Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Encapsulation of FRET-based glucose and maltose biosensors to develop functionalized silica nanoparticles†
G.
Faccio‡
 *a,
M. B.
Bannwarth‡
b,
C.
Schulenburg
a,
V.
Steffen
c,
D.
Jankowska
a,
M.
Pohl
c,
R. M.
Rossi
b,
K.
Maniura-Weber
a,
L. F.
Boesel
b and
M.
Richter§
*a
*a,
M. B.
Bannwarth‡
b,
C.
Schulenburg
a,
V.
Steffen
c,
D.
Jankowska
a,
M.
Pohl
c,
R. M.
Rossi
b,
K.
Maniura-Weber
a,
L. F.
Boesel
b and
M.
Richter§
*a
aEmpa, Swiss Federal Laboratories for Materials Science and Technology, Laboratory for Biointerfaces, Lerchenfeldstrasse 5, 9014 St. Gallen, CH, Switzerland. E-mail: greta.faccio@empa.ch; michael.richter@igb.fraunhofer.de; Tel: +41 58 765 7262, +49 9421 187-353
bEmpa, Swiss Federal Laboratories for Materials Science and Technology, Laboratory for Protection and Physiology, Lerchenfeldstrasse 5, 9014 St. Gallen, CH, Switzerland
cIBG-1: Biotechnology, Forschungszentrum Jülich GmbH, 52425 Jülich, DE, Germany
First published on 19th January 2016
Abstract
Silicate nanoparticles with immobilized FRET-based biosensors were developed for the detection of glucose and maltose. Immobilization of the protein biosensor in the nanoparticle was achieved through specific interaction between the hexa-histidine tag of the protein and a calcium–silicate complex of the silica matrix. Encapsulation of the biosensors preserved the affinity for the respective sugar. Compared to the free biosensors, encapsulation had a stabilizing effect on the biosensor towards chemical and thermal denaturation. The demonstrated immobilization strategy for specific sensing proteins paves the way towards the development of protein-inorganic nanostructures for application in metabolite analyses.
Nanoparticles responsive to changes in environmental conditions allow controlled release of entrapped molecules,1 intracellular and targeted delivery,2 and intracellular sensing.3 Based often on polymers, metals,4 or semiconductors,5 nanoparticles made of silica have also been used for such purposes. Silica particles are characterized by a low cyto- and genotoxicity, and versions functionalized with inorganic dyes have been described to label and identify cells,6 and to sense pH,7,8 oxygen,9 copper ions,10 zinc ions,11 and TNT.12 To our knowledge, we report fluorescent silica–protein nanoparticles incorporating FRET-based protein biosensors for the first time and characterize their performance. FRET-based protein biosensors are established for the detection of intracellular metabolites.13 This study focuses on two FRET-based biosensors for the detection of glucose and maltose.14–16 These biosensors consist of two fluorescent proteins (enhanced cyan fluorescent protein, ECFP, and enhanced yellow fluorescent proteins EYFP and citrine, respectively) representing a FRET pair and flanking the respective central periplasmic sugar binding protein, like a venus-flytrap.
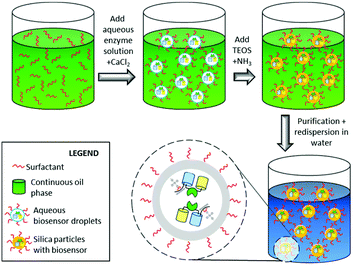
In this study we encapsulated FRET-based glucose and maltose biosensors in silica nanoparticles to evaluate whether their immobilization can widen their field of application, e.g. for extracellular metabolite analysis. Besides the protection against the environment, the immobilization of biomolecular biosensors within a matrix allows facile recovery, reuse in successive processes, and often confers improved robustness towards environmental conditions.17 Key challenge during the immobilization of the biosensor is the preservation of the functionality.18,19 Loss of functionality can be due to the harsh conditions often required for immobilization and by direct interaction of the biomolecules with the matrix that can negatively affect the fluorescent signal. The latter is specifically important in the present case as the function of the FRET-based glucose and maltose biosensors relies on the conformational change of the sugar-binding protein upon binding the sugar ligand, resulting in the alteration of the distance and/or orientation of the flanking fluorescent proteins, and thus in a FRET change. The major challenge addressed in this study is the immobilization of the biosensor to a protective matrix thereby preserving the mobility, which is directly correlated with its functionality. Hence, a permanent, yet orthogonal immobilization is required. Therefore, we made use of a specific interaction between an N-terminal hexahistidine tag fused to the ECFP-part of the biosensor with the silica matrix mediated by calcium cations.20 This specific interaction was achieved during the heterophase encapsulation procedure in which the hexahistidine-tagged biosensor is located inside of aqueous droplets together with calcium ions, which can coordinate to the silica matrix once silica formation is induced. Therefore, the pH is increased, which leads to the hydrolysis of the precursor tetraethyl orthosilicate (TEOS) and subsequent formation of the silica nanohost (Fig. 1).20 This mild approach favours both the site-specific interaction between the protein and the silica nanohost through the hexahistidine tag, and reduces the adsorption of the negatively charged protein e.g. GFP (pI ∼ 6.2)14 and the glucose biosensor (calculated pI = 5.6), to the negatively charged silica.
