Open Access Article
Open Access ArticleAn unsupervised MVA method to compare specific regions in human breast tumor tissue samples using ToF-SIMS†
Blake M.
Bluestein
a,
Fionnuala
Morrish
b,
Daniel J.
Graham
a,
Jamie
Guenthoer
b,
David
Hockenbery
b,
Peggy L.
Porter
b and
Lara J.
Gamble
*a
aUniversity of Washington, Dept. of Bioengineering, MolES Building, Box 351653, Seattle, WA 98195-1653, USA. E-mail: lgamble@uw.edu
bFred Hutchinson Cancer Research Center, Seattle, WA 98109, USA
First published on 15th February 2016
Abstract
Imaging time-of-flight secondary ion mass spectrometry (ToF-SIMS) and principal component analysis (PCA) were used to investigate two sets of pre- and post-chemotherapy human breast tumor tissue sections to characterize lipids associated with tumor metabolic flexibility and response to treatment. The micron spatial resolution imaging capability of ToF-SIMS provides a powerful approach to attain spatially-resolved molecular and cellular data from cancerous tissues not available with conventional imaging techniques. Three ca. 1 mm2 areas per tissue section were analyzed by stitching together 200 μm × 200 μm raster area scans. A method to isolate and analyze specific tissue regions of interest by utilizing PCA of ToF-SIMS images is presented, which allowed separation of cellularized areas from stromal areas. These PCA-generated regions of interest were then used as masks to reconstruct representative spectra from specifically stromal or cellular regions. The advantage of this unsupervised selection method is a reduction in scatter in the spectral PCA results when compared to analyzing all tissue areas or analyzing areas highlighted by a pathologist. Utilizing this method, stromal and cellular regions of breast tissue biopsies taken pre- versus post-chemotherapy demonstrate chemical separation using negatively-charged ion species. In this sample set, the cellular regions were predominantly all cancer cells. Fatty acids (i.e. palmitic, oleic, and stearic), monoacylglycerols, diacylglycerols and vitamin E profiles were distinctively different between the pre- and post-therapy tissues. These results validate a new unsupervised method to isolate and interpret biochemically distinct regions in cancer tissues using imaging ToF-SIMS data. In addition, the method developed here can provide a framework to compare a variety of tissue samples using imaging ToF-SIMS, especially where there is section-to-section variability that makes it difficult to use a serial hematoxylin and eosin (H&E) stained section to direct the SIMS analysis.
Introduction
Mass spectrometry imaging (MSI) is quickly emerging as a key research tool in biological research areas such as neuroscience, drug delivery, and cancer.1–4 The combination of MS chemical and molecular specificity with imaging capabilities has provided a new perspective for biological sample analysis including localization and interactions of drugs in cells and tissues,5–9 proteomics,10,11 and lipidomics.12–14 Specifically, the MS imaging technique time-of-flight secondary ion mass spectrometry (ToF-SIMS) is a label-free method with micron resolution imaging capabilities making it well suited for imaging of cells,15,16 and key tissue regions.17,18 Utilizing the micron lateral resolution of SIMS can be crucial in the process of separating regions of interest within tumor microenvironments for cancer research. These microenvironments can regulate anticancer activities but can also promote cancer progression and provide biological protection which limits therapeutic efficacy and delivery.19 By combining micron resolution imaging with molecular information, it is possible to observe and begin to interpret potential immune response related metabolic events that may associate with cancer progression or regression within the tumor.Breast cancer biopsies can vary in cellular density as well as percent of cancer cell and stroma (connective tissue composed of fat and fibrous tissue) content. Pathological assessment is typically performed with histological staining to determine the location, type and grade of tumors, but does not always predict patient outcome or response to chemotherapeutics.20–25 Stromal heterogeneity and tumor–stroma interactions provide prognostic indicators for invasive growth and metastasis.26–29 Previous studies indicate that stromal-cancer cell metabolite interchange aids tumor growth and progression.30,31 It is hypothesized that the stromal biochemical state may dictate sensitivity to chemotherapy.32 However, it is difficult to acquire metabolic data specifically from cellular and stromal regions, as these regions can be difficult to isolate for metabolic profiling due to the complexity of their spatial distribution. Separating out chemical information specifically from the stromal or cellular region can be useful to compare chemistries from different tissue areas that contain varying amounts of these specific regions.
In this study, a combination of ToF-SIMS and multivariate imaging analysis techniques are used as an analytical tool to identify chemical variation of specific cellular and stromal regions from breast cancer specimens and to compare the chemical variation between pre- and post-chemotherapy. We describe different analysis methods to isolate and interpret metabolic features of cancer cell regions within tissues including pathologist-driven selection of regions of interest (ROIs) using hematoxylin and eosin (H&E) stained tissue sections as well as the use of an unsupervised imaging MVA method to separate out stromal regions in the SIMS images. Herein unsupervised refers both to the fact that principal component analysis (PCA) is an unsupervised MVA method (meaning no input other than peak intensities are used), and to the fact that by using PCA to select ROIs we demonstrate that one can isolate cellular and stromal areas within breast tissue sections and reduce scatter within the resulting scores without introducing human bias through hand-selected regions. This method further provides improvement to isolate and analyze complex regions that consist of either cellular/tumor or stromal regions that cannot be selected by hand or the threshold of just one mass spectrometric image. The MVA method of PLS-DA has been successfully used with InfraRed (IR) imaging data to discern different regions in breast cancer tissue and identify tumor and non-tumor areas within a set of samples.33 However, to our knowledge, the method of using PCA to select ROIs for comparing different regions has not yet been applied to ToF-SIMS imaging data. ToF-SIMS has been used previously to study diseased tissues and cells with a major focus on lipids,18,34–36 which are known to contribute to and also regulate a range of metabolic and biochemical processes within cells. Furthermore, alteration in lipid metabolism is a hallmark of carcinogenesis.37 The imaging data in this study is specifically used to assess how lipid molecules relate to the differences found between tissue samples. One major distinction, however, is that here we compare similar regions (e.g. cellular regions) of four different tissue samples to investigate chemical differences between untreated tumors and those that have been exposed to chemotherapy treatments.33 In addition, the method developed here can provide a framework to compare multiple tissue samples using imaging ToF-SIMS when there is difficulty using a region of interest marked on a serial (consecutive) section to direct the SIMS analysis due to section-to-section variability. This novel approach utilizes the high lateral resolution capability of imaging ToF-SIMS to compare highly specific regions (e.g. just tumor cells) from one tissue to another and uses spectral PCA to highlight chemical differences between those tissue specimens.
