Facile multi-dimensional profiling of chemical gradients at the millimetre scale†
Chih-Lin
Chen
a,
Kai-Ta
Hsieh
a,
Ching-Fong
Hsu
a and
Pawel L.
Urban
 *ab
*ab
aDepartment of Applied Chemistry, National Chiao Tung University, 1001 University Rd, Hsinchu, 300, Taiwan. E-mail: plurban@nctu.edu.tw
bInstitute of Molecular Science, National Chiao Tung University, 1001 University Rd, Hsinchu, 300, Taiwan
First published on 27th October 2015
Abstract
A vast number of conventional physicochemical methods are suitable for the analysis of homogeneous samples. However, in various cases, the samples exhibit intrinsic heterogeneity. Tomography allows one to record approximate distributions of chemical species in the three-dimensional space. Here we develop a simple optical tomography system which enables performing scans of non-homogeneous samples at different wavelengths. It takes advantage of inexpensive open-source electronics and simple algorithms. The analysed samples are illuminated by a miniature LCD/LED screen which emits light at three wavelengths (598, 547 and 455 nm, corresponding to the R, G, and B channels, respectively). On presentation of every wavelength, the sample vial is rotated by ∼180°, and videoed at 30 frames per s. The RGB values of pixels in the obtained digital snapshots are subsequently collated, and processed to produce sinograms. Following the inverse Radon transform, approximate quasi-three-dimensional images are reconstructed for each wavelength. Sample components with distinct visible light absorption spectra (myoglobin, methylene blue) can be resolved. The system was used to follow dynamic changes in non-homogeneous samples in real time, to visualize binary mixtures, to reconstruct reaction-diffusion fronts formed during the reduction of 2,6-dichlorophenolindophenol by ascorbic acid, and to visualize the distribution of fungal mycelium grown in a semi-solid medium.
1. Introduction
Absorption spectroscopy – advanced by many prominent scientists including Joseph Fraunhofer (1787–1826)1 and Gustav Kirchhoff (1824–1887)2 – has widely been used in the physical sciences since the 19th century. The Lambert–Beer's law establishes a linear relationship between the measured value (absorbance, A) and the concentration (C) of the light-absorbing compound present in a sample of a specified thickness (b):3| A = εbC | (1) |
Several instrumental techniques are available for the chemical profiling of samples in the three-dimensional space. For example, confocal microscopy scans fluorophores in micrometre-scale translucent specimens with a focused beam of light, layer-by-layer, in order to reconstruct their three-dimensional distributions.7 While the modern microscopy techniques excel in spatial resolution, they are mainly suitable for studies on micrometre-sized samples, require fluorescent probes, and are costly. If larger objects need to be imaged at a moderate spatial resolution, optical tomography can be implemented. According to the back-projection principle, when several light paths, originating from different directions, are blocked by a non-transparent obstacle, their intensities are decreased. By extending the “shadows”, created by the obstacle, back to the source of radiation, one can identify the real position and physical shape of the obstacle.8 Images can be reconstructed without physical intervention or chemical modification of the specimen. In fact, X-ray tomography is widely used in medical imaging.9 However, apart from X-rays, other forms of electromagnetic waves are utilized in specific applications.10–12 Microscale optical tomography enables imaging specimens as small as individual cells.13–15 Interestingly, the principles of tomography are also applicable to the 3D-profiling of nanoscopic samples using beams of electrons rather than photons.16–18 In chemistry, optical tomography with a blue LED light source was utilized to study organizing centres in a non-homogeneous oscillating reaction mixture.8
Here we develop a simple optical tomography system that enables performing scans of non-homogeneous samples at different wavelengths of visible light. It takes advantage of inexpensive widely accessible pieces of hardware, and can readily be assembled in most chemistry/biochemistry laboratories. We further verify the possibility of implementing this system in the three-dimensional optical profiling of specimens with intrinsic heterogeneities (chemical gradients).