Colloidal analysis of the prepared silica nanohosts containing the sugar biosensors was performed by scanning transmission electron microscopy (STEM) to determine their size and morphology (Fig. 2). The STEM images show spherical nanoparticles with average diameters ∼80 nm and similar sizes for the silica particles with encapsulated maltose (Fig. 2A) or glucose biosensor (Fig. 2B). Hence, the type of encapsulated biosensor did not largely influence the size and morphology of the silica particles. The observed sizes are in good agreement with the macroscopic appearance of the dispersion, which shows only little visual turbidity for such small nanoparticles, allowing an analysis of the optical properties of the encapsulated biosensor proteins without large scattering disturbances.
The surface properties of the nanoparticles were investigated via zeta potential measurements. The silica particles with encapsulated maltose biosensor yielded a potential of −31.3 ± 3.6 mV and for the encapsulated glucose biosensor −31.9 ± 4.6 mV. Thus, the type of encapsulated biosensor does not influence the surface potential of the silica particles. The obtained zeta potentials of ∼−30 mV indicate a moderately stable dispersion not prone to aggregation.
In order to remove free biosensor protein not encapsulated in the silica matrix, the silica nanoparticles were washed several times by centrifugation and redispersion. The presence and integrity of the immobilized biosensors in the silica matrix was then proven by fluorescence microscopy. Therefore, the purified nanoparticles were diluted in 20 mM MOPS (pH 7.4) and images were taken with a fluorescence microscope (Fig. 2) using a YFP fluorescence filter as described in the ESI.† A biosensors-specific YFP fluorescence was detected concentrated in small dots not detectable with visible light. However, a small fraction of aggregated nanoparticles could be found (Fig. 2C and D).
To determine the effect of the immobilization on the fluorescent properties of the biosensor, fluorescence spectroscopy was performed. After immobilization, the fluorescent properties of both fluorescent proteins are preserved. However, the FRET ratio is lower for the encapsulated forms as compared to the free untreated equivalents for both the maltose and glucose biosensor system. The FRET ratio changes from 1.99 to 1.09 for the encapsulated glucose biosensor and from 1.2 to 0.9 for the encapsulated maltose biosensor. This effect can be explained by fluorescence quenching effects inside the nanoparticle. A similar effect has also been observed when the FRET ratios of such biosensors were measured inside E. coli cells (V. Steffen, unpublished). The sensing capabilities of the functionalized nanoparticles were investigated by recording binding isotherms for maltose and glucose, respectively, over a wide range of sugar concentrations covering four orders of magnitude (Fig. 3A and B). The samples were pre-incubated in the presence of different sugar concentrations for two hours to ensure equilibrium conditions. The fluorescence spectrum of the individual solutions were measured and the FRET ratio was calculated as the ratio between the fluorescence intensity emitted at 528 nm and at 485 nm (λex = 428 nm). The affinity (Kd) of the entrapped biosensors was calculated and resulted in the same order of magnitude as determined for the biosensors in free form, e.g. 1.6 vs. 6.4 mM glucose and 0.2 vs. 0.1 mM maltose for the glucose and maltose biosensors in the free and encapsulated forms, respectively. These results demonstrate that the porosity of the nanocapsules is high enough to allow permeation of low-molecular weight metabolites while simultaneously entrapping the proteins.
 | ||
| Fig. 3 Changes in FRET ratio of the maltose (A and C) and glucose (B and D) biosensor when in the presence of 0.0005–100 mM maltose (A) or 0.005–100 mM glucose (B), respectively, or 0–8 M urea (C and D). The encapsulated biosensors (empty dots) and free biosensors (filled dots) are reported and fitted with a Sigmoidal Dose Response tool of SigmaPlot (Systat Software Inc.). More detailed results in Fig. S1 and S2.† Data reported as average ± SEM, n ≥ 3. | ||
In order to assess the effect of encapsulation towards chemical denaturation, both the free sugar biosensor proteins and their encapsulated versions were incubated in the presence of different concentrations of urea. The effect of encapsulation on the biosensor was determined via the change in FRET ratio as a function of the urea concentration. As shown in Fig. 3, encapsulation had a stabilizing effect toward urea. Above 3 M urea, the functionalized nanoparticles containing the maltose biosensor maintained a higher FRET ratio compared to the equivalent free protein (Fig. 3C). Major changes in FRET ratio were detected in the presence of 0–2 M urea for the glucose biosensor for both the free and encapsulated form, with the encapsulated form being more stable (Fig. 3D).
Similarly, the functionalized nanoparticles with the maltose biosensor retained a slightly higher FRET ratio upon thermal treatment by incubation at 70 °C (Fig. S3 and S4†). The functionalized nanoparticles with the glucose biosensor showed the same behaviour as the free biosensor at different pH conditions with increasing FRET ratios at increasing pH and reaching a maximum at pH = 7.5–8 (Fig. S5 and S6†).
