Prenatal diagnosis of single gene disorders using amniotic fluid as the starting material for PCR
Huan
Huang
*a,
Shuo
Li
b,
Shuolian
Lu
a,
Hongshan
Ge
c and
Lizhou
Sun
*a
aDepartment of Gynecology and Obstetrics, First Affiliated Hospital of Nanjing Medical University, Nanjing, 210029, China. E-mail: huanghuan@njmu.edu.cn; lizhou_sun121@hotmail.com; Tel: +86 25 86202116
bInformation Engineering Department, Nanjing Xiaozhuang University, Nanjing, 211171, China
cReproductive Health Center, Second Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325000, China
First published on 12th November 2015
Abstract
A rapid and inexpensive method for fetal genetic diagnosis using amniotic fluid (AF) as the starting material was demonstrated in this study. Raw AF was added directly to polymerase chain reaction (PCR) mixtures with HpH buffer (a high pH buffer), without any pre-treatment. Amplified products were detected by gel electrophoresis and then subjected to Sanger sequencing. The AF from four fetuses, each expressing a single gene disorder (achondroplasia, hypochondroplasia, thanatophoric dysplasia, or X-linked hypohidrotic ectodermal dysplasia), were analyzed. DNA fragments of different lengths were efficiently amplified from 8 μl of AF, allowing each of these single gene disorders to be successfully diagnosed. Although the amplification efficiency of the AF-PCR method is comparable to that of the Chelex method, the amount of the AF sample required was considerably lower than that required for the Chelex method (10 ml). This proposed method of diagnosis is more efficient, simpler, and less expensive, and reduces the chance of cross-contamination relative to the Chelex method, which requires purified DNA or other pre-treatment processes. Our method offers a promising tool that can be used for the diagnosis of various gene disorders in fetuses.
Introduction
Fetal DNA is inherited from both parents, and analysis of fetal DNA in amniotic fluid (AF) is widely used in prenatal diagnosis and paternity testing.1–3 Molecular biological technologies for analyzing DNA have been advancing rapidly, making numerous contributions to prenatal diagnosis in recent years. Indeed, many genetic diseases can be diagnosed during the fetal stage, using fetal DNA extracted from the AF and subjected to molecular biology techniques such as polymerase chain reaction (PCR).4,5 This information helps parents make informed decisions about their babies’ futures. It is important to note that for most genetic tests, the genomic DNA obtained from the AF requires purification before PCR can be performed on it. Purification methods include the phenol/chloroform method,6 the Chelex method,7,8 or commercially available purification kits9–12 for DNA extraction from the AF. However, DNA extraction can be expensive and time consuming, and the isolated DNA can be easily cross-contaminated. Therefore, performing PCR on the AF directly, thereby eliminating the DNA extraction phase, would dramatically improve this technique. Several studies have examined whether or not raw samples (e.g., blood or AF) can be used as the starting material for direct PCR amplification.13–15 Although PCR has been successfully used to amplify small tandem repeat markers from the AF, pretreatment steps involving cell lysis and pH adjustment, or concentration and preheating, are still required before amplification.16–18 In this study, direct PCR amplification was successfully performed using raw AF as the starting material for PCR, without any pretreatment processes that have previously been required when using AF samples.18 Using this simple amplification method, single gene disorders, including achondroplasia,19,20 hypochondroplasia,21,22 thanatophoric dysplasia,23 and X-linked hypohidrotic ectodermal dysplasia,24 were prenatally diagnosed.Experimental section
Chemicals and reagents