2. Experimental section
2.1. Materials and samples
Ascorbic acid, 2,6-dichlorophenolindophenol, methylene blue and myoglobin (from equine skeleton muscle) were purchased from Sigma-Aldrich (St Louis, MO, USA). Agarose (ultra-grade for molecular biology) was purchased from UniRegion Bio-Tech (Hsinchu, Taiwan).The fungal strain, red bread mould, Neurospora crassa (32685; Bioresource Collection and Research Center, Hsinchu, Taiwan) was first cultured in liquid Vogel's medium19 for 3–4 days at 25 °C. Subsequently, the fungus was transferred onto a Petri dish with 0.5% agar Vogel's medium for 1 day culture at 25 °C. The media were sterilized in an autoclave at 121 °C over 15 min. Cells of N. crassa were implanted into the medium with a silica capillary. Then, the test tubes were refrigerated (∼4 °C) for several days to slow down the growth rate of N. crassa mycelium.
2.2. Spectrotomography setup
The developed tomography system incorporates a 2.4 inch LCD screen with LED back-illumination (ITEAD Studio, Shenzhen, China), a diffuser (tracing paper, 45 × 27 mm, thickness: ∼0.06 mm), a servo motor (HS-422; Hitec RCD, Poway, CA, USA) with a holder for the round-cross-section cell (borosilicate glass tube; length: 50 mm; OD: 6 mm; ID: ∼4 mm; Kimble Chase, Vineland, NJ, USA), and a CMOS-based digital camera (OM-D E-M1; Olympus, Tokyo, Japan; Fig. 1). The sample cell was fixed in a holder fabricated using the ABS polymer using a 3D printer (UP Plus 2; Beijing TierTime Technology, Beijing, China). The LCD/LED screen and the motor are connected to an electronic control device (Fig. S1†). It is based on an open-source platform – Arduino.20 The change of screen colour (wavelength of the emitted light) is controlled by Arduino Mega (Torino, Italy) while rotation of the motor spindle (sample cell) is controlled by Arduino Uno. Both operations are synchronized. The printed circuit board (PCB) functions were programmed using custom scripts (written in a C-derived language). Four output pins of the Arduino Uno PCB (9, 10, 11 and 12) were connected to four input pins of the Arduino Mega PCB (9, 10, 11 and 12) to control the rotation of the servo motor. Pins 30, 31, 35, 36 and 37 of the Arduino Mega PCB were used to change the colour of the LCD/LED display alternately among white, red, green, and blue, following subsequent rotations of the motor spindle by ∼180°. When disconnected from the computer, the PCBs were powered by a 9 V adaptor (Hsinchu, Taiwan). Typically, the video files were recorded by the camera as the sample cell rotated from 0° to ∼180°. The time of sample cell rotation was ∼1 s.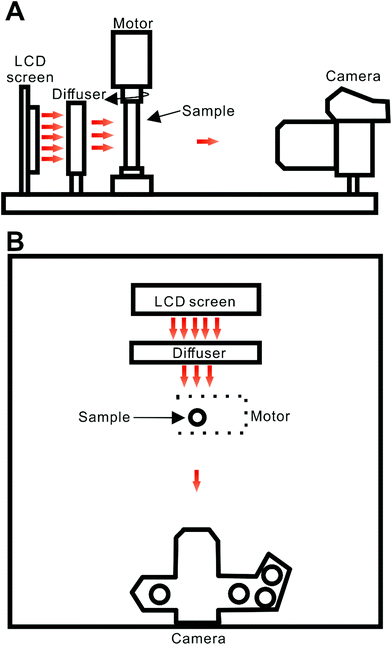 | ||
| Fig. 1 Schematic representation of the experimental system: (A) side view; (B) top view. Distances: LCD screen-diffuser, ∼2.3 cm; diffuser-sample, ∼3.3 cm; sample-lens, ∼19 cm. Red arrows illustrate propagation of light from the light source. Movie S1† shows a typical scan conducted by this system. | ||
In a preliminary test, visible light spectra were obtained for different colour channels of the LCD/LED screen. This measurement was conducted using a miniature fibre optic spectrometer (Ocean Optics, Dunedin, FL, USA; Fig. S2†). The measured wavelengths (maxima) are: 598, 547 and 455 in the case of red, green and blue channels, respectively. The light intensity at the surface of the display – measured with a luxometer (Centenary Materials Company, Hsinchu, Taiwan) – was 48, 12, 25 and 20 Lux in the case of white, red, green and blue channels, respectively.