Methods
Tissue sample preparation
Paired pre- and post-chemotherapy biopsy specimens were obtained from patients consented according to institutional review board protocols. Both patients received standard pre-operative chemotherapy with doxorubicin at 60 mg m−2 IV and cyclophosphamide 600 mg m−2 IV on day 1 every 14 days for 4 cycles and paclitaxel 80 mg m−2 IV weekly × 12 weeks. The patient with ‘Basal Like’ breast cancer received the paclitaxel first while the patient with Luminal A breast cancer received the Doxorubicin and cyclophosphamide combination first, but the order of therapy is known to not substantially affect treatment efficacy. Specimens were immediately embedded in Tissue-Tek® (Fisher Scientific) optimum cutting temperature (OCT) compound, cryopreserved using liquid nitrogen and stored in a −80 °C freezer. Frozen tissue blocks were sectioned in a cryostat-microtome held at −23 °C at the Fred Hutchinson Cancer Research Center (FHCRC). Each of the four tissue samples was serially sectioned three times and each section was ∼5 μm in thickness. The first and third sections were stained for optical imaging using hematoxylin and eosin (H&E), while the second section was analyzed by ToF-SIMS. The second slice of tissue was placed directly on a 2 cm2 silicon wafer that was previously cleaned with two successive sonications in dichloromethane, acetone, and methanol.38 The samples were then placed in a petri dish, sealed with Parafilm® (VWR International), and transported to the University of Washington for immediate ToF-SIMS analysis. Each tissue was sectioned on a different day and the time from tissue cutting to analysis was less than 90 minutes for any sample.Gene expression subtype identification
Six to nine tissue sections were macrodissected to select regions containing the highest proportion of invasive tumor cells to reduce contamination from non-tumor cells. RNA was isolated using the AllPrep DNA/RNA Mini Kit (Qiagen Inc., Valencia, CA) and gene/transcript expression was assessed using the WG-DASL® (HumanHT-12 v4) Assay (Illumina, Inc., San Diego, CA). Data processing and analysis were done in the R environment (v3.0.3). The raw expression data were pre-processed and median normalized using the Bioconductor lumi package,39 and gene expression intrinsic subtypes (Luminal A and Basal-like) were determined using the 50-gene panel described by Parker et al.40 with the software Bioconductor genefu package.41ToF-SIMS
ToF-SIMS experiments were performed using an ION-TOF TOF.SIMS 5-100 (ION-TOF GmbH, Münster, Germany) equipped with a liquid metal ion gun (LMIG) for analysis and an electron flood gun for charge neutralization. The LMIG was used to generate a pulsed 25 keV Bi3+ beam impacting the target at an angle of 45°. The Bi3+ beam was set in spectroscopy mode for high mass resolution (HMR) to acquire spectra in both polarities and fast imaging mode to acquire high spatial resolution (HSR) negative polarity images. The Bi3+ current was typically 0.13–0.15 pA for HMR and 0.05 pA for HSR. Target currents were measured before each data set using a Faraday cup. HSR mode ToF-SIMS images with micron spatial resolution were acquired and compared to features found in H&E images. Large area images of the entire tissue biopsy were created by manually stitching individual optical images of 800 μm × 800 μm from the video camera within the ToF-SIMS before analysis of each tissue. These large optical stitched images were then aligned to H&E images using the tissue borders to aid in selecting areas where analysis patches were to be acquired. For all data collection, HMR positive ion data was acquired followed immediately by HMR negative ion data on the same area. X and Y sample stage coordinates were saved in the software to ensure data acquired was from the same region in both polarities. HSR images were obtained from each sample region after all HMR spectra were completed. In HMR mode, mass resolution (m/Δm) for the C2H3+ ion was greater than 4500. Positive ion spectra were calibrated to CH3+, C2H3+, and C4H5+. Negative ion spectra were calibrated to CH−, OH−, and C2H−. Spectra were acquired from 1 mm × 1 mm or 1.6 mm × 0.6 mm “patches” comprising of 25 or 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 μm × 200 μm “tiles” on each tissue, an example of positions is shown in Fig. 1. Each tile contains 256 × 256 pixels, giving the patches a total pixel count of 1280 × 1280. Selecting to analyze three large patches rather than the entire tissue biopsy sample was chosen due to the time intensiveness required to analyze the samples. Long analysis times can lead to degradation of lipid signals or lipid migration.38,42 Thus, analyzing three patches results in a timely analysis, providing the most relevant and reproducible data of the tissue's native chemical composition. The Bi3+ dose was limited to ≤5.0 × 1011 ions per cm2 for each tile in both positive and negative ion modes, resulting in a total Bi3+ dose ≤1.0 × 1012 ions per cm2 per tile. SurfaceLab 6 software (ION-TOF GmbH, Münster, Germany) was used for all analyses.
200 μm × 200 μm “tiles” on each tissue, an example of positions is shown in Fig. 1. Each tile contains 256 × 256 pixels, giving the patches a total pixel count of 1280 × 1280. Selecting to analyze three large patches rather than the entire tissue biopsy sample was chosen due to the time intensiveness required to analyze the samples. Long analysis times can lead to degradation of lipid signals or lipid migration.38,42 Thus, analyzing three patches results in a timely analysis, providing the most relevant and reproducible data of the tissue's native chemical composition. The Bi3+ dose was limited to ≤5.0 × 1011 ions per cm2 for each tile in both positive and negative ion modes, resulting in a total Bi3+ dose ≤1.0 × 1012 ions per cm2 per tile. SurfaceLab 6 software (ION-TOF GmbH, Münster, Germany) was used for all analyses.
