Serum protein biomarkers relevant to hepatocellular carcinoma and their detection
Eric
Waidely
a,
Abdul-Rahman Obaid
Al-Yuobi
b,
A. S.
Bashammakh
b,
Mohammad S.
El-Shahawi
b and
Roger M.
Leblanc
*a
aDepartment of Chemistry, University of Miami, 1301 Memorial Drive, Cox Science Center, Coral Gables, FL 33146, USA. E-mail: rml@miami.edu; Tel: +1 (305) 284-2194
bDepartment of Chemistry, King Abdulaziz University, P.O. Box 80200, Jeddah 21589, Kingdom of Saudi Arabia
First published on 11th November 2015
Abstract
Hepatocellular carcinoma (HCC) is one of the most recurrent and lethal cancers worldwide. The low survival rate of this particular strain of carcinoma is largely due to the late stages at which it is diagnosed. Tumorigenesis of hepatocellular carcinoma is most frequently detected through ultrasonography, magnetic resonance imaging and computerized tomography scans, however, these methods are poor for detection of early tumor development. This review presents alternative hepatocellular carcinoma detection techniques through the use of protein and enzyme/isozyme biomarkers. The detection methods used to determine the serum levels of α-fetoprotein (AFP), glypican-3 (GPC3), Golgi protein 73 (GP73), α-L-fucosidase (AFU), des-γ-carboxyprothrombin (DCP), γ-glutamyl transferase (GGT) and squamous cell carcinoma antigen (SCCA) are presented and each marker's respective validity in the diagnosis of hepatocellular carcinoma is evaluated.
Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of cancer related death worldwide, killing a majority of people affected within a year of diagnosis.1 Approximately 90–95% of worldwide liver cancers are HCC.2 The main risk factors involved in HCC onset are hepatitis and cirrhosis.3 In countries where hepatitis and cirrhosis are frequent, HCC is not only one of the most prevalent cancers but is also one of the most recurrent causes of death.4 Neoplasm formation in the liver is generally detected through ultrasonography, magnetic resonance imaging (MRI) or an abdominal computerized tomography (CT) scan.5 While these methods are accurate in the detection of HCC, they often fail to discover tumor formations until the later stages. It is therefore of interest to establish ways to detect tumor formation in the earlier stages to screen the populations at risk.The use of biosensing has shown promising results in early cancer detection and in some cases can even help determine the prognosis of the disease.6–8 Biosensors are developed by combining a biological component with a physiochemical detector. The biological component is an analyte found within the body which is referred to as a biomarker. Biomarkers are any measurable substance found within an organism whose presence can be indicative of a disease or infection. The use of biomarkers has greatly advanced early cancer detection, which generates better survival rates. The physiochemical detectors chosen vary by the analyte being detected. Some of the more common methods of biosensing for cancers involve the use of spectroscopy,9,10 ELISA11,12 or potentiometric techniques.13
HCC biomarkers can be divided into four categories: embryonic and glycoprotein antigens; enzymes and isozymes; genes; and cytokines.2 Recent data suggests that genes and cytokines could potentially be the most sensitive for detection, however, further research needs to be performed to defend these claims.14 As such, this review will focus on the more thoroughly investigated protein and enzyme biomarkers. Serum alpha-fetoprotein (AFP) is the most universally used antigen. AFP is an embryonic protein that is typically only produced by the fetal liver. Research has shown that serum AFP levels can be drastically elevated in individuals expressing HCC, leading to potential cancer detection.14 However, AFP is not a perfect diagnostic test as 40% of individuals with early HCC development express normal or acceptable AFP levels.15,16 Alpha-L-fucosidase (AFU), a lysosomal enzyme present in all mammalian cells, may offer a new scaffold for HCC detection. It has been found that alpha-L-fucosidase can diagnose 85% of HCC patients up to 6 months before it is detectable by ultrasonography.17
Developing a standard biosensor for HCC has proven to be a difficult task. While several biomarkers appear to be relevant in HCC diagnosis, a consistent detection method has yet to be determined, which has limited the clinical use of HCC biomarkers. The most widely explored ways to detect early tumor growth for HCC have been through protein biomarkers, predominantly AFP. This review will discuss the current protein and enzyme biomarkers found to be relevant to HCC, with a focus on the methods used to quantify each biomarker's serum levels and what these levels mean with regard to neoplasm formation.
Embryonic and glycoprotein antigens
Consisting of 591 amino acids, AFP is a 70 kDa oncofetal glycoprotein21 that contains a single asparagine-linked carbohydrate chain.22 Being a glycoprotein, AFP exists as a heterogeneous entity with several different glycoforms found based upon affinity towards different lectins or electrophoretic separation.23,24 Lens culinaris agglutinin (LCA) is able to distinguish AFP into three different isoforms: AFP-L1 (LCA nonreactive), AFP-L2 (intermediate reactive) and AFP-L3 (LCA-affinitive).25 The AFP-L3 glycoform appears to be produced by cancer cells, potentially allowing for discrimination between chronic liver diseases and HCC at lower serum levels (≥400 ng mL−1).26
Elevated AFP levels were first associated with tumor development in mice by Abelev et al. in 1963.27 Tatarinov later found AFP in the serum of patients with hepatoma, leading to its current use as a biomarker for HCC.28 The first quantitative serum assay for AFP was developed in 197120 and has since been extensively studied.29–31 The best analysis methods for AFP are all based on modified immunoassays and can be found in Table 1.