2.3 Data processing
The raw video files were uploaded to the Free Video to JPG Converter (version 5.0.55.113; Softonic Internacional, Barcelona, Spain), and converted to JPG files. Every scan (one wavelength) produces 24 images. The resolution of these images was further reduced down to 768 × 432 using the PhotoCap software (version 6.0) in order to make them compatible with the algorithm used for further processing. The image analysis algorithm – used in this study – is a modified version of the one previously developed in our group for 2D imaging of chemical reactions.21 Briefly, following the selection of the rectangular area of interest, the program analysed horizontal rows of pixels from top to bottom. Absorbance values corresponding to these pixels were then exported, normalized, and further processed in the Excel software (2010; Microsoft, Albuquerque, NM, USA). The raw absorbance values (A) were calculated using the formula:22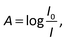 | (2) |
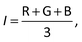 | (3) |
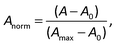 | (4) |
3. Results and discussion
3.1. Multidimensional imaging
Standard spectroscopic methods used in chemistry provide zero-dimensional (0D) results that are not attributed to any specific position within the sample. In such cases, it is of paramount importance to assure high homogeneity of the sample. However, in the case of non-homogeneous samples, the concentration/quantity data need to be linked with spatial position within the sample. For example, chromatographic and electrophoretic separations as well as flow injection analysis protocols lead to the formation of one-dimensional (1D) gradients in the flow lines. They are subsequently scanned by single-point28 detectors – recording signals in the time domain – or imaging detectors29–31 – recording signals in the distance domain. However, a single spatial dimension (x) is not always sufficient to follow complex physical and chemical phenomena. Thus, methods of two-dimensional (2D) imaging,21,32–34 attributing two co-ordinates (x, y) to every data point, have been developed. Nonetheless, all processes occur in three-dimensional (3D) space (characterized by three co-ordinates: x, y, and z), and it is appealing to avoid treating 3D heterogeneous systems as quasi-1D or quasi-2D matrices. In some previous studies, multi-dimensional heterogeneous systems were digitized (aliquoted) to enable measurements at arbitrary points using conventional (0D) detectors.35 However, such procedures are cumbersome and not readily applicable to 3D systems. Optical tomography provides a facile way to visualize distributions of chemical species within the 3D space. The 3D system described here (Fig. 1) is particularly simple and inexpensive. Thus, it may readily be replicated by chemists who are less familiar with optical and electronic technology.The tomography system developed here was tested by recording high-aspect-ratio objects suspended in agarose hydrogel medium. Objects with diameters below 200 μm produce dark features in tomograms (Fig. S4†). However, imaging light-reflecting objects leads to artefacts, thus degrading spatial resolution. Please note that the imaging system used to collect individual snapshots provides a resolution of at least ∼160 μm (verified using negative 1951 USAF and NBS 1963A resolution targets). However, the overall resolution of tomography depends on various other factors, including: precision of sample rotation (enabled by the motor), number of snapshots taken per half-rotation, quality of sample preparation, optical artefacts, and limitations of the data processing algorithms (e.g.ref. 36). The temporal resolution of the system was in the order of ∼2 s which corresponds to the duration of one full scan (single wavelength, including return to the original position, cf. Movie S1†). The scan rate could be increased by implementing a faster display (to change the light wavelength at a high speed), a faster motor, and a faster camera (taking more snapshots per second). However, the anticipated improvement of temporal resolution would be associated with a higher cost of the device.