Principal component analysis
Principal component analysis (PCA) was applied to ToF-SIMS images acquired from the tissues using all pixels in the data set (herein referred to as image data and displayed as images) and to summed spectral data from individual patches (herein referred to as spectral data and displayed as individual data points).Data used in this study were pre-processed for PCA as follows: (1) ToF-SIMS image data were Poisson scaled and mean centered, and (2) summed spectral data from individual tile images were normalized to the sum of the intensities of all of the peaks in the peak list, square-root transformed, and mean centered. Regions of exposed silicon substrate and OCT (e.g. holes or tears from cutting tissue and embedding medium surrounding tissue) were excluded from all analyses by applying a threshold to the pixels with a Si+ signal, where m/z 27.9 was used to detect silicon and m/z 332.2 (C14H29+, a fragment of the benzalkonium additive in OCT)38 is used to detect OCT areas. All PCA was performed using the NBToolbox SpectraGUI and ImageGUI (Daniel Graham Ph.D., NESAC/BIO, University of Washington), that operate within MATLAB (MathWorks, Natick, MA). Peaks were chosen whose maximum intensity was twice or more than that of the average background intensity. The spectra from all tissues were overlaid and then peaks were manually selected and integrated to full width half max. All peaks below m/z 920 were selected, excluding known salt, salt adducts, substrate and inorganic peaks. A total peak list of 846 and 807 peaks were chosen from the positive and negative ion modes, respectively. All peaks in the list were used for image PCA analysis while spectral PCA was limited to peaks with m/z above 200 resulting in 391 and 329 peaks for the positive and negative ions peak lists, respectively.
PCA, using the ImageGUI, is first applied to image data formatted as .bif6 files from SurfaceLab 6. No peaks are excluded from this data except salts, substrate (Si and Si containing peaks), and embedding medium (OCT). Normalization was not applied to imported image patches; image data were pre-processed by Poisson scaling and mean centering before PCA.
The presence of large fatty acid droplets, observed as well defined high intensity areas of C16:1 (C16H29O2−, palmitoleic acid, m/z 253.2),43,44 C16:0 (C16H31O2−, palmitic acid, m/z 255.2),44,45 C18:2 (C18H29O2−, linoleic acid, m/z 279.2),44,46 C18:1 (C18H33O2−, oleic acid, m/z 281.2),44,45 and C18:0 (C18H35O2−, stearic acid, m/z 283.2),13,44,45,47 were occasionally observed in different tissue sections. The strong signal from the fatty acid droplets would dominate PCA and the main variability between the samples would then be related to fatty acid droplets present in that particular tissue slice. Therefore, the fatty acid droplets were removed prior to PCA as manually selected ROIs of the tissue image data, using the polyline function within SurfaceLab 6, so sample comparison could be focused on specific tissue regions. Droplets were easily identified in images as localized areas with characteristic fatty acid peak intensities at least 5 times that of the fatty acids distributed within the remaining tissue section. It is important to note that breast tissue is a fatty tissue, therefore the prevalence of fatty acid droplets within tissue sections can vary. Analysis of other types of tissues (i.e. brain, heart, and liver) may or may not contain these droplets.44,45,48,49 When analyzing chemical variances between patients or within one patient, including the fatty acid droplets present in breast tissue could cause misinterpretation of the data due to the variability of droplet presence (i.e. if there happened to be a droplet in that particular biopsy sample and/or section).
Scores images that corresponded to cellular and stromal areas from serial H&E images were selected to be used as masks. Using SurfaceLab 6, scores images were imported and spectra reconstructed by applying a 10% minimum, 90% maximum signal threshold to the pixels within the selected score image. The resulting data, in .bif6 format, were imported back into ImageGUI and the patch parsed into individual 200 μm × 200 μm tiles, shown in Fig. 1, where each tile represents one data point in the spectral PCA plots. The parsed data was then imported in .xlsx format into SpectraGUI for spectral analysis, where each individual tile represents one data point in the PCA scores plots. Imported data were normalized to the sum of the intensities of all of the peaks in the peak list, square-root transformed, and mean centered prior to spectral PCA.
Results and discussion
Paired pre- and post-chemotherapy tissue samples from two patients were acquired for a total of four tissue samples. The hematoxylin and eosin (H&E) stained sections for the four tissue samples are shown in Fig. 1A and B. The hematoxylin stain, blue color, indicates the cellular nuclei while eosin stains the stromal or connective tissue in a lighter pink color. Three stitched patches (as described in Fig. 1) were taken per tissue for a total of 12 stitched patches. Each of the breast cancer specimens were classified into gene expression intrinsic subtypes.40 The first pre/post-therapy specimen pair was classified as a Luminal A breast cancer, characterized by having genetic expression patterns similar to the luminal epithelial component of the normal breast.20,50 Generally, the Luminal A cancers are estrogen receptor (ER) and progesterone receptor (PgR) positive, and human epidermal growth factor receptor 2 (HER2) negative with lower expression of proliferative genes. The second pre/post-therapy tissue pair was classified as a Basal-like breast cancer. The Basal-like subtype typically lacks expression of ER, PgR, and shows low or no HER2 expression. Basal-like tumors are characterized by an expression pattern corresponding to that of the basal epithelial cells in the normal breast and body and highly express proliferation genes.20,50In order to ascertain chemical differences between pre- and post-chemotherapy tissues, PCA was used to analyze ToF-SIMS spectral and image data. Three different methods are used to acquire information from different ROIs from different tissue slices. Specifically, (1) using the spectral data from all patches within the region imaged from a given tissue, (2) using regions indicated by pathologist on a H&E stained slice image and (3) using the spectral data from all patches after generating ROIs using imaging PCA. The negative polarity ion data showed the best correlation between the pre and post chemotherapy treatment samples and thus is used to compare the spectral PCA results generated from the three different ROI methods. The positive ion PCA results did not show separation that correlated with pre- and post-chemotherapy treatment regardless of the method used. The positive polarity data are presented and discussed using the last method only.