| Immunoassay | Antibody-label | Detection limit (μg L−1) | Inter-assay CV | Intra-assay CV | Ref. |
|---|---|---|---|---|---|
| Radioimmunoassay | Radioactive isotope (125I) | 1–16 | 3.4–8.8% | 2.7–5.2% | 20,32 |
| Nephelometry | Latex particle | 5.0 | 8.4–10% | 5.1–10% | 33 |
| ELISA | Enzyme | 3.0 | 3.5–16% | 6.9–15% | 34–37 |
| Microparticle enzyme immunoassay | Microparticle + enzyme | 0.5 | 6.8% | 4.9% | 38 |
| Electrochemiluminescence | Ruthenium(II)-tris(bipyridyl) complex | 0.4 | 3.5–4.6% | 2.3–4.0% | 39 |
| Immunoradiometric assay | Radioactive isotope (125I) | 0.2 | 3.0–10% | 3.0–6.0% | 40 |
As Table 1 suggests, radioimmunoassays for AFP offer the lowest detection limit. The radioimmunoassay involves labeling known quantities of AFP with Na125I. The radioactive iodine isotope binds to tyrosine residues located on AFP and the labeled antigen is then incubated with known quantities of AFP specific antibodies and loaded into a column. Upon addition of serum samples, the unlabeled AFP found in the sera will displace the radioactive labeled AFP, causing it to elute from the column. The quantity of labeled AFP eluted can be quantified with a gamma counter and allow for indirect measurements of the concentration of AFP found in the serum samples.20 While radioimmunoassays for AFP offer great sensitivity, they suffer from long preparation times, large amounts of required dilutions and separations, and radiation damage. A more commonly implemented detection technique for serum AFP is ELISA. Although Table 1 portrays ELISA as one of the weaker methods with regard to the detection limit, yet it has benefits such as quick detection time, longer shelf life, and simpler detection techniques. The exact ELISA format varies but most employ isolation of the target antigen onto a plastic bead or plate followed by incubation with the enzyme-labeled antibody. The enzyme labels chosen are most commonly horseradish peroxidase36 and alkaline phosphatase.34 Upon binding of the enzyme-labeled antibody, excess antibody is washed from the plastic surface and an enzyme specific chromogen substrate is added. The enzyme interacts with the chromogen substrate, producing a fluorescent or colored product which can be used to quantify the amount of antigen present. Another factor to be taken into account when determining which assay to use is the variability that can arise from experiment to experiment. As shown in Table 1, the inter-assay and intra-assay CV for all the immunoassays listed shows little variance between each assay type.
Although Table 1 would suggest that using an immunoradiometric scaffold is the best for serum AFP measurements, there are many variables to consider beyond the detection limit for the assay: including time, cost, cut-off point and ease of measurement. The cut-off point is used as an indicator to show that serum levels are above the norm and is the set point at which the distinction between the healthy and diseased occurs. It is therefore not entirely relevant to obtain the lowest detection limit, but rather to obtain the highest sensitivity and specificity for the disease being detected. In diagnosing cases of HCC, the serum concentration of 20 ng mL−1 of AFP is the most universally used cut-off point; however, this cut-off point has been found to fluctuate between ethnic groups.41,42Table 2 lists the most common AFP serum concentrations used in HCC diagnosis along with the respective sensitivity and specificity as found from a statistical study by Trevisani et al.43 The sensitivity of the assay measures the true positive rate while the specificity measures the false positive rate caused by other disorders expressing increased serum levels. As Table 2 depicts, higher AFP cut-off values become more specific for HCC but the measurement loses its sensitivity. The sensitivity of detection decreases at higher cut-off values due to the low serum levels of AFP expressed. Many individuals with HCC express only slight elevation of AFP while 80% of the smaller cases (tumors <3 cm) show no elevation whatsoever, causing higher cut-off values to be less effective.44 The specificity increases due to false positives from alternative viral etiology and other liver diseases that cause elevation in AFP becoming less relevant at such high serum levels.45
| AFP cut-off (ng mL−1) | Sensitivity (%) | Specificity (%) |
|---|---|---|
| 16 | 62.4 | 89.4 |
| 20 | 60.0 | 90.6 |
| 100 | 31.2 | 98.8 |
| 200 | 22.4 | 99.4 |
| 400 | 17.1 | 99.4 |
The heterogeneous nature of AFP has shown promising results in increasing the sensitivity and specificity of HCC diagnosis. As noted, total AFP levels can be divided into three different types, based upon each isoforms affinity towards LCA. The LCA-bound glycoform, AFP-L3, is able to detect HCC at the cut-off value of 15% with a sensitivity of 96.9% and a specificity of 92%.46 However, the most promising results come not from using AFP-L3 alone, but rather combining AFP-L3 percentages with the total serum levels of AFP. Leerapun et al.47 found that using a cut-off value between 10–200 ng mL−1 of the total AFP and a cut-off of 35% AFP-L3, the specificity of HCC detection reaches 100%.
Hsu et al.51 found that the mRNA of GPC3 was detectable in 74.8% of HCC patients while only 3.2% expression was observed in noncancerous livers. Soon after, it was found that GPC3 mRNA was expressed in 75% of HCC cases while negligible expression was detected in normal, cirrhotic and focal nodular hyperplasia livers.52 These findings led future groups to analyze GPC3 levels and assess its relevance as a novel HCC biomarker. Up to now, the most commonly used detection technique for GPC3 serum levels is ELISA with cut-off values ranging from 3.9 pg mL−1 to 300 ng mL−1.53
Capurro et al.54 presented the first assay analyzing GPC3 levels in the serum of healthy and HCC patients using a sandwich ELISA with horseradish peroxidase labeled antibodies and hydrogen peroxide with o-phenylenediamine as the substrate. The sandwich ELISA used involved placing monoclonal antibodies specific for GPC3 in 96-well plates followed by 24 hour incubation with serum samples. After thorough washing, a polyclonal antibody labeled with horseradish peroxidase is then added and the enzymatic activity is used to produce a calibration curve that can determine GPC3 serum levels. It was found that GPC3 was expressed in 72% of HCC patients while AFP was only expressed in 59% at the cut-off of 20 ng mL−1, and an even lower 32% of patients when using the more sensitive cut-off of 100 ng mL−1. As is the case for AFP detection, various cut-off points have been used in correlating GPC3 serum levels with HCC diagnosis. However, in the case of GPC3, there has yet to be a standard cut-off point determined that maximizes sensitivity and specificity. Currently, the cut-off values used vary from as low as 3.9 pg mL−1 all the way to 300 ng mL−1, with the sensitivity of GPC3 ranging from 36–65% and specificities ranging from 65–100%.50,54–57 While GPC3 shows promise in being more sensitive and specific than serum AFP, a meta-analysis performed by Liu et al.53 concludes that the current data presented is flawed, presenting issues in sample size, patient selection, serum control and the heterogeneous nature in experimental techniques. Future research will need to be conducted to determine the merit of GPC3 as a replacement or supplement to AFP in HCC diagnostics.