Two kinds of Taq polymerases were purchased from TaKaRa (Dalian, China) and Promega (Madison, WI, USA). Ethidium bromide was purchased from Promega (Madison, WI, USA). Agarose was purchased from Oxoid Limited (Basingstoke Hampshire, England) and a dNTP mix was purchased from Genebase Gene-Tech (Shanghai, China). DNA marker I (100–600 bp) and marker II (100–1000 bp) were purchased from Tiangen (Beijing, China). All solutions were prepared with deionized and sterilized water. All other chemicals used in this study were of extra pure commercial grade.Gene targets and primer sequences
The nucleotide sequences for the fibroblast growth factor receptor 3 (FGFR3) gene and the ectodysplasin-A (EDA) gene are available in GenBank (http://www.ncbi.nlm.nih.gov/) under the accession numbers NG_012632.1 and NG_009809.1, respectively. Primer sequences used to diagnose achondroplasia, hypochondroplasia, thanatophoric dysplasia, and X-linked hypohidrotic ectodermal dysplasia are listed in Table 1. Primer Premier software (version 5.0; http://www.premierbiosoft.com/) was used to calculate the melting point and to analyze the risk of primer-dimer formation and secondary structures. The primer names and sequences of nine fragments with different lengths are listed in Table 2.| Gene | Primer | Sequence (5′ → 3′) | Single gene disorder |
|---|---|---|---|
| FGFR3 | Forward | AGGAGCTGGTGGAGGCTGA | Achondroplasia |
| Reverse | GGAGATCTTGTGCACGGTGG | ||
| Forward | ACTGACAAGGACCTGTCGGACC | Hypochondroplasia | |
| Reverse | TTGCAGGTGTCGAAGGAGTAGTC | ||
| Forward | CCCTGCCCTGAGATGCTG | Thanatophoric dysplasia | |
| Reverse | TGGGAAGGCGGTGTTGG | ||
| EDA | Forward | CAGGCCTGGCAGCTGCTTTAC | X-linked hypohidrotic ectodermal dysplasia |
| Reverse | TGGCCCCCTCTCTCTTTCCTC |
| Gene | Primer | Sequence (5′ → 3′) | Product size (bp) |
|---|---|---|---|
| AZF | Forward | CAGGCAGGACAGCTTAAAAG | 109 |
| Reverse | ACTGTGGCAAAGTTGCTTTC | ||
| Forward | CATTCATCTTCCCTTTTGAAGG | 201 | |
| Reverse | ATGCAAGTCRCAGGAAATCT | ||
| ALOX12B | Forward | TTTGCAAAACGGTGAGAGC | 309 |
| Reverse | ATAATCCCTGGTCCAGCCTC | ||
| Forward | CAGAGACCTAAGTGGATGGGG | 401 | |
| Reverse | GTTTTCCTCCCCGACTCC | ||
| Forward | CAGCTTTTCCAGAATTTGGCT | 496 | |
| Reverse | CAGCTCTCCAGCCTCTCCT | ||
| Forward | GGCACAGAAGCTGAACTCC | 700 | |
| Reverse | TGGGGCTGTCTTGGAGG | ||
| Forward | CTTAGGAGTGTTTTCACCTTCTCATACT | 801 | |
| Reverse | ATGAAGTTGGTTCTCCTGTCTAGAAAG | ||
| Forward | GGGAGTCCATCTCAGCGAC | 900 | |
| Reverse | CACCGCGATTCCGTTCTC | ||
| TGM | Forward | CTCCTGCTTCTTCCTCCTGA | 558 |
| Reverse | GTGTGGCATGGACTGAAAGA |
Sample collection and processing
Samples were collected from patients who underwent amniocentesis because of a family history of genetic disease or an abnormality indicated by ultrasound. Sample collection was approved by the ethics committee of the First Affiliated Hospital of Nanjing Medical University, and the participants provided written informed consent for their participation in the study. In the amniocentesis procedure, a needle is inserted through the mother's abdominal wall, then the wall of the uterus, and finally the amniotic sac. With the aid of ultrasound guidance, a physician punctures the sac in an area away from the fetus and extracts approximately 12 ml of AF for fetal genetic diagnosis. In this study, 200 μl of AF was collected from each patient and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 3 min. After removing 180 μl of the supernatant, the remaining sample was mixed evenly and heated at 94 °C for 2 min. The treated (concentration and heating) AF (T-AF) was stored at 4 °C until PCR was performed.
000g for 3 min. After removing 180 μl of the supernatant, the remaining sample was mixed evenly and heated at 94 °C for 2 min. The treated (concentration and heating) AF (T-AF) was stored at 4 °C until PCR was performed.