MVA of all patches
As detailed in the methods section, each analysis patch was separated into individual tiles after removal of substrate, embedding medium, and large fatty acid deposits, followed by the application of PCA. The spectral PCA results from the negative ion polarity data with peaks above m/z 200 (key m/z values, deviations, and proposed biological molecule are shown in ESI Table 1†) of the entire stitched patches (both pre and post for both tissue types) are shown in Fig. 2. The lines above and below the data points indicate 95% confidence intervals. PC1 data (not shown) indicates slight separation between tissues from the two patients, rather than by pre- and post-therapy possibly identifying person-to-person chemical variation. In PC2, it is noticeable that the post-therapy tissues do indicate a trend with higher scores, which correspond to a high loading value of fatty acid C16:0 (C16H31O2−, palmitic acid, m/z 255.2), while the pre-therapy tissues trend with lower score values, corresponding to the strongest negative loading peak of Vitamin E (C29H49O2−, m/z 429.3)46 and fatty acid C18:1 (C18H33O2−, oleic acid, m/z 281.2). Using the entire patch as a region of interest, the scores exhibit a large spread between the 95% confidence intervals signifying a high amount of variability within each patch as well as for different patches within the tissue. It is possible that the large variability may be due to comparison of the entire tissue areas rather than comparison of specific tumor features within the analysis patch.MVA analysis of H&E driven ROI selections
Due to the heterogeneity and complexity of the sample, a second type of ROI selection was performed to focus on pathologist-recommended regions, e.g. high density of cancer cells, from the serial H&E slice. These regions are outlined in light blue in the H&E and black in the ToF-SIMS summed image of CN− and CNO− (Fig. 3A and B), to provide a guide for a more accurate ToF-SIMS spectral comparison between cancer cell regions or “cancer cell nests” and stromal regions. Fig. S1† shows the pathologist-selected areas on the H&E images for all four tissues.It can be noted (in Fig. 3) that the H&E image of the tissue that was examined by the pathologist is not identical to the ToF-SIMS image. This is not surprising since there is at least a 5 μm difference between these serial sections (including the section thickness and frost buildup on the tissue sample prior to the slicing of the next section). In an effort to develop a minimally time consuming process, a rough estimate of the pathologist-selected areas were used, meaning that only tiles that primarily corresponded with pathologist-selected areas were selected for the analysis (Fig. 3C). The results using PCA on this more selective region of interest are shown in ESI Fig. 2.† Comparing the method of a selective region to whole spot analysis (Fig. 2), it can be seen that the percentage of variance of PC2 as well as the loadings peaks look similar for both types of ROI analyses. However, the scores separation between the pre- and post-therapy specimens from the Basal-like cancer are no longer observed while the pre and post specimens from the Luminal A cancer now show a clearer separation. While a hand drawn ROI to attempt to directly select the regions selected by the pathologist may have slightly improved these results, the section-to-section variability noted during the analysis likely plays the major role in incorrectly selecting the ROIs on the ToF-SIMS image (since the regions of interest on the pathology section may not match those on the ToF-SIMS section). For example, the image shown in Fig. 3 has a large, obvious stromal feature (large pink region), while other tissue slices such as the Basal-like post chemotherapy tissue (ESI Fig. 1†) had very small, well separate regions that were difficult to correlate and identify on the ToF-SIMS image. This difference in separation could be due the following; (1) large stromal areas excluded by the pathologist in the Basal-like post-chemotherapy tissue, (2) small distributed cellular areas included by the pathologist within the Basal-like pre-chemotherapy tissue, (3) lowering the number of tiles used in PCA, and (4) larger cellular areas comprised of more tiles included by the pathologist within the Luminal A cancer tissues.
Variations in serial tissue slices
Composition and localization of cellular areas within a tumor vary as you move serially through a tissue block. This variability increases the complexity of choosing the ToF-SIMS analysis region of a tissue section by comparing to a region from a serial section. Fig. 4 illustrates an example of the heterogeneity between the three serial 5 μm sections. For accurate spot location on each section, images were aligned by overlay at the same magnification and approximate spot location boxed. In Fig. 4A and D, the initial H&E section reviewed by the pathologist, a circular structure, noted by a black arrow, is present within the 1 mm × 1 mm analysis area. It can also be seen that in Fig. 4A there is not a substantial amount of cell nuclei (blue stain) populating the region. Fig. 4B and E shows the ToF-SIMS stitched microscope camera image and the summed CN− and CNO− HSR image acquired. Fig. 4C is the serial section cut after the ToF-SIMS section. The higher intensity region in the center of the ion image (Fig. 4E) is indicative of the stromal region whereas the lower intensity regions indicate cellular regions (with the exception of the area with no signal in the lower left area of the image which is due to fatty acid droplets). Comparing all the images in Fig. 4 it can be seen that the ToF-SIMS image in 4E is more similar to the H&E image in 4F, but still has some differences highlighting the difficulties in using serial sections to choose analysis regions as certain structures vary in depth.MVA using unsupervised selection of region specific ROIs by PCA
H&E-stained serial tissue sections do not necessarily show the same areas of interest as the ToF-SIMS analysis section and, as shown in the previous two methods, the amount of cellular and stromal area in the analysis area may dominate the PCA separation. Therefore, a different method must be developed to focus on tissue areas of interest that can provide tissue to tissue slice comparisons. Additionally, there is interest in comparing specific regions from different tissue samples (for example comparing only cellular regions from two different patient biopsies). While the tissue shown in Fig. 3 had a rather prominent stromal feature, making it relatively simple to separate out major stromal areas from the cellular areas, many of the tissue sections had less prominent stromal features making it more difficult to use a coarse method to precisely separate out the stromal and cellular areas. In order to more precisely separate the stromal and cellular areas of the tissue, PCA was applied to the image data of each patch with an m/z 0–920 peak list. The low mass peaks, such as CN− and CNO−, were previously shown to be indicative of stromal regions (e.g.Fig. 3B) and would aid in separation of these areas.In Fig. 5, principal component 2 image scores (A and C) and loadings (B and D) produced from image PCA demonstrate the separation between cellular (Fig. 5A and B) and stromal regions (Fig. 5C and D), which can be visually seen to correspond with the cellular and stromal structures visible in adjacent H&E-stained sections as seen in Fig. 3A. Again, the tissue section with the most prominent stromal feature is chosen to demonstrate the utility of this method to separate out stromal features. In this particular sample, PC1 separates the presence of a fatty acid droplet and regions exhibiting vitamin E (shown in Fig. 6C) from the remainder of the image, which is dominated by high loading peaks of CN− and CNO− and other low mass fragments (not shown). In tissue samples where fatty acid droplets are not present, the separation between cellular and stromal is found in PC1.