The primary detection techniques for GP73 include western blot, immunoblotting and ELISA. Marrero et al.62 presented one of the first human serum assays of GP73 using an immunoblot assay. Zhou et al.63 performed a meta-analysis on GP73 assays compared with AFP. GP73 sensitivities range from 69–95% while the specificities range from 35–97%. These values are in agreement with the results from all three assay types used and suggest a large amount of heterogeneity between each assay type. While higher sensitivity and specificity has been reported for GP73 which might suggest that it is a possible replacement to AFP, further research needs to be done to confirm the consistency of this marker.
Enzymes and isozymes
Due to the heterogeneous nature of α-L-fucosidases, an exact molecular weight is not available and as such other means of quantifying serum levels must be employed. In contrary to the previous biomarkers mentioned, AFU serum values are typically given with regard to the enzymes catalytic activity using the SI unit katal or more commonly the enzymatic unit (U). Avila and Convit67 developed the first assay on AFU in 1974 by means of measuring the enzymatic activity with the colorimetric substrate p-nitrophenyl-α-L-fucopyranoside (PNFP) as depicted in Fig. 1.64 In brief, the enzymatic activity is measured at a pH of 5.5 in an acetate buffer and the absorbance of the cleaved p-nitrophenol is observed at 400 nm. The connection between AFU serum levels and HCC onset was not made until ten years later when Deugnier et al.68 used a previously described AFU assay69 to establish a correlation between high AFU levels and carcinoma.
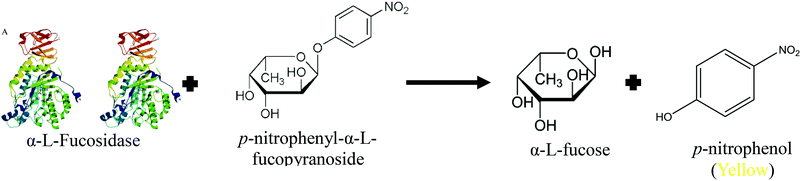 | ||
| Fig. 1 Schematic of the assay developed through the hydrolysis of p-nitrophenyl-α-L-fucopyranoside by AFU to produce α-L-fucose and the colored product p-nitrophenol. | ||
To date, the most commonly used detection technique is the above mentioned assay using the colorimetric substrate PNFP. Many research groups have used this approach to determine the onset of HCC with sensitivities ranging from 60–90% and specificities ranging from 55–98%.4,70–74 An alternative method was proposed by Wang and Cao75 by chlorinating the 2 position of the standard substrate p-nitrophenyl-α-L-fucopyranoside to produce 2-chloro-4-nitrophenyl-α-L-fucoside. The addition of a chlorine group to the substrate leads to an increased cleavage rate and produces a faster assay with sensitivity for HCC detection reported at 81.8% and specificity of 85.4%. While the standard approach for AFU detection appears to show promise in HCC detection, it is still affected by incubation times, indirect measurements via catalytic rates and interference caused by the overlap between the yellow color of the substrate collected and that of the inherently yellow color of serum samples.13
Blanchard et al.78 developed the first assay for DCP detection using a radioimmunoassay technique with 125I labeling. The assay developed was a competitive assay, involving the addition of 125I labeled DCP to polyclonal antibody specific for abnormal prothrombin. As serum is added with the native unlabeled DCP, the 125I-DCP is displaced from the antibody and eluted for detection. Liebman et al.79 used this radioimmunoassay to measure the plasma DCP levels in patients with primary HCC and found significant elevation in 91% of the patients, leading to further research on this potential new biomarker. These findings led Motohara et al.80 to develop an ELISA to investigate the validity of DCP as a new biomarker for HCC. The ELISA was established using a monoclonal antibody specific for PIVKA-II as a capture antibody and plates containing this antibody were incubated with plasma samples. After proper incubation, the plates are washed and treated with a second antibody labeled with horseradish peroxidase. After incubating the second antibody followed by thorough washing, 2,2′-azino-di-[3-ethylbenzthiazoline sulfonate] was added to measure the activity of the attached enzyme. The ELISA technique proved to be quite sensitive with a detection limit of 0.13 U mL−1.80 Fujiyama et al.81 then set out to use the above mentioned ELISA technique to study the correlation between DCP serum levels and HCC. It was found that 63% of patients with HCC show DCP levels >0.1 U mL−1 while 48% of patients expressed values >0.3 U mL−1.