3.2. Temporal monitoring in three dimensions
In the first demonstration of the presented setup, we applied it to follow redistribution of a coloured substance in the hydrogel matrix (Fig. 2). In this experiment, a small amount of methylene blue powder was inserted into the test tube filled with 2% agarose. Subsequently, the sample was loaded into the device (Fig. 1), and a scan was performed. It was repeated after 30 and 60 min incubation at ∼24 °C. Processing the collected raw data led to obtaining 3D projections of the sample contents (Fig. 2). The powder particles were initially “immobilized” at an arbitrary location within the sample vial – due to the high viscosity of agarose hydrogel. As time went by, methylene blue was solubilized in the water-rich matrix, and redistributed – due to diffusion and convection – from the zone of high concentration (origin) toward the surrounding zones of low concentration. Thus, the concentration was equalized. Consequently, the reconstructed 3D image obtained for 60 min-incubation is paler than the one recorded at the start (Fig. 2). Due to the relatively short scan time (∼1 s), one can assume that the sample is quasi-steady in this time scale.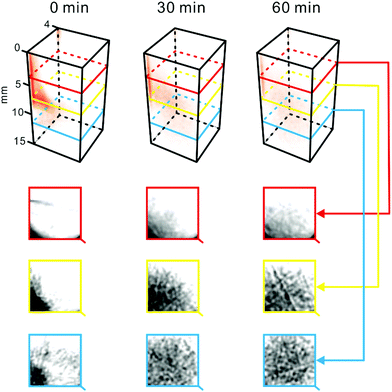 | ||
| Fig. 2 Dispersion of methylene blue in agarose. Methylene blue crystals (∼0.8 mg) were introduced to the sample cell filled with 2% agarose. Temperature: ∼24 °C. Top: 3D representations of the sample reconstructed based on the tomograms recorded at different time points. Bottom: Tomograms of the sample obtained at different heights. Dark areas indicate high concentration of methylene blue. For a replicate result obtained while scanning the sample at different wavelengths, see Fig. S6.† | ||
The grey value of some tomograms was analysed for a selected intersect line (Fig. S5†) pointing to the movement of the methylene blue front due to diffusion and convection. This analysis shows that – using the presented approach – it is possible to observe spatial and temporal gradients of light-absorbing molecules in any direction of interest within the 3D space.
3.3. Spectral tomography of heterogeneous binary mixtures
To verify the utility of the system in the chemical analysis of multi-component optically heterogeneous mixtures, two different compounds – methylene blue and myoglobin – were dispersed in 2% agarose medium. The specimen was scanned by means of the constructed tomography setup at different wavelengths (one by one; cf. Movie S1†). Methylene blue strongly absorbs light within the range 580–740 nm,37 while myoglobin absorbs light at ∼409 nm.38 Thus, the zones of these compounds can readily be distinguished in the series of tomograms obtained at different wavelengths (λ = 598, 547, and 455 nm; Fig. 3). It is striking to note that gas bubbles were accidentally trapped in the agarose medium. Due to their ability to scatter light, these bubbles caused a slight distortion of tomogram images corresponding to the middle section of the sample cell (Fig. 3, yellow rectangle).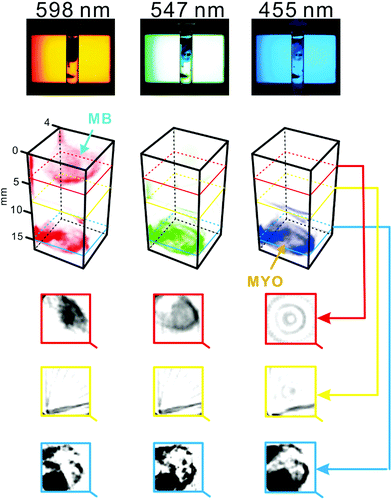 | ||
| Fig. 3 Spectral tomography of a heterogeneous sample containing two substances with distinct optical properties. The sample cell was filled with 2% agarose. Subsequently, ∼0.7 mg of crystalline myoglobin and ∼0.3 mg of methylene blue powder were inserted with a metal spatula. The sample was scanned at three wavelengths individually, and with the combination of the three wavelengths (white light, not shown). Top: original snapshots (raw data). Middle: 3D representations of the sample reconstructed based on the tomograms recorded at different wavelengths. Bottom: Tomograms of the sample obtained at different heights. Dark areas indicate high concentration of the dispersed substances: myoglobin (MYO) and methylene blue (MB). For a replicate result and an alternative representation, see Fig. S7.† | ||