 | ||
| Fig. 5 Negative ion image PCA results with a variance of 0.66% from an example patch. (a) Positive scores image showing the isolation of cellular regions. (b) Positive loadings that display the chemical species identified within (a) the cellular scores image. (c) Negative scores image or the stromal region. (d) Negative loadings showing the chemical species identified within the stromal region. The cellular and stromal regions can be distinguished from the H&E stained image presented in Fig. 3a. Image contrast was enhanced for clearer presentation for publication. Scale bars = 200 μm. | ||
The PC2 loadings plot for this patch exhibit high negative loadings for CN−, CNO−, and the fatty acids C18:2 (C18H29O2−, linoleic acid, m/z 279.2) and C18:1 (C18H33O2−, oleic acid, m/z 281.2). These negative loading masses can be associated with the negative PC2 scores image analogous to the stromal region of the tissue. The high negative loading of the fatty acid peaks could be due to the fatty acid droplet located at the left edge of the patch, visible as a high intensity region in the negative PC2 scores image. The composition of these fatty acid droplet regions, which appear as white (un-stained) ‘holes’ in the histology image, are easily identified with ToF-SIMS imaging. Consistent trends generated by image PCA observed from the loadings plot show CN−, CNO−, and PO3H− (m/z 79.9) loading in the direction of the stromal region scores.
The positive loading masses can be associated with the positive PC2 scores image, which is indicative of the cellular regions within the tissue sample. General trends observed while using image PCA indicated that cellular areas consistently had higher relative intensities of fragments related to vitamin E (C10H11O2−, m/z 163.1 and C29H49O2−) and phosphoinositol (C9H16PO9−, m/z 299.05).51 Due to the section-to-section variability in the samples, several cellular domains were revealed in the imaging PCA analysis of the tissues that were not visible in the histology images.
The scores images that were representative of cellular and stromal areas were used to create an ROI threshold “mask”, to extract the imaging mass spectral information specifically from stromal and cellular regions. Fig. 6 demonstrates the process used to create specific cellular and stromal ROIs using the image PCA scores as masks. First, the representative cellular and stromal scores images are scaled to equal the same number of pixels acquired in the ToF-SIMS image (Fig. 6A and B). As previously discussed in the methods section, breast tissue is a fatty tissue and the presence of fatty acid droplets may vary section-to-section and between patient specimens. When comparing the chemical variance between one patient or many, it is important that the data is not misinterpreted by the presence or absence of fatty acid droplets in a particular specimen. Before the mask is applied, fatty acid droplets must be removed from the scores image or the results from spectral PCA will be heavily influenced by their presence or absence when comparing sample chemistries. The droplet can be verified by viewing the raw data images. Fatty acid droplets generally separated from the tissue sample and are typically visible in PC1 scores (Fig. 6C, white arrow). In order to remove the fatty acid droplets from these data sets, PC2 can be overlaid with PC1 and the fatty acid droplet area can be subtracted from the PC2 scores image. Once the droplet area has been removed new cellular or stromal images can be generated. These new images can then be applied as ROI masks as previously described. An example of completed masks are shown in teal in Fig. 6D and E.
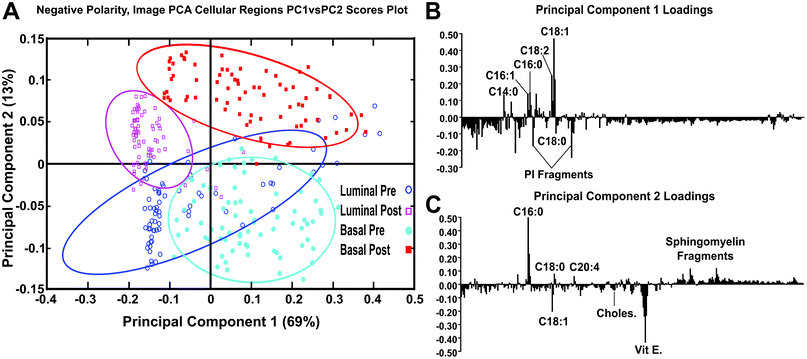
After spectra were reconstructed using either the cellular or stromal ROI masks, the ROI patch areas were subjected to the same process, parsing the patch into tiles for individual data points and followed by spectral PCA of the tiles as was done in the previous datasets. Fig. 7 shows the resulting spectral PCA scores and corresponding loadings plots for PC2 of the reconstructed tiles of the cellular regions identified by image PCA. The scores trends are similar to those seen in PC2 scores of the pathologist-selected areas (ESI Fig. 2B†) as well as the analysis of the entire patch (Fig. 2). As with the previous analysis methods, the post-therapy tissues have positive scores values and the pre-therapy tissues have negative scores. However, when data specifically from the cellular regions of the images are compared, the spread of the data between the confidence intervals is reduced. The loadings plots for PC2 in Fig. 2B and ESI Fig. 2B† are also similar to the loadings plot seen in Fig. 7B, where saturated fatty acids C16:0, C18:0 (C18H35O2−, stearic acid, m/z 283.2) and fragments of possible sphingomyelin (C34H67NO6P−, C36H69NO6P−, C38H76N2O6P−, SM (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), m/z 616.5, 642.5, 687.6 respectively)44 have positive loadings, while vitamin E and the unsaturated fatty acid C18:1 load negatively, indicating different chemical profiles for cellular regions within pre/post-therapy tissues. Key differences, however, include little to no contribution from PI fragments or C14. The PC1 vs. PC2 scores plot and corresponding loading plots (Fig. 8) for the cellular specific ROIs shows both the separation between pre- and post-chemotherapy tissues on PC2 as well as some separation between the Basal-like subtype and Luminal A subtype tissues across PC1. There is large variability in the Luminal A pre-chemotherapy tumor tissue, where the confidence interval slightly overlaps both post-chemotherapy tissues. However, both the pre and post-chemotherapy Luminal tissues have largely negative scores on PC1, while the Basal-like tissues have largely positive scores. Phosphoinositol fragments (C6H1−PO8−, m/z 241.01, and C6H12PO9−, m/z 259.02) and C18:0 trend with Luminal type tissues while C14:0 (C14H27O2−, m/z 227.2), C16:0, and C18:1 fragments are correlated with the Basal-like type tissues.