The future of DCP as a biomarker for HCC detection relies on its effectiveness when compared to the standard marker AFP.82 Mita et al.83 compared the detection capability of DCP with AFP using an ELISA approach for both measurements. Of the 91 patients with HCC measured, 62% had DCP levels above the cutoff 40 mU mL−1 while only 47% had AFP levels above the standard cutoff of 20 ng mL−1. While these results are promising for the future of using DCP as an HCC biomarker, there are complications that could limit its use as a universal marker for all cases of HCC. While the exact cause of the production of abnormal prothrombin is unclear, it has been found to be elevated not only in the cases of HCC but also in patients treated with coumarin anticoagulants, which could limit its effectiveness in detecting HCC universally.79
Goldbarg et al.91 developed the first assay for GGT using a colorimetric set up. Applying a process developed by Bratton and Marshall,92 the colorimetric assay utilizes the chromogenic substrate N-(DL-γ-glutamyl)aniline to measure GGT. GGT transfers the γ-glutamyl moiety to an acceptor amino acid, releasing free aniline. The solution is then treated with sodium nitrite and N-(1-naphthyl)ethylenediamine dihydrochloride leading to diazotization of the released aniline substrate, producing a blue azo dye which can then be quantified using a photoelectric colorimeter to determine the enzymatic activity.
While the colorimetric assay was able to measure serum GGT levels in various human tissues, the values produced combine all isoforms of GGT which could hinder the sensitivity and specificity for HCC diagnosis. In fact, it is the GGT-II isoform that has been found as the diagnostic standard for cases of hepatoma. To overcome the issues that arise from GGT heterogeneity, many groups have set out to separate the various isoforms through electrophoretic methods with agarose gel,93 starch gel,94 cellulose acetate,95 polyacrylamide gel96–98 and paper99 prior to measuring enzymatic activity. Xu et al.100 utilized vertical slab stage electrophoresis on polyacrylamide gel to separate GGT into 9–11 activity bands. They then adopted a modified version on the aforementioned colorimetric assay using γ-glutamyl p-nitroanilide as the enzyme substrate, glycyl glycine as an amino acid acceptor for the γ-glutamyl moiety, and N-(1-naphthyl)ethylenediamine dihydrochloride to produce a red azo dye through diazotization with free p-nitroanilide.90 GGT-II appears as the second separation band and was found to have a sensitivity of 90% and a specificity of 97.1% for hepatoma cases, while it is expressed in only 3.1% of other liver diseases.100 Cui et al.101 set out to replicate these findings and while they observed a lower sensitivity and specificity, 74% and 82.2% respectively, their findings still show promise for GGT-II as a supplemental biomarker in HCC detection.
Kato and Torigoe108 presented the first isolation and immunoassay for SCCA using a radioimmunoassay approach for associating SCCA with the onset of cervical squamous cell carcinoma. Pontisso et al.109 were the first to establish serum SCCA as a biomarker for HCC using an ELISA with horseradish peroxidase and found SCCA was expressed in 85% of HCC cases. Further groups investigated serum SCCA levels using a similar method with sensitivities ranging from 18–84% and specificities ranging from 27–73%.110–114 While serum SCCA may prove to be a valid supplementary biomarker for HCC diagnosis, more promising results have been obtained by quantifying serum SCCA–IgM immunocomplexes found in circulation. Beneduce et al.115 found that by combining SCCA–IgM with AFP the sensitivity of HCC detection was 70% while the specificity reached 100%. Using an ELISA technique, SCCA–IgM produces sensitivities ranging from 52–89% and specificities from 49–100%.111,115–118
Combined tests
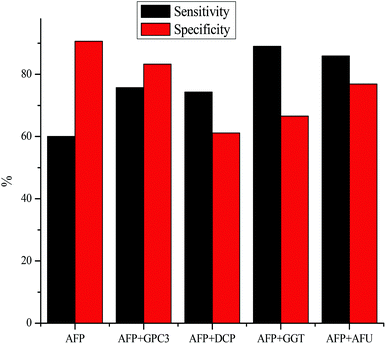
While promising results have been shown in testing for one biomarker at a time, many groups have found that the sensitivity of detection can be increased by detecting multiple biomarkers at a time. Fig. 2 portrays the sensitivity and specificity of HCC detection by using the serum marker AFP combined with GPC3,119 DCP,101,120 GGT,101,121 and AFU.71,121 In each case, combining an additional biomarker with AFP for HCC diagnosis leads to an increase in the sensitivity of detection, however, the specificity of detection takes a hit. While the lowered specificity is not ideal, it is possible that the specificity could be increased by varying the cut-off values used. As shown in Table 2, the specificity of AFP detection increased as the cut-off value is raised. Each of the combined tests is done at the cut-off value of 20 ng mL−1, so it is possible that the specificity of detection could be raised by using a slightly higher AFP cut-off value.Conclusion
In summary, the detection techniques and the relevance of protein and enzyme biomarkers for hepatocellular carcinoma has been evaluated. While radioimmunoassays typically produce the lowest detection limits for protein biomarkers for HCC, the complications and difficulty that arise from these measurements has limited its use in serum biomarker measurements. Instead, the most commonly used technique for biomarker evaluation is an ELISA approach with horseradish peroxidase. AFP remains the gold standard for HCC diagnosis but cancer detection proves to be too complicated to be limited to a single marker. Variables such as geographical location, ethnicity, and preexisting viral etiology cause variations for each biomarker. The future of HCC diagnosis will likely lie in combination measurements by evaluating several of the above mentioned biomarkers simultaneously, rather than simply using one.References
- L. Dai, N. Lei, M. Liu and J.-Y. Zhang, Exp. Hematol. Oncol., 2013, 2, 15 CrossRef CAS PubMed.
- Y. Zhao, Q. Ju and G. Li, Mol. Clin. Oncol., 2013, 1, 593 CAS.
- H. Shiraha, K. Yamamoto and M. Namba, Int. J. Oncol., 2013, 42, 1133 CAS.
- P. Tangkijvanich, P. Tosukhowong, P. Bunyongyod, S. Lertmaharit, O. Hanvjvatvong, P. Kullavanijaya and Y. Poovorawan, Southeast Asian J. Trop. Med. Public Health, 1999, 30, 110 CAS.