Genomic DNA extraction from AF by the Chelex method
Genomic DNA was extracted from 10 ml of uncultured AF using the Chelex method. AF was centrifuged at 2000 rpm for 8 min. After removing the supernatant, the remaining sample was mixed evenly with 250 μl of 5% Chelex-100 (Bio-Rad, Hercules, CA, USA) and 10 μl of 20 mg ml−1 proteinase K and incubated at 56 °C for 30 min on a heat block. The tube was then briefly vortexed and incubated at 98 °C for 8 min. After centrifuging the AF at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min, the supernatant was transferred into a 0.2 ml Eppendorf tube and stored at 4 °C until PCR was performed.
000 rpm for 3 min, the supernatant was transferred into a 0.2 ml Eppendorf tube and stored at 4 °C until PCR was performed.
Direct PCR amplification
The PCR reaction using AF as the starting material was carried out in a 25 μl-mixture containing 2.7 μl of HpH buffer (a high pH buffer), 200 μM of dNTPs, 2.5 U of Taq DNA polymerase, 0.4 μM of each primer, and varying amounts of AF (8 μl of AF per 25 μl of the mixture, unless specified). The cycling reaction was performed using a PTC-225 Peltier Thermal Cycler (MJ Research, Watertown, MA, USA). The mixtures were subjected to an initial cell lysis step at 96 °C for 3 min, followed by 35 amplification cycles (96 °C for 30 s, 55–65 °C for 30 s, and 72 °C for 30 s), and a final extension at 72 °C for 7 min.Gel electrophoresis and sequencing of PCR products
PCR products were analyzed by agarose gel electrophoresis followed by Sanger sequencing. Electrophoresis of a 2% agarose gel stained with 0.5 μg ml−1 ethidium bromide in 1× TAE buffer was performed at 100 V for 10 min. Gel images were captured using the GeneGenius BioImaging System (Syngene, England). Sanger sequencing was performed by Invitrogen (Shanghai, China).Results and discussion
Investigation of PCR inhibitors in AF
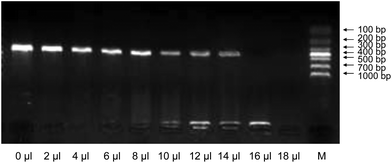
In early pregnancy, AF is largely composed of water with electrolytes.25 At the beginning of the 8th week of pregnancy, when the fetal kidneys begin to function, fetal urine is also present in the AF.26 After the 12–14th week of pregnancy, AF contains proteins, carbohydrates, lipids, phospholipids, and urea, all of which aid in the growth of the fetus.27 These AF components may interfere with cell lysis and/or inactivate thermostable DNA polymerases during PCR if AF is used directly as the starting material.18 Since the amount of inhibitors varies with the quantity of AF, it was necessary to investigate this effect on PCR yields. Various volumes (2 μl, 4 μl, 6 μl, 8 μl, 10 μl, 12 μl, 14 μl, 16 μl, and 18 μl) of blank-AF, which were prepared by removing the precipitate from fetal cells after centrifugation of the raw AF, were included in the PCR mixtures with HpH buffer and the same amount of template DNA. The results are shown in Fig. 1. We show that the suppression of PCR by blank-AF and the amplification efficiency of PCR decreased as the volume of blank-AF in the PCR reaction mixture increased. However, with less than 8 μl of blank-AF, the PCR yields did not vary greatly, whereas more than 8 μl of blank-AF significantly suppressed PCR amplification, and PCR failed with 16 μl blank-AF. These findings indicate that in order to obtain high amplification efficiency, 8 μl of AF may be optimal.Effect of different volumes of AF on the amplification efficiency of AF-PCR
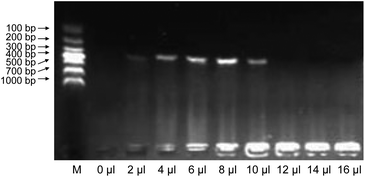
Although blank-AF suppressed PCR amplification, increased AF volumes offered more targets from the fetal cells for PCR. Therefore, the effect of different volumes of AF on AF-PCR was investigated. As shown in Fig. 2, distinct bands in the agarose gels were observed when the volumes of AF ranged from 2 to 10 μl, and the optimal volume of AF for successful PCR was 8 μl. These results suggested that more targets (less than 8 μl) would provide a higher PCR yield when the suppressive effect of AF with a volume of less than 8 μl can be ignored. If the volume of AF was greater than 10 μl, the suppressive effect dominated and