing the selectivity and improving efficient miRNA separation. The predominant interactions of pre-miR-29b with L-arginine and L-lysine ligands are ionic between the amino positive groups of the L-arginine and L-lysine and the negative phosphate groups of the pre-miR-29b and hydrogen bonds with the more exposed bases, mainly with guanosine. Overall, the present study showed that SPR can be used as a first technique to screen, manipulate, determine and establish the best binding/elution conditions that can lead to an improved performance and selectivity of the chromatographic process. In addition, it was demonstrated that although some conditions used in chromatographic experiments favor multiple interactions between the pre-miR-29b and the respective ligand (L-arginine and L-lysine), these conditions can affect the structural stability of the target molecule. From CD, it was possible to conclude that the biorecognition observed by the pre-miR-29b can be related to the higher availability and base exposition, which confers a different conformational structure to this molecule. Moreover, the highest equilibrium dissociation constants determined by SPR ranged from 10−6 to 10−7 M to L-arginine and from 10−5 to 10−7 M to L-lysine, indicating that pre-miR-29b showed high binding affinity to these surfaces. In addition, the results obtained in this study provide valuable and relevant information that can be further applied in unveiling the miRNA chromatographic behavior in these systems, in order to improve the binding of the pre-miR-29b onto the amino acid-based supports, preserving their structural stability, integrity and activity. In conclusion, it was proved that the association of these two techniques might be a powerful strategy to define the best conditions to be applied in the miRNA purification.Conflict of interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.Acknowledgements
This work was supported by the Portuguese Foundation for Science and Technology (FCT), through the projects EXPL/BBB-BIO/1056/2012, Pest-OE/SAU/UI0709/2014 and FCOMP-01-0124-FEDER-041068 – EXPL/QEQ-MED/1068/2013. Patrícia Pereira and Carla Cruz acknowledge the fellowships, SFRH/BD/81914/2011 and SFRH/BPD/100015/2014, respectively from FCT. The authors also acknowledge the program Fundo Europeu de Desenvolvimento Regional (FEDER) [COMPETE [FCOMP-01-0124-FEDER-027560]]. The authors would like to thank Prof. Yo Kikuchi (Division of Life Science and Biotechnology, Department of Ecological Engineering, Toyohashi University of Technology) for kindly providing pBHSR1-RM plasmid.References
- D. P. Bartel, Cell, 2004, 116, 281–297 CrossRef CAS PubMed.
- J. A. Broderick and P. D. Zamore, Gene Ther., 2011, 18, 1104–1110 CrossRef CAS PubMed.
- A. Eulalio and M. Mano, J. Biomol. Screening, 2015, 20, 1003–1017 CrossRef PubMed.
- C. L. Bartels and G. J. Tsongalis, Clin. Chem., 2009, 55, 623–631 CAS.
- M. V. Latronico and G. Condorelli, Nat. Rev. Cardiol., 2009, 6, 419–429 Search PubMed.
- P. T. Nelson, W. X. Wang and B. W. Rajeev, Brain Pathol., 2008, 18, 130–138 CrossRef CAS PubMed.
- L. Du and A. Pertsemlidis, J. Mol. Cell Biol., 2011, 3, 176–180 CrossRef CAS PubMed.
- I. Vomelova, Z. Vanickova and A. Sedo, Folia Biol., 2009, 55, 243–251 CAS.
- M. A. Dineva, L. MahiLum-Tapay and H. Lee, Analyst, 2007, 132, 1193–1199 RSC.
- L. Ponchon and F. Dardel, Methods, 2011, 54, 267–273 CrossRef CAS PubMed.
- S. C. Walker, F. H. Scott, C. Srisawat and D. R. Engelke, Methods Mol. Biol., 2008, 488, 23–40 CAS.
- R. Martins, C. J. Maia, J. A. Queiroz and F. Sousa, J. Sep. Sci., 2012, 35, 3217–3226 CrossRef CAS PubMed.
- R. Martins, J. A. Queiroz and F. Sousa, J. Mol. Recognit., 2010, 23, 519–524 CrossRef CAS PubMed.
- P. Pereira, A. Sousa, J. Queiroz, I. Correia, A. Figueiras and F. Sousa, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2014, 951–952, 16–23 CrossRef CAS PubMed.