1), m/z 616.5, 642.5, 687.6 respectively)44 have positive loadings, while vitamin E and the unsaturated fatty acid C18:1 load negatively, indicating different chemical profiles for cellular regions within pre/post-therapy tissues. Key differences, however, include little to no contribution from PI fragments or C14. The PC1 vs. PC2 scores plot and corresponding loading plots (Fig. 8) for the cellular specific ROIs shows both the separation between pre- and post-chemotherapy tissues on PC2 as well as some separation between the Basal-like subtype and Luminal A subtype tissues across PC1. There is large variability in the Luminal A pre-chemotherapy tumor tissue, where the confidence interval slightly overlaps both post-chemotherapy tissues. However, both the pre and post-chemotherapy Luminal tissues have largely negative scores on PC1, while the Basal-like tissues have largely positive scores. Phosphoinositol fragments (C6H1−PO8−, m/z 241.01, and C6H12PO9−, m/z 259.02) and C18:0 trend with Luminal type tissues while C14:0 (C14H27O2−, m/z 227.2), C16:0, and C18:1 fragments are correlated with the Basal-like type tissues.
ESI Fig. 3† shows the PCA scores and corresponding loadings plots of PC2 comparing the stromal regions of the samples in the negative polarity. Phosphoinositol fragments, C18:1, and vitamin E trend to be associated with the pre-chemotherapy samples. Where C14:0, C16:0, C18:0, and sphingomyelin fragments are correlated with the post-chemotherapy samples. The 95% confidence intervals have a wider spread for stromal region data than was seen for the cellular region data, however, a trend is still noticeable within the stroma data indicating that differences between pre- and post-chemotherapy samples can be found in the stromal as well cellular regions.
As was noted previously for ToF-SIMS investigation of breast cancer cells, the negative polarity ions provide the ability to observe distributional changes of fatty acids and intact lipids, while the positive data has been shown to provide the ability to observe changes in mono and diacylglycerides.47 While PCA analysis of the pre- and post-chemotherapy tissues using negative polarity ions similarly shows a trend in the scores related to fatty acids and lipids, the positive ion data results in more overlap of the scores 95% confidence intervals than found for the negative ion data. This is best shown in ESI Fig. 4† where PC2 vs. PC4 scores and corresponding loadings are shown for the positive ion data. There is near separation between the pre/post Basal-like samples in PC2, but separation between the Luminal A pre/post samples is not observed until PC4. In summary, there does not appear to be a trend in the loadings peaks for the pre- and post-therapy tissues using positive ions, indicating that any changes in the mono and diacylglyceride content as a result of chemotherapy are not consistent across tissue types.
Conclusions
Current cancer research has indicated, primarily through gene expression data, that specific microenvironments in breast tumors may provide signals and nutrients to promote cancer cell survival and/or chemoresistance.27,28,52 However, due to the heterogeneity of human breast cancer tissues, it remains difficult to acquire supportive metabolic data to aid in understanding tumor growth and treatment efficacy. The regions of interest (ROIs) selected for such molecular characterization require micron-level lateral resolution. Here we show that imaging ToF-SIMS can be used to chemically identify distinct tissue regions in tumors with high lateral resolution. In this work, we have presented an unsupervised methodology for isolating and analyzing specific tissue regions providing a way to compare similar regions in multiple tissue slices. These results demonstrate that the combination of imaging ToF-SIMS and image principal component analysis (PCA) can be used as an unsupervised method to select distinct ROIs within tissues. Comparisons are made using the entire analysis regions as well as hand-selected ROIs. When different tissue samples are compared using imaging PCA-driven ROIs there is less spread in the PCA scores. An advantage of using the imaging PCA-directed method is that it allows for like regions to be compared in spectral PCA and thereby improves chemical separation when multiple tissue samples are compared. Here four different tissue sections from two different patients before and after chemotherapy were compared using the negative ion ToF-SIMS data and PCA-driven ROI selection. Trends are found for tissues breast cancer specimens that were taken before chemotherapy treatment (pre) and those taken from the same patient after treatment (post). From the spectral PCA results it is seen that the unsaturated fatty acids C16:0 and C18:0 and sphingomyelin correspond with the post-chemotherapy tissues. Deficiency of sphingomyelin is thought to be related to the disruption of apoptosis in highly invasive cancer cells,53 therefore an increase in intensity in the post-treated samples compared to the pre-treated may correlate with treatment response of the patient. C16:0 has been shown to generate apoptotic signals, some related to sphingolipids.54 Conversely, overexpression of fatty acid synthase in breast cancer, which is responsible for the synthesis of C16:0, has been shown to contribute to drug resistance.55Vitamin E and the unsaturated fatty acid C18:1 correspond with the pre-chemotherapy tissues in the PCA analysis. Vitamin E may provide strong antioxidant protection of cancer cells from lipid peroxidation, facilitating tumor growth when in the presence of reactive oxygen species (ROS).56 That the vitamin E signal corresponds to the pre-treated and not the post-treated tissues is consistent with several studies that have shown that chemotherapy and radiation therapy are associated with increased formation of reactive oxygen species and depletion of critical plasma and tissue antioxidants.57,58 The C18:1 oleic acid is known to prevent cytotoxicity and decrease mitochondrial superoxide production induced by C16:0 palmitate.59 This provides a possible explanation for the trends seen within the pre-chemotherapy treated tissues for both C18:1 and vitamin E. While these findings require further experimental investigation to gain a concrete understanding in cancer biology, the results demonstrate the utility of PCA-driven ROI selection of ToF-SIMS data to compare metabolic trends of specific regions across multiple patients and tissue sections.
Acknowledgements
We would like to thank Kelly Wirtala at FHCRC for expert sectioning of tissue samples and helping with the development of sample preparation protocols. We would also like to thank Michael Robinson for scientific discussions and data acquisition. We would also like to acknowledge our funding sources: NESACBIO NIH P41 EB002027, SPORE NIH P50 CA138293, and NSF-GRFP DGE-0718124/1256082.References
- F. Benabdellah, A. Seyer, L. Quinton, D. Touboul, A. Brunelle and O. Laprévote, Anal. Bioanal. Chem., 2009, 396, 151–162 CrossRef PubMed.
- L. A. McDonnell and R. M. Heeren, Mass Spectrom. Rev., 2007, 26, 606–643 CrossRef CAS PubMed.