amplification efficiency decreased. Therefore, to obtain the highest PCR yield, a volume of 8 μl of AF in a 25 μl PCR reaction was used routinely. The amplicons resulting from the amplification of DNA in 8 μl of AF were sufficient for Sanger sequencing to diagnose single gene disorders. Usually, 10 ml of AF is required when using the Chelex method, and 1 ml of raw AF is needed for the method reported by Ye et al.18 Therefore, our method requires only one-hundredth of the amount of raw AF used in the method described by Ye et al. and only one-thousandth of the amount of raw AF required in the Chelex method.Comparison of Chelex-PCR and AF-PCR with or without pre-treatment processes
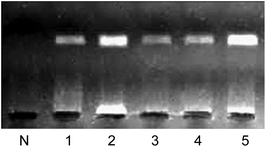
The Chelex method was developed for extracting DNA from many different types of samples, including whole blood, bloodstains, hair, and AF, for use in PCR, and especially for paternity testing.28,29 Although DNA extraction using Chelex 100 is as efficient, or more efficient, than methods using proteinase K and phenol–chloroform extraction,30,31 the Chelex 100 method requires multiple steps for DNA extraction, which can result in cross-contamination and is costly. In our previous work,18 we proposed a method using AF as the starting material for PCR to overcome the drawbacks of other extraction methods. However, the concentration and heating processes performed on AF before it was added to the PCR mixture may have caused cross-contamination of the sample. Because pre-treatment processes were necessary, our previously proposed method was not a genuine AF-PCR method. In this study, we developed an AF-PCR method with reliable amplification efficiency using AF as the starting material for PCR, thereby eliminating the pretreatment processes. The amplification efficiencies of the methods mentioned above are shown in Fig. 3. The amplification efficiency of the AF-PCR method is much higher than that of the methods using either pre-concentration or pre-heating processes, and equal to that of the Chelex method. Although the amplification efficiency of the AF-PCR method is a little lower than the efficiency following pre-treatment, the amplicon yield is sufficient for downstream analysis. Therefore, the target DNA sequence can be efficiently amplified from the AF directly, without any pre-treatment processes.Applicability of AF-PCR to the amplification of fragments of various lengths and from different samples
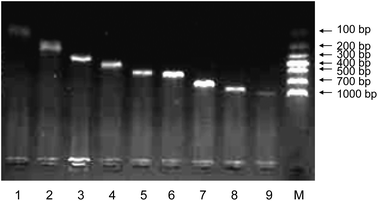
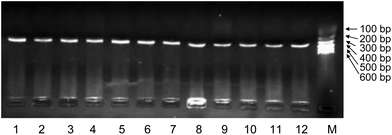
To investigate the applicability of AF-PCR, nine DNA fragments of different lengths ranging from 109 bp to 900 bp were tested. As shown in Fig. 4, the electrophoresis results showed that all the fragments were effectively amplified, indicating that AF-PCR can be used for the amplification of fragments with lengths ≤900 bp and that it may be suitable for detecting single gene disorders, which usually requires the analysis of short DNA fragments. The AF from 12 pregnant women at various times throughout gestation (from the 18th week to the 23rd week) was used as the starting material to amplify DNA fragments of 309 bp using the AF-PCR method. The successful amplification of these 12 samples is shown in Fig. 5. We thus show that DNA fragments of different lengths can be amplified directly from various AF samples, and specifically from the samples collected at 18–23 weeks of gestation.Analyzing single gene disorders by AF-PCR
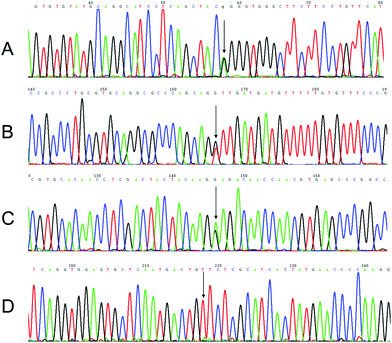