- P. Pereira, A. Sousa, J. Queiroz, A. Figueiras and F. Sousa, J. Chromatogr. A, 2014, 1331, 129–132 CrossRef CAS PubMed.
- R. Martins, J. A. Queiroz and F. Sousa, Biomed. Chromatogr., 2012, 26, 781–788 CrossRef CAS PubMed.
- R. Martins, J. A. Queiroz and F. Sousa, J. Chromatogr. A, 2014, 1355, 1–14 CrossRef CAS PubMed.
- F. Sousa, D. M. Prazeres and J. A. Queiroz, Trends Biotechnol., 2008, 26, 518–525 CrossRef CAS PubMed.
- C. Cruz, E. J. Cabrita and J. A. Queiroz, Anal. Bioanal. Chem., 2011, 401, 983–993 CrossRef CAS PubMed.
- C. Cruz, S. D. Santos, E. J. Cabrita and J. A. Queiroz, Int. J. Biol. Macromol., 2013, 56, 175–180 CrossRef CAS PubMed.
- C. Cruz, A. Sousa, E. Mota, F. Sousa and J. A. Queiroz, Int. J. Biol. Macromol., 2014, 70, 131–137 CrossRef CAS PubMed.
- C. Cruz, A. Sousa, F. Sousa and J. A. Queiroz, Anal. Methods, 2013, 5, 1682–1686 RSC.
- C. T. Campbell and G. Kim, Biomaterials, 2007, 28, 2380–2392 CrossRef CAS PubMed.
- P. S. Katsamba, S. Park and I. A. Laird-Offringa, Methods, 2002, 26, 95–104 CrossRef CAS PubMed.
- B. Nguyen, F. A. Tanious and W. D. Wilson, Methods, 2007, 42, 150–161 CrossRef CAS PubMed.
- J. Hwang and S. Nishikawa, J. Biomol. Screening, 2006, 11, 599–605 CrossRef CAS PubMed.
- H. Suzuki, S. Umekage, T. Tanaka and Y. Kikuchi, J. Biosci. Bioeng., 2011, 112, 458–461 CrossRef CAS PubMed.
- P. Chomczynski and N. Sacchi, Nat. Protoc., 2006, 1, 581–585 CrossRef CAS PubMed.
- D. P. Bartel, Cell, 2009, 136, 215–233 CrossRef CAS PubMed.
- F. V. Rivas, N. H. Tolia, J. J. Song, J. P. Aragon, J. Liu, G. J. Hannon and L. Joshua-Tor, Nat. Struct. Mol. Biol., 2005, 12, 340–349 CAS.
- S. Griffiths-Jones, Methods Mol. Biol., 2006, 342, 129–138 CAS.
- J. D. Puglisi, J. R. Wyatt and I. Tinoco Jr., Biochemistry, 1990, 29, 4215–4226 CrossRef CAS PubMed.
- R. E. Farrell, RNA Methodologies - Laboratory Guide for Isolation and Characterization, Academic Press, York, USA, 4th edn, 2005 Search PubMed.
- B. J. Calnan, B. Tidor, S. Biancalana, D. Hudson and A. D. Frankel, Science, 1991, 252, 1167–1171 CrossRef CAS PubMed.
- T. Janas, J. J. Widmann, R. Knight and M. Yarus, RNA, 2010, 16, 805–816 CrossRef CAS PubMed.
- A. Sousa, F. Sousa and J. A. Queiroz, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2011, 879, 3507–3515 CrossRef CAS PubMed.
- A. Sousa, F. Sousa and J. A. Queiroz, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 3257–3260 CrossRef CAS PubMed.
- D. S. Jones, H. K. Lundgren and F. T. Jay, Nucleic Acids Res., 1976, 3, 1569–1576 CrossRef CAS PubMed.
- J. J. Ellis, M. Broom and S. Jones, Proteins, 2007, 66, 903–911 CrossRef CAS PubMed.
- M. M. Hoffman, M. A. Khrapov, J. C. Cox, J. Yao, L. Tong and A. D. Ellington, Nucleic Acids Res., 2004, 32, D174–D181 CrossRef CAS PubMed.
- B. Ranjbar and P. Gill, Chem. Biol. Drug Des., 2009, 74, 101–120 CAS.
| This journal is © The Royal Society of Chemistry 2016 |