- L. S. Eberlin, A. L. Dill, A. B. Costa, D. R. Ifa, L. Cheng, T. Masterson, M. Koch, T. L. Ratliff and R. G. Cooks, Anal. Chem., 2010, 82, 3430–3434 CrossRef CAS PubMed.
- R. Van de Plas, J. Yang, J. Spraggins and R. M. Caprioli, Nat. Methods, 2015, 12, 366–372 CrossRef CAS PubMed.
- S. S. Rubakhin, J. C. Jurchen, E. B. Monroe and J. V. Sweedler, Drug Discovery Today, 2005, 10, 823–837 CrossRef CAS PubMed.
- D. S. Cornett, S. L. Frappier and R. M. Caprioli, Anal. Chem., 2008, 80, 5648–5653 CrossRef CAS PubMed.
- M. K. Passarelli, C. F. Newman, P. S. Marshall, A. West, I. S. Gilmore, J. Bunch, M. R. Alexander and C. T. Dollery, Anal. Chem., 2015, 87, 6696–6702 CrossRef CAS PubMed.
- J. M. Wiseman, D. R. Ifa, Y. X. Zhu, C. B. Kissinger, N. E. Manicke, P. T. Kissinger and R. G. Cooks, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18120–18125 CrossRef PubMed.
- B. Prideaux and M. Stoeckli, J. Proteomics, 2012, 75, 4999–5013 CrossRef CAS PubMed.
- E. H. Seeley and R. M. Caprioli, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18126–18131 CrossRef CAS PubMed.
- H. Nygren and P. Malmberg, Proteomics, 2010, 10, 1694–1698 CrossRef CAS PubMed.
- N. Goto-Inoue, T. Hayasaka, N. Zaima and M. Setou, Biochim. Biophys. Acta, 2011, 1811, 961–969 CrossRef CAS PubMed.
- M. K. Passarelli and N. Winograd, Biochim. Biophys. Acta, 2011, 1811, 976–990 CrossRef CAS PubMed.
- L. S. Eberlin, A. L. Dill, A. B. Costa, D. R. Ifa, L. Cheng, T. Masterson, M. Koch, T. L. Ratliff and R. G. Cooks, Anal. Chem., 2010, 82, 3430–3434 CrossRef CAS PubMed.
- J. Brison, M. A. Robinson, D. S. Benoit, S. Muramoto, P. S. Stayton and D. G. Castner, Anal. Chem., 2013, 85, 10869–10877 CrossRef CAS PubMed.
- M. A. Robinson, D. J. Graham and D. G. Castner, Anal. Chem., 2012, 84, 4880–4885 CrossRef CAS PubMed.
- J. W. Park, H. Jeong, B. Kang, S. J. Kim, S. Y. Park, S. Kang, H. K. Kim, J. S. Choi, D. Hwang and T. G. Lee, Sci. Rep., 2015, 5, 11077 CrossRef CAS PubMed.
- B. Cillero-Pastor, G. Eijkel, A. Kiss, F. J. Blanco and R. M. Heeren, Anal. Chem., 2012, 84, 8909–8916 CAS.
- M. R. Junttila and F. J. de Sauvage, Nature, 2013, 501, 346–354 CrossRef CAS PubMed.
- C. M. Perou, T. Sorlie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, C. A. Rees, J. R. Pollack, D. T. Ross, H. Johnsen, L. A. Akslen, O. Fluge, A. Pergamenschikov, C. Williams, S. X. Zhu, P. E. Lonning, A. L. Borresen-Dale, P. O. Brown and D. Botstein, Nature, 2000, 406, 747–752 CrossRef CAS PubMed.
- T. Sorlie, C. M. Perou, R. Tibshirani, T. Aas, S. Geisler, H. Johnsen, T. Hastie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, T. Thorsen, H. Quist, J. C. Matese, P. O. Brown, D. Botstein, P. E. Lonning and A. L. Borresen-Dale, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 10869–10874 CrossRef CAS PubMed.
- S. Badve, D. J. Dabbs, S. J. Schnitt, F. L. Baehner, T. Decker, V. Eusebi, S. B. Fox, S. Ichihara, J. Jacquemier, S. R. Lakhani, J. Palacios, E. A. Rakha, A. L. Richardson, F. C. Schmitt, P. H. Tan, G. M. Tse, B. Weigelt, I. O. Ellis and J. S. Reis-Filho, Mod. Pathol., 2011, 24, 157–167 CrossRef PubMed.
- K. R. Bauer, M. Brown, R. D. Cress, C. A. Parise and V. Caggiano, Cancer, 2007, 109, 1721–1728 CrossRef PubMed.
- B. Weigelt and J. S. Reis-Filho, Nat. Rev. Clin. Oncol., 2009, 6, 718–730 CrossRef CAS PubMed.
- M. L. Doria, C. Z. Cotrim, C. Simoes, B. Macedo, P. Domingues, M. R. Domingues and L. A. Helguero, J. Cell. Physiol., 2013, 228, 457–468 CrossRef CAS PubMed.
- A. H. Beck, A. R. Sangoi, S. Leung, R. J. Marinelli, T. O. Nielsen, M. J. van de Vijver, R. B. West, M. van de Rijn and D. Koller, Sci. Transl. Med., 2011, 3, 108ra113 Search PubMed.
- Y. Mao, E. T. Keller, D. H. Garfield, K. Shen and J. Wang, Cancer Metastasis Rev., 2013, 32, 303–315 CrossRef PubMed.
- J. A. Joyce and J. W. Pollard, Nat. Rev. Cancer, 2009, 9, 239–252 CrossRef CAS PubMed.
- M. Egeblad, E. S. Nakasone and Z. Werb, Dev. Cell, 2010, 18, 884–901 CrossRef CAS PubMed.
- U. E. Martinez-Outschoorn, R. M. Balliet, D. B. Rivadeneira, B. Chiavarina, S. Pavlides, C. Wang, D. Whitaker-Menezes, K. M. Daumer, Z. Lin, A. K. Witkiewicz, N. Flomenberg, A. Howell, R. G. Pestell, E. S. Knudsen, F. Sotgia and M. P. Lisanti, Cell Cycle, 2010, 9, 3256–3276 CAS.