A single gene disorder is the result of a single mutated gene. Over 4000 human diseases are caused by single gene defects.32 Almost all genetic disorders result in decreased quality of life or result directly in death. Since there are no known cures for most genetic disorders, couples with one or both partners suffering from a genetic disorder or that carry a single gene mutation should undergo prenatal genetic testing to determine whether the fetus is carrying the mutated gene. The most common sample used for prenatal genetic diagnosis is AF.33 To achieve a simple and rapid clinical diagnostic, we utilized AF-PCR without any pre-treating process. We analyzed four single gene disorders, including achondroplasia, hypochondroplasia, thanatophoric dysplasia, and X-linked hypohidrotic ectodermal dysplasia, in order to validate the utility of AF-PCR in the field of prenatal diagnosis.Achondroplasia and hypochondroplasia are the most common forms of chondrodysplasia, characterized by rhizomelia, exaggerated lumbar lordosis, brachydactyly, and macrocephaly with frontal bossing and midface hypoplasia.34,35 These disorders arise from mutations in the FGFR3 gene, which encodes a transmembrane receptor that is important in regulating linear bone growth, among other functions.36 A mutation from G to A at nucleotide position 1138 (G380R) and a mutation from G to T (or C) at nucleotide position 1620 (N540K) in the FGFR3 gene were analyzed by AF-PCR followed by sequencing. The results are shown in Fig. 6A and B. A mutation resulting in the G380R substitution was identified in the AF DNA of one fetus, whereas mutations resulting in the N540K substitution were identified in the AF DNA of another fetus. In addition, thanatophoric dysplasia, a severe and generally lethal skeletal dysplasia presenting in the prenatal period and characterized by micromelia, macrocephaly, narrow thorax, and distinctive facial features,37 which contains a mutation at nucleotide position 1948 in the FGFR3 gene, was diagnosed in another fetus by our method. This mutation from A to G is shown in Fig. 6C. A mutation in the EDA gene in X-linked hypohidrotic ectodermal dysplasia was also successfully detected by AF-PCR and sequencing (Fig. 6D). This c.822G>T mutation causes characteristic facial features, including hypotrichosis (sparseness of scalp and body hair), hypohidrosis (reduced ability to sweat), and hypodontia (congenital absence of teeth).38
Conclusion
Prenatal diagnostics are a long-sought goal in human genetics. In the diagnosis of congenital fetal abnormalities, the sooner the diagnosis is made, the better the outcome. Developing a genetic diagnostic method that does not require DNA extraction would significantly improve the speed of diagnosis and wide applicability of the test. However, using AF directly for PCR amplification has proven to be difficult, as some substances in the AF may interfere with cell lysis and/or inactivate thermostable DNA polymerases. Accordingly, inhibition of PCR by AF was evaluated, and the HpH buffer for direct PCR from the AF samples was successfully applied. Since this method eliminates the DNA extraction process, PCR with AF is less expensive and reduces the risk of sample contamination relative to other amplification methods. Moreover, AF-PCR is highly sensitive, with less than 8 μl of AF being sufficient to obtain successful sequencing results. Therefore, coupling HpH buffer with AF-PCR for the rapid diagnosis of single gene disorders was successfully realized in this study. An additional observation from the study was that storing AF at 4 °C for 2 months up to 1 year did not affect the AF-PCR yield. Although AF-PCR is a simple, fast, and inexpensive method that can be used for prenatal diagnosis, the method still requires invasive sampling. With the development of high throughput sequencing technologies, free fetal DNA in maternal peripheral blood is now widely used in noninvasive prenatal diagnosis of trisomy 21, trisomy 18, and trisomy 13. It is hoped that the AF-PCR method can be improved and used in noninvasive prenatal diagnosis of single gene disorders in fetuses, providing another diagnostic tool for use during pregnancy.Acknowledgements
This work was funded by the National Natural Science Foundation of China (21305069, 81470065), the Project for Training of Young Talents at the Jiangsu Province Maternal and Child Health Hospital (FRC201302, FXK201217) and the Project for the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801).Notes and references
- A. Emad, J. Lamoureux, A. Ouellet and R. Drouin, Fetal Diagn. Ther., 2015, 38, 65–76 CrossRef PubMed.