- J. Tejeda-Maldonado, I. García-Juárez, J. Aguirre-Valadez, A. González-Aguirre, M. Vilatobá-Chapa, A. Armengol-Alonso, F. Escobar-Penagos, A. Torre, J. Sánchez-Ávila and D. Luis Carrillo-Pérez, World J. Hepatol., 2015, 7, 362 CrossRef PubMed.
- M. Ijichi, T. Takayama, M. Matsumura, Y. Shiratori, M. Omata and M. Makuuchi, Hepatology, 2002, 35, 853 CrossRef CAS PubMed.
- H. Okuda, T. Nakanishi, K. Takatsu, A. Saito, N. Hayashi, M. Yamamoto, K. E. N. Takasaki and M. Nakano, J. Gastroenterol. Hepatol., 2002, 17, 772 CrossRef CAS PubMed.
- W. Tang, N. Kokudo, Y. Sugawara, Q. Guo, H. Imamura, K. Sano, H. Karako, X. Qu, M. Nakata and M. Makuuchi, Oncol. Rep., 2005, 13, 25 CAS.
- Z. Chen, X. Ren, X. Meng, Y. Zhang, D. Chen and F. Tang, Anal. Chem., 2012, 84, 4077 CrossRef CAS PubMed.
- G. C. Tsay and G. Dawson, Anal. Biochem., 1976, 78, 423 CrossRef.
- H. Tjalsma, Expert Rev. Proteomics, 2010, 7, 879 CrossRef CAS PubMed.
- B. Nilsson, Curr. Opin. Immunol., 1989, 2, 898 CrossRef CAS PubMed.
- A. M. Othman, M. E. El-Houseini, M. S. El-Sofy and H. Y. Aboul-Enein, Anal. Bioanal. Chem., 2011, 400, 787 CrossRef PubMed.
- L. Zhou, J. Liu and F. Luo, World J. Gastroenterol., 2006, 12, 1175 CrossRef CAS PubMed.
- J. Y. Choi, S. W. Jung, H. Y. Kim, M. Kim, Y. Kim, D. G. Kim and E.-J. Oh, World J. Gastroenterol., 2013, 19, 339 CrossRef CAS PubMed.
- M. S. Attia, A. M. Othman, M. M. Aboaly and M. S. A. Abdel-Mottaleb, Anal. Chem., 2010, 82, 6230 CrossRef CAS PubMed.
- M. E. El-Houseini, M. E. Sherbiny, M. M. El- Din Awad, M. A. Amer, A. H. Saad El Din and T. D. Hussein, J. Egypt. Natl. Cancer Inst., 2001, 13, 277 Search PubMed.
- C. G. Bergstrand and B. Czar, Scand. J. Clin. Lab. Invest., 1956, 8, 174 CrossRef CAS PubMed.
- E. N. Debruyne and J. R. Delanghe, Clin. Chim. Acta, 2008, 395, 19 CrossRef CAS PubMed.
- E. Ruoslahti and M. Seppälä, Int. J. Cancer, 1971, 8, 374 CrossRef CAS.
- G. J. Mizejewski, Exp. Biol. Med., 1997, 215, 333 CrossRef CAS.
- H. Yoshima, T. Mizuochi, M. Ishii and A. Kobata, Cancer Res., 1980, 40, 4276 CAS.
- J. BrĘBorowicz, A. Mackiewicz and D. BrĘBorowicz, in Scand. J. Immunol., Blackwell Publishing Ltd, 1981, vol. 14, p. 15 Search PubMed.
- E. Alpert, J. W. Drysdale, K. J. Isselbacher and P. H. Schur, J. Biol. Chem., 1972, 247, 3792 CAS.
- P. J. Johnson, T. C. W. Poon, N. M. Hjelm, C. S. Ho, C. Blake and S. K. W. Ho, Br. J. Cancer, 2000, 83, 1330 CrossRef CAS PubMed.
- M.-Q. Du, W. L. Hutchinson, P. J. Johnson and R. Williams, Cancer, 1991, 67, 476 CrossRef CAS PubMed.
- G. I. Abelev, S. D. Perova, N. I. Khramkova, Z. A. Postnikova and I. S. Irlin, Transplantation, 1963, 1, 174 CrossRef CAS PubMed.
- Y. Tatarinov, Vopr. Med. Khim., 1964, 10, 90 CAS.
- M. E. Alpert, J. Uriel and B. de Nechaud, N. Engl. J. Med., 1968, 278, 984 CrossRef CAS PubMed.
- E. Alpert, R. Hershberg, P. H. Schur and K. J. Isselbacher, Gastroenterology, 1971, 61, 137 CAS.
- S. Gupta, S. Bent and J. Kohlwes, Ann. Intern. Med., 2003, 139, 46 CrossRef CAS PubMed.
- J. A. Chayvialle and P. C. Ganguli, Lancet, 1973, 1, 1355 CrossRef CAS.
- D. R. Bernard, J. R. Delanghe, M. L. De Buyzere and G. G. Leroux-Roels, Eur. J. Clin. Chem. Clin. Biochem., 1996, 34, 847 CAS.
- D. W. Chan, M. Kelsten, R. Rock and D. Bruzek, Clin. Chem., 1986, 32, 1318 CAS.
- L. Belanger, D. Hamel, D. Dufour and M. Pouliot, Clin. Chem., 1976, 22, 198 CAS.
- P. Mancal, M. Srámek, I. Malbohan and L. Simek, J. Hyg., Epidemiol., Microbiol., Immunol., 1988, 32, 209 CAS.
- R. Maiolini and R. Masseyeff, J. Immunol. Methods, 1975, 8, 223 CrossRef CAS PubMed.