In conclusion, silica nanoparticles responsive to glucose or maltose were assembled by incorporating two specific FRET-based protein biosensors. The functionalized nanoparticles had affinity for glucose or maltose similar to the free biosensor protein and showed an enhanced performance towards denaturation by urea and temperature. Moreover, they were highly permeable to small molecules. Functional nanoparticles converting the local concentration of a metabolite, e.g. glucose or maltose, into a fluorescent signal could be prepared by combining an inorganic support such as silica with the specificity provided by FRET-based protein biosensors. Our concept developed here is very promising not only for in vitro sensing metabolites, e.g. in biological fluids, but also for in vivo metabolite analysis.
This research was supported by a grant from the Swiss Confederation and funded by Nano-Tera.ch within the Nano-Tera project “Fabrication of fluorescence biosensors in a Textile Dressing for Non-invasive Lifetime Imaging-based Wound Monitoring”, FLUSITEX (RTD 2013) that was scientifically evaluated by the SNSF.
Notes and references
- M. B. Bannwarth, S. Ebert, M. Lauck, U. Ziener, S. Tomcin, G. Jakob, K. Münnemann, V. Mailänder, A. Musyanovych and K. Landfester, Macromol. Biosci., 2014, 14, 1205–1214 CrossRef CAS PubMed.
- I. I. Slowing, B. G. Trewyn and V. S. Lin, J. Am. Chem. Soc., 2007, 129, 8845–8849 CrossRef CAS PubMed.
- M. Motornov, Y. Roiter, I. Tokarev and S. Minko, Prog. Polym. Sci., 2010, 35, 174–211 CrossRef CAS.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- L. Shang and G. U. Nienhaus, Mater. Today, 2013, 16, 58–66 CrossRef CAS.
- H. Liu, S. Wu, C. Lu, M. Yao, J. Hsiao, Y. Hung, Y. Lin, C. Mou, C. Yang, D. Huang and Y. Chen, Small, 2008, 4, 619–626 CrossRef CAS PubMed.
- T. Ye, Y. Du, C. He, B. Qiu, Y. Wang and X. Chen, Anal. Methods, 2012, 4, 1001–1004 RSC.
- S. Widmer, M. J. Reber, P. Muller, C. E. Housecroft, E. C. Constable, R. M. Rossi, D. Brühwiler, L. J. Scherer and L. F. Boesel, Analyst, 2015, 140, 5324–5334 RSC.
- M. J. Ruedas-Rama, J. D. Walters, A. Orte and E. A. H. Hall, Anal. Chim. Acta, 2012, 751, 1–23 CrossRef CAS PubMed.
- C. Zong, K. Ai, G. Zhang, H. Li and L. Lu, Anal. Chem., 2011, 83, 3126–3132 CrossRef CAS PubMed.
- P. Teolato, E. Rampazzo, M. Arduini, F. Mancin, P. Tecilla and U. Tonellato, Chem. – Eur. J., 2007, 13, 2238–2245 CrossRef CAS PubMed.
- D. Gao, Z. Wang, B. Liu, L. Ni, M. Wu and Z. Zhang, Anal. Chem., 2008, 80, 8545–8553 CrossRef CAS PubMed.
- W. B. Frommer, M. W. Davidson and R. E. Campbell, Chem. Soc. Rev., 2009, 38, 2833–2841 RSC.
- R. Moussa, A. Baierl, V. Steffen, T. Kubitzki, W. Wiechert and M. Pohl, J. Biotechnol., 2014, 191, 250–259 CrossRef CAS PubMed.
- K. Deuschle, S. Okumoto, M. Fehr, L. L. Looger, L. Kozhukh and W. B. Frommer, Protein Sci., 2005, 14, 2304–2314 CrossRef CAS PubMed.
- M. Fehr, W. B. Frommer and S. Lalonde, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9846–9851 CrossRef CAS PubMed.
- C. Mateo, J. M. Palomo, G. Fernandez-Lorente, J. M. Guisan and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2007, 40, 1451–1463 CrossRef CAS.
- A. Sassolas, L. J. Blum and B. Leca-Bouvier, Biotechnol. Adv., 2012, 30, 489–511 CrossRef CAS PubMed.
- G. Faccio, M. M. Kämpf, C. Piatti, L. Thöny-Meyer and M. Richter, Sci. Rep., 2014, 4, 5370 CAS.
- A. Cao, Z. Ye, Z. Cai, E. Dong, X. Yang, G. Liu, X. Deng, Y. Wang, S. Yang, H. Wang, M. Wu and Y. Liu, Angew. Chem., Int. Ed., 2010, 49, 3022–3025 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5an02573g |
| ‡ These authors contributed equally. |
| § Current address: Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB, Bio-, Electro and Chemocatalysis BioCat, Straubing Branch, Schulgasse 11a, 94315 Straubing (DE). |
| This journal is © The Royal Society of Chemistry 2016 |