- J. L. Callaway, S. Huang, E. Karampetsou and J. A. Crolla, Mol. Biotechnol., 2014, 56, 312–318 CrossRef CAS PubMed.
- M. S. Pollack, S. D. Heagney, D. Braun Jr. and G. J. O'Neill, Prenatal Diagn., 1981, 1, 183–195 CrossRef CAS.
- M. T. Rebello, G. Hackett, J. Smith, F. E. Loeffler, S. Robson, N. MacLachlan, R. W. Beard, C. H. Rodeck, R. Williamson and D. V. Coleman, et al. , Prenatal Diagn., 1991, 11, 41–46 CrossRef CAS.
- M. L. Santoro, L. Mobili, A. Mesoraca and C. Giorlandino, Prenatal Diagn., 2003, 23, 1083–1085 CrossRef CAS PubMed.
- R. Hua, W. Yang, N. H. Sun and X. Zhang, Chin. J. Ophthalmol., 2011, 47, 801–805 CAS.
- Y. Rong, J. Gao, X. Jiang and F. Zheng, Int. J. Mol. Sci., 2012, 13, 5972–5981 CrossRef CAS PubMed.
- Y. Li, B. T. Hao, Y. L. Yang, W. Y. Zhu, Y. M. Si and Y. T. Wang, Yichuan/Hereditas, 2002, 24, 639–642 CAS.
- H. J. Lee, S. Park, H. J. Kang, J. K. Jun, J. A. Lee, D. S. Lee, S. S. Park and M. W. Seong, Ann. Lab. Med., 2012, 32, 380–384 CrossRef CAS PubMed.
- I. Papoulidis, E. Siomou, A. Sotiriadis, G. Efstathiou, A. Psara, E. Sevastopoulou, E. Anastasakis, S. Sifakis, T. Tsiligianni, M. Kontodiou, C. Malamaki, M. Tzimina, M. B. Petersen, E. Manolakos and A. Athanasiadis, Prenatal Diagn., 2012, 32, 680–685 CrossRef CAS PubMed.
- W. Bi, A. M. Breman, S. F. Venable, P. A. Eng, T. Sahoo, X. Y. Lu, A. Patel, A. L. Beaudet, S. W. Cheung and L. D. White, Prenatal Diagn., 2008, 28, 943–949 CrossRef CAS PubMed.
- B. Nagy, Z. Ban, L. Lazar, R. G. Nagy, C. Papp, E. Toth-Pal and Z. Papp, Prenatal Diagn., 2005, 25, 1138–1141 CrossRef CAS PubMed.
- C. M. Connelly, L. R. Porter and J. R. TerMaat, BMC Med. Genet., 2014, 15, 130–138 CrossRef PubMed.
- M. Ohhara, Y. Kurosu and M. Esumi, BioTechniques, 1994, 17, 726–728 CAS.
- M. Hayashida, K. Iwao-Koizumi, S. Murata and K. Kinoshita, Anal. Sci., 2009, 25, 1487–1489 CrossRef CAS PubMed.
- C. M. Ogilvie, C. Donaghue, S. P. Fox, Z. Docherty and K. Mann, J. Histochem. Cytochem., 2005, 53, 285–288 CrossRef CAS PubMed.
- L. J. Levett, S. Liddle and R. Meredith, Ultrasound Obstet. Gynecol., 2001, 17, 115–118 CrossRef CAS PubMed.
- H. Ye, H. Wu, H. Huang, Y. Liu, B. Zou, L. Sun and G. Zhou, Analyst, 2013, 138, 2443–2448 RSC.