- M. Fiore, J. Mitchell, T. Doan, R. Nelson, G. Winter, C. Grandone, K. Zeng, R. Haraden, J. Smith, K. Harris, J. Leszczynski, D. Berry, S. Safford, G. Barnes, A. Scholnick and K. Ludington, Clin. Chem., 1988, 34, 1726 CAS.
- G. F. Blackburn, H. P. Shah, J. H. Kenten, J. Leland, R. A. Kamin, J. Link, J. Peterman, M. J. Powell, A. Shah, D. B. Talley, S. K. Tyagi, E. Wilkins, T. G. Wu and R. J. Massey, Clin. Chem., 1991, 37, 1534 CAS.
- M. Suzuki, Radioisotopes, 1988, 37, 225 CrossRef CAS PubMed.
- K. Taketa, S. Okada, N. Win, N. K. T. Hlaing and K. M. Win, Acta Med. Okayama, 2002, 56, 317 Search PubMed.
- M. Soresi, C. Magliarisi, P. Campagna, G. Leto, G. Bonfissuto, A. Riili, A. Carroccio, R. Sesti, S. Tripi and G. Montalto, Anticancer Res., 2003, 23, 1747 CAS.
- F. Trevisani, P. E. D'Intino, A. M. Morselli-Labate, G. Mazzella, E. Accogli, P. Caraceni, M. Domenicali, S. De Notariis, E. Roda and M. Bernardi, J. Hepatol., 2001, 34, 570 CrossRef CAS PubMed.
- R. Saffroy, P. Pham, M. Reffas, M. Takka, A. Lemoine and B. Debuire, Clin. Chem. Lab. Med., 2007, 45, 1169 CrossRef CAS PubMed.
- M. H. Nguyen, R. T. Garcia, P. W. Simpson, T. L. Wright and E. B. Keeffe, Hepatology, 2002, 36, 410 CrossRef CAS PubMed.
- A. Singhal, M. Jayaraman, D. N. Dhanasekaran and V. Kohli, Crit. Rev. Oncol. Hematol., 2012, 82, 116 CrossRef PubMed.
- A. Leerapun, S. V. Suravarapu, J. P. Bida, R. J. Clark, E. L. Sanders, T. A. Mettler, L. M. Stadheim, I. Aderca, C. D. Moser, D. M. Nagorney, N. F. LaRusso, P. C. de Groen, K. V. N. Menon, K. N. Lazaridis, G. J. Gores, M. R. Charlton, R. O. Roberts, T. M. Therneau, J. A. Katzmann and L. R. Roberts, Clin. Gastroenterol. Hepatol., 2007, 5, 394 CrossRef CAS PubMed.
- S. K. Wang, D. L. Zynger, O. Hes and X. J. Yang, Adv. Anat. Pathol., 2014, 21, 450 CrossRef CAS PubMed.
- J. Filmus and S. B. Selleck, J. Clin. Invest., 2001, 108, 497 CrossRef CAS PubMed.
- T. Nakatsura, Y. Yoshitake, S. Senju, M. Monji, H. Komori, Y. Motomura, S. Hosaka, T. Beppu, T. Ishiko, H. Kamohara, H. Ashihara, T. Katagiri, Y. Furukawa, S. Fujiyama, M. Ogawa, Y. Nakamura and Y. Nishimura, Biochem. Biophys. Res. Commun., 2003, 306, 16 CrossRef CAS PubMed.
- H.-C. Hsu, W. Cheng and P.-L. Lai, Cancer Res., 1997, 57, 5179 CAS.
- Z. Zhu, H. Friess, L. Wang, M. Abou-Shady, A. Zimmermann, A. Lander, M. Korc, J. Kleeff and M. Buchler, Gut, 2001, 48, 558 CrossRef CAS PubMed.
- X.-F. Liu, Z.-D. Hu, X.-C. Liu, Y. Cao, C.-M. Ding and C.-J. Hu, Clin. Biochem., 2014, 47, 196 CrossRef CAS PubMed.
- M. Capurro, I. R. Wanless, M. Sherman, G. Deboer, W. Shi, E. Miyoshi and J. Filmus, Gastroenterology, 2003, 125, 89 CrossRef CAS.
- H. Liu, P. Li, Y. Zahai, C. F. Qu, L. J. Zhang, Y. F. Tan, N. Li and G. D. Ding, World J. Gastroenterol., 2010, 16, 4410 CrossRef CAS PubMed.
- P. Tangkijvanich, T. Chanmee, S. Komtong, V. Mahachai, N. Wisedopas, P. Pothacharoen and P. Kongtawelert, J. Gastroenterol. Hepatol., 2010, 25, 129 CrossRef CAS PubMed.
- E. Yasuda, T. Kumada, H. Toyoda, Y. Kaneoka, A. Maeda, S. Okuda, N. Yoshimi and O. Kozawa, Hepatol. Res., 2010, 40, 477 CrossRef CAS PubMed.
- C. J. Fimmel and L. Wright, Hepatology, 2009, 49, 1421 CrossRef CAS PubMed.
- R. D. Kladney, G. A. Bulla, L. Guo, A. L. Mason, A. E. Tollefson, D. J. Simon, Z. Koutoubi and C. J. Fimmel, Gene, 2000, 249, 53 CrossRef CAS PubMed.
- R. D. Kladney, X. Cui, G. A. Bulla, E. M. Brunt and C. J. Fimmel, Hepatology, 2002, 35, 1431 CrossRef CAS PubMed.
- T. M. Block, M. A. Comunale, M. Lowman, L. F. Steel, P. R. Romano, C. Fimmel, B. C. Tennant, W. T. London, A. A. Evans, B. S. Blumberg, R. A. Dwek, T. S. Mattu and A. S. Mehta, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 779 CrossRef CAS PubMed.