Interestingly, in the work by Bánsági et al.,39 the authors used a lamp with a 532 nm interference filter in order to match the absorption spectrum of ferroin which was the main reactant in the studied process. However, in various processes, the relevant components might absorb light at different wavelengths; thus, it is appealing to enhance the selectivity of optical tomography by monitoring several wavelengths. In the work by Hsieh et al.,21 an LCD/LED screen was used as the light source to conduct 2D imaging of the Belousov–Zhabotinsky reaction at three wavelengths. This feature enabled detection of the transient reactants. Due to the limitations of that system, processes occurring in the vertical (z) direction of the reaction cell (e.g. vertical waves) were ignored. The current work combines the advantages of using the LCD/LED light source with the benefits gained by implementing optical tomography (i.e. 3D imaging); thus providing a clear advantage in the studies of non-homogeneous multi-component samples. In one interesting report, dissolution of an insulin formulation in a hydrogel has been examined by an innovative approach of UV absorption imaging detection.32 In that case, 2D gradients of insulin at different pH values could be visualized in real time. While the presented spectrotomography method enables constructing 3D projections of light-absorbing molecules, its limitation is that it only covers a portion of the visible light spectrum. Thus, there is still room for further development to extend the covered spectral range toward shorter and longer wavelengths.
3.4. Three-dimensional monitoring of reaction fronts
We further aimed to demonstrate the usefulness of this system in the monitoring of non-homogeneous chemical reactions in the hydrogel matrix. For this purpose, we implemented the reduction of Tillman's reagent (2,6-dichlorophenolindophenol) by ascorbic acid as the model reaction:| C12H7Cl2NO2 + C6H8O6 → C12H9Cl2NO2 + C6H6O6 | (5) |
Notably, this reaction is frequently used to quantify vitamin C40 and as a model reaction41 in kinetic studies. The solution of 2,6-dichlorophenolindophenol in agarose hydrogel has a blue appearance. The above reaction (eqn (5)) leads to decolouration of the blue matrix. Thus, it can readily be followed at white light (a mixture of red, green, and blue light emitted by the LCD/LED screen; Fig. 4). However, when it is scanned with light at individual wavelengths of the LCD/LED screen (598, 547, and 455 nm), the darkest features can be seen at 598 nm (red light; Fig. S8†). The experiment was conducted by dispersing either pure crystals of ascorbic acid or a commercial acetaminophen powder (containing: acetaminophen, caffeine, phenylephrine hydrochloride, dextromethorphan hydrobromide, and ascorbic acid) in the 2,6-dichlorophenolindophenol/agarose matrix (Fig. S8† and Fig. 4, respectively). Tomography scans reveal propagation of the reaction front – marked as an abrupt signal change due to decolouration of the reagent – spreading in the three dimensional space, from the locations where particles of ascorbic acid powder were initially embedded. Any of the 112 tomograms (stacked vertically) can be selected and analysed in order to observe propagation of the reaction front in a defined direction, for example an arbitrary line that is approximately normal to a tangent of the reaction front (cf.Fig. 4, bottom). In this particular case, the initial speed of the reaction front movement was estimated to be ∼0.43 mm min−1 (∼7.2 × 10−6 m s−1). This value is very low when compared with the high rate of the reaction in eqn (5), reported elsewhere (e.g.ref. 41). This observation is explained with the fact that the highly viscous reaction medium (hydrogel) is not homogenized (stirred), thus the reaction rate is limited by very slow diffusion and convection processes.