- E. Perez-Carbajo, I. Zapardiel, L. Sanfrutos-Llorente, S. Cruz-Melguizo, C. Martinez-Payo and E. Iglesias-Goy, Case Rep. Obstet. Gynecol., 2015, 2015, 980749 Search PubMed.
- S. Boulet, M. Althuser, F. Nugues, J. P. Schaal and P. S. Jouk, Prenatal Diagn., 2009, 29, 697–702 CrossRef PubMed.
- C. Karadimas, S. Sifakis, P. Valsamopoulos, C. Makatsoris, V. Velissariou, G. Nasioulas, M. B. Petersen, E. Koumantakis and A. Hatzaki, Am. J. Med. Genet., Part A, 2006, 140, 998–1003 CrossRef CAS PubMed.
- C. P. Chen, Y. N. Su, T. H. Lin, T. Y. Chang, J. W. Su and W. Wang, Taiwan. J. Obstet. Gynecol., 2013, 52, 580–585 CrossRef PubMed.
- G. Tonni, D. Azzoni, A. Ventura, B. Ferrari, C. D. Felice and M. Baldi, Fetal Pediatr. Pathol., 2010, 29, 314–322 CrossRef CAS PubMed.
- J. Zonana, A. Clarke, M. Sarfarazi, N. S. Thomas, K. Roberts, K. Marymee and P. S. Harper, Am. J. Hum. Genet., 1988, 43, 75–85 CAS.
- L. Belkacemi, M. Desai, M. H. Beall, Q. Liu, J. T. Lin, D. M. Nelson and M. G. Ross, J. Matern.–Fetal Neonat. Med., 2011, 24, 752–759 CrossRef CAS PubMed.
- A. Goncalves, W. G. Franca, S. G. Moraes, L. A. Pereira and L. Sbragia, Int. Braz. J. Urol., 2004, 30, 508–513 CrossRef PubMed.
- K. Kike, Pract. Gynecol. Obstet., 1966, 15, 993–999 CAS.
- B. Grskovic, G. Mrsic, O. Polasek, A. Vrdoljak, S. Merkas and S. Andelinovic, Forensic Sci., Med., Pathol., 2011, 7, 155–161 CrossRef PubMed.
- A. Manamperi, C. Hapuarachchi, N. S. Gunawardene, A. Bandara, D. Dayanath and W. Abeyewickreme, Ceylon Med. J., 2009, 54, 85–89 Search PubMed.
- C. Zandotti, X. De Lamballerie, C. Guignole-Vignoli, C. Bollet and P. De Micco, Acta Virol., 1993, 37, 106–108 Search PubMed.
- P. S. Walsh, D. A. Metzger and R. Higuchi, BioTechniques, 1991, 10, 506–513 CAS.
- H. Galjaard, Bailliere's Clin. Obstet. Gynaecol., 1987, 1, 547–567 CrossRef CAS.
- P. Miny, S. Tercanli, D. Ganshirt and W. Holzgreve, Ther. Umsch. Rev. Ther., 1995, 52, 792–800 CAS.
- Y. L. Shin, J. H. Choi, G. H. Kim and H. W. Yoo, J. Pediatr. Endocrinol. Metab., 2005, 18, 999–1005 CAS.
- A. A. Kaissi, S. Farr, R. Ganger, J. G. Hofstaetter, K. Klaushofer and F. Grill, Open Orthop. J., 2013, 7, 33–39 CrossRef PubMed.
- M. B. Bober, M. Taylor, R. Heinle and W. Mackenzie, Am. J. Med. Genet., Part A, 2012, 158A, 2336–2341 CrossRef PubMed.
- R. Marquis-Nicholson, S. Aftimos and D. R. Love, Sultan Qaboos Univ. Med. J., 2013, 13, 80–87 CrossRef PubMed.
- M. Callea, A. Vinciguerra, C. E. Willoughby, L. Deroma and G. Clarich, Ophthalmic Genet., 2013, 34, 58–60 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2016 |