- J. A. Marrero, P. R. Romano, O. Nikolaeva, L. Steel, A. Mehta, C. J. Fimmel, M. A. Comunale, A. D'Amelio, A. S. Lok and T. M. Block, J. Hepatol., 2005, 43, 1007 CrossRef CAS PubMed.
- Y. Zhou, X. Yin, J. Ying and B. Zhang, BMC Cancer, 2012, 12, 17 CrossRef CAS PubMed.
- G. Sulzenbacher, C. Bignon, T. Nishimura, C. A. Tarling, S. G. Withers, B. Henrissat and Y. Bourne, J. Biol. Chem., 2004, 279, 13119 CrossRef CAS PubMed.
- O. J. Cordero, A. Merino, M. P. dela Cadena, B. Bugía, M. Nogueira, J. E. Viñuela, V. S. Martínez-Zorzano, A. de Carlos and F. J. Rodríguez-Berrocal, Eur. J. Biochem., 2001, 268, 3321 CrossRef CAS PubMed.
- G. A. Levvy and A. McAllan, Biochem. J., 1961, 80, 435 CrossRef CAS PubMed.
- J. Avila and J. Convit, Biochim. Biophys. Acta, 1974, 358, 308 CrossRef CAS.
- Y. Deugnier, V. David, P. Brissot, P. Mabo, D. Delamaire, M. Messner, M. Bourel and J.-Y. Legall, Hepatology, 1984, 4, 889 CrossRef CAS PubMed.
- J. Troost, M. C. M. van der Heijden and G. E. J. Staal, Clin. Chim. Acta, 1976, 73, 329 CrossRef CAS.
- S. Bukofzer, P. M. Stass, M. C. Kew, M. de Beer and H. T. Groeneveld, Br. J. Cancer, 1989, 59, 417 CrossRef CAS PubMed.
- M. E. El-Houseini, M. S. Mohammed, W. M. Elshemey, T. D. Hussein, O. S. Desouky and A. A. Elsayed, Cancer Control, 2005, 12, 248 Search PubMed.
- S. F. El-Tayeh, T. D. Hussein, M. E. El-Houseini, M. A. Amer, M. El-Sherbini and W. M. Elshemey, Dis. Markers, 2012, 32, 255 CrossRef CAS PubMed.
- M. G. Giardina, M. Matarazzo, R. Morante, A. Lucariello, A. Varriale, V. Guardasole and G. De Marco, Cancer, 1998, 83, 2468 CrossRef CAS PubMed.
- M. F. Montaser, M. A. Sakr and M. O. Khalifa, Arab J. Gastroenterol., 2012, 13, 9 CrossRef PubMed.
- J.-J. Wang and E.-H. Cao, Clin. Chim. Acta, 2004, 347, 103 CrossRef CAS PubMed.
- P. Stefaniuk, J. Cianciara and A. Wiercinska-Drapalo, World J. Gastroenterol., 2010, 16, 418 CrossRef CAS PubMed.
- H. A. Liebman, B. C. Furie and B. Furie, Hepatology, 1982, 2, 488 CrossRef CAS PubMed.
- R. A. Blanchard, B. C. Furie, M. Jorgensen, S. F. Kruger and B. Furie, N. Engl. J. Med., 1981, 305, 242 CrossRef CAS PubMed.
- H. A. Liebman, B. C. Furie, M. J. Tong, R. A. Blanchard, K.-J. Lo, S.-D. Lee, M. S. Coleman and B. Furie, N. Engl. J. Med., 1984, 310, 1427 CrossRef CAS PubMed.
- K. Motohara, F. Endo and I. Matsuda, Lancet, 1985, 326, 242 CrossRef.
- S. Fujiyama, T. Morishita, O. Hashiguchi and T. Sato, Cancer, 1988, 61, 1621 CrossRef CAS PubMed.
- M. Abu El Makarem, Hepat. Mon., 2012, 12, e6122 Search PubMed.
- Y. Mita, Y. Aoyagi, M. Yanagi, T. Suda, Y. Suzuki and H. Asakura, Cancer, 1998, 82, 1643 CrossRef CAS PubMed.
- M. H. Hanigan and H. C. Pilot, Carcinogenesis, 1985, 6, 165 CrossRef CAS PubMed.
- S. Horiuchi, M. Inoue and Y. Morino, Eur. J. Biochem., 1978, 87, 429 CrossRef CAS PubMed.
- Y. Matsuda, A. Tsuji and N. Katunuma, J. Biochem., 1983, 93, 1427 CAS.
- A. Meister, Science, 1973, 180, 33 CAS.
- N. P. Curthoys and R. P. Hughey, Enzyme, 1979, 24, 383 CAS.
- C. S. Hanes, F. J. R. Hird and F. A. Isherwood, Biochem. J., 1952, 51, 25 CrossRef CAS PubMed.
- G. Szasz, Clin. Chem., 1969, 15, 124 CAS.
- J. A. Goldbarg, O. M. Friedman, E. P. Pineda, E. E. Smith, R. Chatterji, E. H. Stein and A. M. Rutenburg, Arch. Biochem. Biophys., 1960, 91, 61 CrossRef CAS PubMed.
- A. C. Bratton and E. K. Marshall, J. Biol. Chem., 1939, 128, 537 CAS.
- Ö. Hetland, T. R. Andersson and T. Gerner, Clin. Chim. Acta, 1975, 62, 425 CrossRef.
- M. Orłowski and A. Szczeklik, Clin. Chim. Acta, 1967, 15, 387 CrossRef.
- X. Y. Meng, K. C. Xu, Z. H. Yang, Y. J. Huang, Z. H. Rong, Y. H. Liu, Z. J. Ge, M. Y. Cheng, J. L. Zhang and H. M. Wan, Jiangsu Med. J., 1982, 8, 8 Search PubMed.