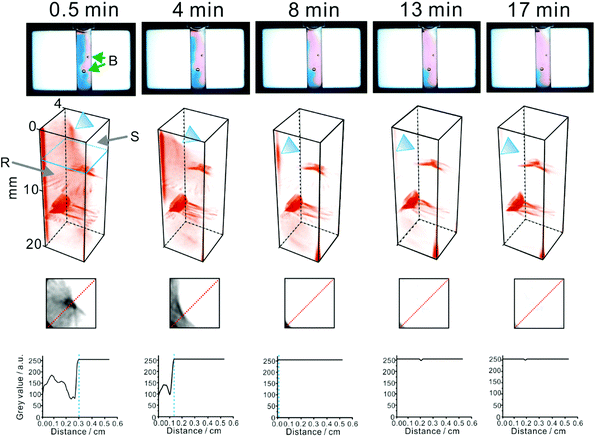 | ||
| Fig. 4 Tomography-aided monitoring of a non-homogeneous chemical reaction mixture (cf.eqn (5)). A small amount of a commercial acetaminophen formulation (∼30 mg), containing ascorbic acid (0.67 wt%), was dispersed in 0.5% agarose, containing 0.3 mM 2,6-dichlorophenolindophenol reagent. Temperature: ∼24 °C. B: bubbles (accidentally introduced into the agarose matrix); R: zone of the 2,6-dichlorophenolindophenol reagent; S: zone of ascorbic acid (substrate). The time values (indicated in the upper row) correspond to the time interval between the introduction of the sample powder and the tomography scan. The blue triangle indicates the reaction front, where the reagent is reduced. The lower images show tomograms obtained at an arbitrary height of the vial (see the blue marker in the 3D projection on the left). The bottom plots relate grey values of the pixels along the red dotted lines on the above tomograms with distance. Blue dashed lines indicate the position of the reaction front (advancing toward the lowere left corner of the tomograms). | ||
3.5. Pilot study on imaging fungal growth
We also aimed to verify the suitability of the presented system for imaging microbial growth in the 3D space. The growth of red bread mould (Neurospora crassa) was stimulated inside the sample cell filled with the Vogel's medium. The resulting reconstructions (Fig. S9†) reveal the intricacies of mycelial growth. The signals arise from light scattering and refraction at the cells’ surface as well as absorption of light due to pigmentation of the biological material. In fact, N. crassa cells contain substantial amounts of carotenoid compounds which are responsible for the reddish pigmentation of the mycelium.42,43 One can speculate that – in future, following further refinements – its applications may extend beyond those presented in this article. For example, the method might also be applicable to the imaging of cultured animal cells and tissues, providing insights into interactions between cells. Perhaps it could also be used to track chemotaxis of nematodes in 3D chemical gradients (cf.ref. 44).4. Conclusions
In this study, we have developed a simple optical tomography setup which enables rudimentary 3D imaging of non-homogeneous translucent samples. The important advantage of the device is that it can obtain tomograms at three different wavelengths almost simultaneously (within several seconds). The total cost of the device is ∼£200 (∼£1500, including the digital reflex camera). It can readily be constructed and operated by chemists who do not have much background knowledge in optics, electronics and computer science. In fact, it incorporates open-source electronic modules which are widely accessible. The customized elements can easily be fabricated using a low-end 3D-printer. The system can enable studies on physical, chemical and biological phenomena occurring in space and time (dispersion, reactions, cell growth). Performance of the system is limited by the quality of elements used to construct it. It may be upgraded by increasing the total cost of the platform. The method can currently be used to scan specimens at three wavelengths within the visible part of the electromagnetic spectrum. However, only some compounds absorb in this wavelength range. Moreover, absorption bands of different compounds can overlap; thus, the method is vulnerable to spectral interferences. In further developmental work, it would be interesting to extend the wavelength range of the tomographic scans (to UV and IR), and enable time-resolved spectroscopic measurements.Acknowledgements
We would like to thank the Ministry of Science and Technology of Taiwan (formerly, National Science Council; grant number: NSC 102-2113-M-009-004-MY2) for the financial support of this work. Thanks are also due to Prof. Yu-Chie Chen for lending us the Ocean Optics spectrometer.Notes and references
- I. Howard-Duff, J. Br. Astron. Assoc., 1987, 97, 339–347 Search PubMed.