- G. Bonuccelli, A. Tsirigos, D. Whitaker-Menezes, S. Pavlides, R. G. Pestell, B. Chiavarina, P. G. Frank, N. Flomenberg, A. Howell, U. E. Martinez-Outschoorn, F. Sotgia and M. P. Lisanti, Cell Cycle, 2010, 9, 3506–3514 CrossRef CAS PubMed.
- P. Farmer, H. Bonnefoi, P. Anderle, D. Cameron, P. Wirapati, V. Becette, S. Andre, M. Piccart, M. Campone, E. Brain, G. Macgrogan, T. Petit, J. Jassem, F. Bibeau, E. Blot, J. Bogaerts, M. Aguet, J. Bergh, R. Iggo and M. Delorenzi, Nat. Med., 2009, 15, 68–74 CrossRef CAS PubMed.
- M. Verdonck, A. Denayer, B. Delvaux, S. Garaud, R. De Wind, C. Desmedt, C. Sotiriou, K. Willard-Gallo and E. Goormaghtigh, Analyst, 2016, 141, 606–619 RSC.
- N. Tahallah, A. Brunelle, S. De La Porte and O. Laprevote, J. Lipid Res., 2008, 49, 438–454 CrossRef CAS PubMed.
- Y. K. Magnusson, P. Friberg, P. Sjovall, J. Malm and Y. Chen, Obesity, 2008, 16, 2745–2753 CrossRef CAS PubMed.
- M. J. Baker, E. Gazi, M. D. Brown, N. W. Clarke, J. C. Vickerman and N. P. Lockyer, Appl. Surf. Sci., 2008, 255, 1084–1087 CrossRef CAS.
- C. R. Santos and A. Schulze, FEBS J., 2012, 279, 2610–2623 CrossRef CAS PubMed.
- L. J. Gamble, D. J. Graham, B. Bluestein, N. P. Whitehead, D. Hockenbery, F. Morrish and P. Porter, Biointerphases, 2015, 10, 019008 CrossRef PubMed.
- P. Du, W. A. Kibbe and S. M. Lin, Bioinformatics, 2008, 24, 1547–1548 CrossRef CAS PubMed.
- J. S. Parker, M. Mullins, M. C. Cheang, S. Leung, D. Voduc, T. Vickery, S. Davies, C. Fauron, X. He, Z. Hu, J. F. Quackenbush, I. J. Stijleman, J. Palazzo, J. S. Marron, A. B. Nobel, E. Mardis, T. O. Nielsen, M. J. Ellis, C. M. Perou and P. S. Bernard, J. Clin. Oncol., 2009, 27, 1160–1167 CrossRef PubMed.
- D. M. Gendoo, N. Ratanasirigulchai, M. S. Schroder, L. Pare, J. S. Parker, A. Prat and B. Haibe-Kains, genefu: Computation of Gene Expression-Based Signatures in Breast Cancer. R package version 2.3.2, 2015, http://www.pmgenomics.ca/bhklab/software/genefu.
- P. Sjövall, B. Johansson and J. Lausmaa, Appl. Surf. Sci., 2006, 252, 6966–6974 CrossRef.
- P. Malmberg, H. Nygren, K. Richter, Y. Chen, F. Dangardt, P. Friberg and Y. Magnusson, Microsc. Res. Tech., 2007, 70, 828–835 CrossRef CAS PubMed.
- D. Debois, M. P. Bralet, F. Le Naour, A. Brunelle and O. Laprevote, Anal. Chem., 2009, 81, 2823–2831 CrossRef CAS PubMed.
- Y. Magnusson, P. Friberg, P. Sjovall, F. Dangardt, P. Malmberg and Y. Chen, Clin. Physiol. Funct. Imaging, 2008, 28, 202–209 CrossRef CAS PubMed.
- D. Touboul, A. Brunelle, F. Halgand, S. De La Porte and O. Laprevote, J. Lipid Res., 2005, 46, 1388–1395 CrossRef CAS PubMed.
- M. A. Robinson, D. J. Graham, F. Morrish, D. Hockenbery and L. J. Gamble, Biointerphases, 2015, 11, 02A303 CrossRef PubMed.
- P. Sjovall, J. Lausmaa and B. Johansson, Anal. Chem., 2004, 76, 4271–4278 CrossRef PubMed.
- P. Malmberg, K. Borner, Y. Chen, P. Friberg, B. Hagenhoff, J. E. Mansson and H. Nygren, Biochim. Biophys. Acta, 2007, 1771, 185–195 CrossRef CAS PubMed.
- J. D. Brenton, L. A. Carey, A. A. Ahmed and C. Caldas, J. Clin. Oncol., 2005, 23, 7350–7360 CrossRef CAS PubMed.
- M. L. Heien, P. D. Piehowski, N. Winograd and A. G. Ewing, in Mass Spectrometry Imaging, Springer, 2010, pp. 85–97 Search PubMed.
- G. Kroemer and J. Pouyssegur, Cancer Cell, 2008, 13, 472–482 CrossRef CAS PubMed.
- S. Wang, X. Chen, H. Luan, D. Gao, S. Lin, Z. Cai, J. Liu, H. Liu and Y. Jiang, Rapid Commun. Mass Spectrom., 2016, 30, 533–542 CrossRef CAS PubMed.
- A. Kourtidis, R. Srinivasaiah, R. D. Carkner, M. J. Brosnan and D. S. Conklin, Breast Cancer Res. Treat., 2009, 11, R16–R16 CrossRef PubMed.
- H. Liu, Y. Liu and J.-T. Zhang, Mol. Cancer Ther., 2008, 7, 263–270 CrossRef CAS PubMed.
- Y. Ni and C. Eng, Clin. Cancer Res., 2012, 18, 4954–4961 CrossRef CAS PubMed.
- N. Weijl, F. Cleton and S. Osanto, Cancer Treat. Rev., 1997, 23, 209–240 CrossRef CAS PubMed.
- E. J. Ladas, J. S. Jacobson, D. D. Kennedy, K. Teel, A. Fleischauer and K. M. Kelly, J. Clin. Oncol., 2004, 22, 517–528 CrossRef CAS PubMed.
- B. Kwon, H.-K. Lee and H. W. Querfurth, Biochim. Biophys. Acta, Mol. Cell Res., 2014, 1843, 1402–1413 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5an02406d |
| This journal is © The Royal Society of Chemistry 2016 |