- N. Sawaby, M. Nakagen, M. Yoneda, H. Makino, S. Kameda, K. Kobayashi, G. Sugioka, N. Hattori and M. Ishii, Acta Hepatil. Jap., 1977, 18, 101 CrossRef.
- H. Suzuki, Jpn. J. Gastroenterol., 1981, 16, 422 Search PubMed.
- J. Kojima, M. Kantani, N. Nakamura, T. Kashiwagi, F. Tohijoh and M. Akiyama, Clin. Chim. Acta, 1980, 106, 165 CrossRef CAS.
- F. Kokot and J. Kuska, Clin. Chim. Acta, 1965, 11, 118 CrossRef CAS.
- K.-C. Xu, X.-Y. Meng, Y.-C. Shi, Z.-J. Ge, L. Ye, Z.-J. Yu and D.-M. Yang, Int. J. Cancer, 1985, 36, 667 CrossRef CAS.
- R. Cui, J. He, F. Zhang, B. Wang, H. Ding, H. Shen, Y. Li and X. Chen, Br. J. Cancer, 2003, 88, 1878 CrossRef CAS PubMed.
- Y. Suminami, F. Kishi, K. Sekiguchi and H. Kato, Biochem. Biophys. Res. Commun., 1991, 181, 51 CrossRef CAS PubMed.
- G. Giannelli and S. Antonaci, Dig. Liver Dis., 2006, 38, 854 CrossRef CAS PubMed.
- H. Kato, Y. Suehiro, H. Morioka, T. Torigoe, A. Myoga, K. Sekiguchi and I. Ikeda, Jpn. J. Cancer Res., 1987, 78, 1246 CAS.
- S. S. Schneider, K. E. Schick, E. Fish, J. C. Miller, D. S. Pena, S. M. Treter, G. A. Hui and A. Silverman, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 3147 CrossRef CAS.
- Y. Suminami, S. Nagashima, N. L. Vujanovic, K. Hirabayashi, H. Kato and T. L. Whiteside, Br. J. Cancer, 2000, 82, 981 CrossRef CAS PubMed.
- Y. Suminami, N. Shigeki, A. Murakami, S. Nawata, T. Gondo, H. Hirakawa, F. Numa, G. A. Silverman and H. Kato, Cancer Res., 2001, 61, 176 Search PubMed.
- H. Kato and T. Torigoe, Cancer, 1977, 40, 1621 CrossRef CAS PubMed.
- P. Pontisso, F. Calabrese, L. Benvegnù, M. Lise, C. Belluco, M. G. Ruvoletto, S. De Falco, M. Marino, M. Valente, D. Nitti, A. Gatta and G. Fassina, Br. J. Cancer, 2004, 90, 833 CrossRef CAS PubMed.
- G. Giannelli, F. Marinosci, P. Trerotoli, A. Volpe, M. Quaranta, P. Dentico and S. Antonaci, Int. J. Cancer, 2005, 117, 506 CrossRef CAS PubMed.
- G. Giannelli, E. Fransvea, P. Trerotoli, M. Beaugrand, F. Marinosci, L. Lupo, G. Nkontchou, P. Dentico and S. Antonaci, Clin. Chim. Acta, 2007, 383, 147 CrossRef CAS PubMed.
- M. Hussein, A. Ibrahim, H. Abdella, I. Montasser and M. Hassan, Indian J. Cancer, 2008, 45, 167 CrossRef CAS PubMed.
- G. Beale, D. Chattopadhyay, J. Gray, S. Stewart, M. Hudson, C. Day, P. Trerotoli, G. Giannelli, D. Manas and H. Reeves, BMC Cancer, 2008, 8, 200 CrossRef PubMed.
- O. Soyemi, J. Otegbayo, S. Ola, A. Akere and T. Soyemi, BMC Res. Notes, 2012, 5, 403 CrossRef PubMed.
- L. Beneduce, F. Castaldi, M. Marino, S. Quarta, M. Ruvoletto, L. Benvegnù, F. Calabrese, A. Gatta, P. Pontisso and G. Fassina, Cancer, 2005, 103, 2558 CrossRef CAS PubMed.
- P. Pontisso, S. Quarta, C. Caberlotto, L. Beneduce, M. Marino, E. Bernardinello, N. Tono, G. Fassina, L. Cavalletto, A. Gatta and L. Chemello, Int. J. Cancer, 2006, 119, 735 CrossRef CAS PubMed.
- C. Pozzan, R. Cardin, M. Piciocchi, N. Cazzagon, G. Maddalo, V. Vanin, A. Giacomin, P. Pontisso, U. Cillo and F. Farinati, J. Gastroenterol. Hepatol., 2014, 29, 1637 CrossRef CAS PubMed.
- N. Mossad, E. Mahmoud, E. Osman, S. Mahmoud and H. Shousha, Tumor Biol., 2014, 35, 11559 CrossRef CAS PubMed.
- X. Jia, J. Liu, Y. Gao, Y. Huang and D. Zhi, Arch. Med. Res., 2014, 45, 580 CrossRef CAS PubMed.
- R. K. Sterling, L. Jeffers, F. Gordon, A. P. Venook, K. R. Reddy, S. Satomura, F. Kanke, M. E. Schwartz and M. Sherman, Clin. Gastroenterol. Hepatol., 2009, 7, 104 CrossRef CAS PubMed.
- J. Zhu, F. Jiang, H.-B. Ni, M.-B. Xiao, B.-Y. Chen, W.-K. Ni, C.-H. Lu and R.-Z. Ni, Exp. Ther. Med., 2013, 5, 89 CAS.
| This journal is © The Royal Society of Chemistry 2016 |