- G. Kirchhoff, Verh. Naturhist. Med. Ver. Heidelberg, 1859, 1, 251–255 Search PubMed.
- D. F. Swinehart, J. Chem. Educ., 1962, 39, 333–335 CrossRef CAS.
- A. B. Kostinski, J. Opt. Soc. Am. A, 2001, 18, 1929–1993 CrossRef CAS PubMed.
- F. Siegert and C. J. Weijer, Curr. Biol., 1995, 5, 937–943 CrossRef CAS PubMed.
- E. K. Lin, C. L. Soles, D. L. Goldfarb, B. C. Trinque, S. D. Burns, R. L. Jones, J. L. Lenhart, M. Angelopoulos, C. G. Willson, S. K. Satija and W.-L. Wu, Science, 2002, 297, 372–375 CrossRef CAS PubMed.
- M. Minsky, Scanning, 1988, 10, 128–138 CrossRef.
- A. T. Winfree, S. Caudle, G. Chen, P. McGuire and Z. Szilagyi, Chaos, 1996, 6, 617–626 CrossRef CAS PubMed.
- R. Cierniak, X-Ray Computed Tomography in Biomedical Engineering, Springer, Berlin, 2011 Search PubMed.
- C. Westbrook and C. K. Roth, MRI in Practice, Wiley-Blackwell, Hoboken, 4th edn, 2011 Search PubMed.
- R. Cabeza and L. Nyberg, J. Cognit. Neurosci., 2000, 12, 1–47 CrossRef CAS.
- C. Wang, J. Kim, C. T. Jin, P. H. W. Leong and A. McEwan, J. Near Infrared Spectrosc., 2012, 20, 237–247 CrossRef CAS.
- B. Simon, M. Debailleul, V. Georges, V. Lauer and O. Haeberlé, Eur. Phys. J.: Appl. Phys., 2008, 44, 29–35 CrossRef.
- M. Debailleul, V. Georges, B. Simon, R. Morin and O. Haeberlé, Opt. Lett., 2009, 34, 79–81 CrossRef CAS PubMed.
- B. Simon, M. Debailleul, A. Beghin, Y. Tourneur and O. Haeberlé, J. Biophotonics, 2010, 3, 462–467 CrossRef PubMed.
- P. A. Midgley and M. Weyland, Ultramicroscopy, 2003, 96, 413–431 CrossRef CAS PubMed.
- T. Segal-Peretz, J. Winterstein, M. Doxastakis, A. Ramírez-Hernández, M. Biswas, J. Ren, H. S. Suh, S. B. Darling, J. A. Liddle, J. W. Elam, J. J. de Pablo, N. J. Zaluzec and P. F. Nealey, ACS Nano, 2015, 9, 5333–5347 CrossRef CAS PubMed.
- J. Loos, E. Sourty, K. Lu, B. Freitag, D. Tang and D. Wall, Nano Lett., 2009, 9, 1704–1708 CrossRef CAS PubMed.
- H. J. Vogel, Microb. Genet. Bull., 1956, 13, 42–43 Search PubMed.
- P. L. Urban, Analyst, 2015, 140, 963–975 RSC.
- K.-T. Hsieh and P. L. Urban, RSC Adv., 2014, 4, 31094–31100 RSC.
- D. A. Skoog, F. J. Holler and S. R. Crouch, Principles of Instrumental Analysis, Brooks/Cole, Belmont, 6th edn, 2007 Search PubMed.
- A. Abbaspour, M. A. Mehrgardi, A. Noori, M. A. Kamyabi, A. Khalafi-Nezhad and M. N. S. Rad, Sens. Actuators, B, 2006, 113, 857–865 CrossRef CAS.
- A. Baghel and S. Amlathe, J. Chem. Pharm. Res., 2012, 4, 1546–1552 CAS.
- T. Schwaebel, O. Trapp and U. H. F. Bunz, Chem. Sci., 2013, 4, 273–281 RSC.
- Y. Nievergelt, SIAM Rev., 1986, 28, 79–84 CrossRef.
- Wolfram Mathematica, http://mathworld.wolfram.com/ RadonTransform.html (viewed on 09/10/2015).
- Agilent Technologies, http://www.agilent.com/en-us/products/liquid-chromatography/lc-detectors/1260-infinity-diode-array-detector (viewed on 01/09/2015).
- J. Wu and J. Pawliszyn, Anal. Chem., 1992, 64, 224–227 CrossRef CAS.
- S.-H. Lin, T. Yu, A. Sheu, D.-J. Yang and S.-C. Pai, J. Chromatogr., A, 2008, 1201, 129–131 CrossRef PubMed.
- P. L. Urban, D. M. Goodall, E. T. Bergström and N. C. Bruce, J. Biotechnol., 2006, 126, 508–518 CrossRef CAS PubMed.
- S. S. Jensen, H. Jensen, C. Cornett, E. H. Møller and J. Østergaard, Eur. J. Pharm. Sci., 2015, 69, 26–36 CrossRef CAS PubMed.
- V. K. Vanag and I. R. Epstein, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14635–14638 CrossRef CAS PubMed.
- C. Almarcha, P. M. J. Trevelyan, P. Grosfils and A. D. Wit, Phys. Rev. Lett., 2010, 104, 044501 CrossRef CAS PubMed.
- P.-H. Li, H. Ting, Y.-C. Chen and P. L. Urban, RSC Adv., 2012, 2, 12431–12437 RSC.
- W. H. Press, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19249–19254 CrossRef CAS PubMed.
- J. Cenens and R. A. Schoonheydt, Clays Clay Miner., 1988, 36, 214–224 CAS.
- P. A. Sykes, H.-C. Shiue, J. R. Walker and R. C. Bateman Jr., J. Chem. Educ., 1999, 76, 1283–1284 CrossRef CAS.
- T. Bánsági Jr., V. K. Vanag and I. R. Epstein, Science, 2011, 331, 1309–1312 CrossRef PubMed.
- S.-H. Chiu and P. L. Urban, Analyst, 2015, 140, 5145–5151 RSC.
- D. N. Mortensen and E. R. Williams, Anal. Chem., 2014, 86, 9315–9321 CrossRef CAS PubMed.
- Taxonomic classification: an overview, http://www.fgsc.net/neurospora/sectionb1.htm (viewed on 01/09/2015).
- G. E. Bartley, T. J. Schmidhauser, C. Yanofsky and P. A. Scolnik, J. Biol. Chem., 1990, 265, 16020–16024 CAS.
- G. A. Cooksey and J. Atencia, Lab Chip, 2014, 14, 1665–1668 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Text file (including Fig. S1–S9), and a video file (Movie S1). See DOI: 10.1039/c5an01807b |
| This journal is © The Royal Society of Chemistry 2016 |
