Open Access Article
Open Access ArticleBioluminescence readout lateral flow immunoassay using nanobody targeting aflatoxin B1†
Shun
Takahashi
,
Yuki
Hiruta
 and
Daniel
Citterio
and
Daniel
Citterio
 *
*
Department of Applied Chemistry, Faculty of Science and Technology, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan. E-mail: citterio@applc.keio.ac.jp; Tel: +81 45 566 1568
First published on 10th March 2025
Abstract
Multiple signal detection methods are known for lateral flow immunoassays (LFIAs), with colorimetric approaches dominating the field. However, their limited sensitivity is a remaining challenge. Fluorescence-based signaling is regarded as a more sensitive method, but it comes at the cost of partial sacrifice of the user-friendliness of LFIAs due to the requirement of an excitation light source. In this context, bioluminescence providing an inherently high signal to noise ratio without the need of excitation light could be an attractive alternative. But only a few studies have demonstrated the application of bioluminescence signaling in LFIAs. This work aimed at the development of a simple bioluminescence-based LFIA for the detection of aflatoxin B1 (AFB1), used as a model target in a competitive LFIA format. Signal transduction was achieved by nanobody-nanoluciferase (Nluc) fusion proteins. These small-sized recombinant heavy-chain-only antibodies derived from the camelidae family directly linked with the Nluc enzyme produce high intensity glow-type bioluminescence in combination with the furimazine substrate. LFIA devices consisting of a sample pad, nitrocellulose membrane and absorbent pad with AFB1-BSA conjugate deposited at the test line on the nitrocellulose membrane, achieved an LOD of 0.26 ng mL−1 for aqueous AFB1 solutions pre-mixed with Nanobody-Nluc and bioluminescence emission observed on an imaging system. More user-friendly LFIA devices with integrated conjugate pad and pre-deposited Nanobody-Nluc provided clear AFB1 concentration-dependent bioluminescence signals with low background and enabled readout with a standard digital camera, resulting in an LOD of 1.12 ng mL−1. Finally, the LFIA strips have been applied in AFB1-spiked oat milk samples. The LOD of 4.09 ng mL−1 achieved in the real sample matrix is well below the maximum allowable residual concentration of AFB1 in the U.S. (20 ng mL−1).
Introduction
Because of various advantages such as low cost, short assay time, and simplicity requiring no professional equipment or skills, lateral flow immunoassays (LFIAs) are now routinely used analytical devices for clinical diagnostics, environmental analysis, drug abuse testing and food safety monitoring, among others.1,2 Gold nanoparticles (AuNPs) represent the most commonly used colorimetric labels for detection antibodies in LFIAs, because their high extinction coefficients resulting in intense red color enable not only quantitative analysis when used in combination with ordinary optical devices like digital cameras or smartphones, but also direct detection by the naked eye, in addition to ease of synthesis and surface modification.3–5 However, the detection sensitivity of conventional AuNP-based LFIAs is insufficient for the measurement of low-concentration samples. To achieve higher sensitivity, various techniques including the use of surface-enhanced Raman scattering (SERS) nanoparticles, magnetic nanoparticles, electrochemical methods, silver enhancement, chemiluminescence, fluorescence and other detection methods have been reported.6–8 Because of their high signal-to-noise ratio, luminescence-based LFIAs using fluorescent materials such as fluorescent organic dyes, nanoparticles and quantum dots (QDs) are regarded as promising methodologies to enhance sensitivity.9,10 However, these fluorescence techniques require excitation light sources to obtain signals, which comes at the cost of partial sacrifice of the user-friendliness of LFIAs. On the other hand, bioluminescence, which is generated by the reaction between bioluminescent enzymes and their respective substrate, has attracted attention. Since bioluminescence is generated from a dark state by an enzymatic reaction, it achieves higher signal-to-noise ratio compared to fluorescence and does not require any special external excitation light source. In addition, the enzymatic reaction cycle leads to signal amplification further improving the detection sensitivity.11 However, despite these promising properties for sensitivity enhancement, the application of bioluminescence as a signaling method to LFIAs has been limited by the low stability and large molecular weight of bioluminescent enzymes that cause challenges for detection antibody modification and reduce the mobility of antibody-enzyme conjugates on paper substrates.12 For these reasons, there are only relatively few reports on bioluminescence-based LFIAs.12Recently, the artificial luciferase Nanoluc (Nluc) based on the Oluc luciferase isolated from the deep-sea shrimp (Oplophorus gracilirostris) has been developed.13 Compared to the commonly used sea pansy Renilla reniformis luciferase (Rluc) and the firefly luciferase (Fluc), Nluc is physically more stable and more resistant to heat and pH. Our group has for example previously shown that the Nluc/FMZ pair maintains its activity when stored on paper substrates packed under inert gas atmosphere at −20 °C for two months.14 Importantly, Nluc is only of 19 kDa size, making it significantly smaller than Rluc (36 kDa) and Fluc (61 kDa). At the same time, furimazine (FMZ), an optimally designed luminescent substrate for Nluc, was also developed. The Nluc-FMZ combination emits high-intensity glow-type bioluminescence that is about 150 times stronger than that of the Fluc system, in addition to not requiring any cofactors such as ATP and Mg2+.13 Furthermore, the small molecular weight of Nluc and its monomeric nature enable it to be easily employed as a transcriptional reporter or fusion partner. These advantages are promising for fusion to antibodies and other binding proteins/peptides in immunoassay development.15,16
Since their first reporting in 1993, nanobodies, which are a new type of recombinant antibodies derived from camelids and related species, have attracted much attention.17 Camelids produce unique antibodies without light chains, called heavy-chain antibodies.18 Heavy chain antibodies recognize antigens at the variable domain (VHH) of the heavy chain. These VHHs can be isolated and expressed to produce single-domain heavy-chain antibodies, which are called nanobodies. These recombinant antibodies retain the antigen-binding capacity of conventional antibodies, but are much smaller in size, averaging about 15 kDa. Expression of nanobodies is much easier than conventional recombinant antibodies. Nanobodies are also more water soluble and stable against heat and organic solvents compared to conventional antibodies and their recombination properties allow them to be expressed as fusion proteins with reporter proteins such as alkaline-phosphatase, green fluorescent protein (GFP) and nanoluciferase.16,19–22 These advantageous properties make nanobodies promising reagents in next-generation immunoassays, and they have been widely applied not only in the therapeutic field but also in the diagnosis of infectious diseases and detection of toxic substances.23–27 In previous studies, a newly synthesized fusion protein in which a nanobody and Nanoluc were directly linked was used as a detection probe in microplate-based enzyme-linked immunosorbent assays (ELISA).16,28 The fusion protein was easily expressed while maintaining its small size, the binding ability of the nanobody, and the enzymatic activity of Nanoluc. These properties motivated us to develop a bioluminescence-based LFIA based on a Nanobody-Nluc fusion protein. The mycotoxin Aflatoxin B1 was chosen as an analytical target to confirm the feasibility of this concept. Aflatoxins are highly poisonous compounds produced as a secondary metabolite of Aspergillus fungal species mainly found in hot and humid environments and are known to cause hepatocellular carcinoma.29 Among the more than 20 types of existing aflatoxins, six species are known as problematic when present in foods (aflatoxin B1, B2, G1, G2, M1, and M2). Aflatoxin B1 (AFB1) is classified by the International Agency for Research on Cancer (IARC) as a class 1 carcinogen, because of its highest toxicity in nature.30 In addition, since AFB1 is very stable and does not denature during cooking or food processing, its existence must be tested not only in raw products such as grains but also in bakery foods and daily consumables. Accordingly, maximum allowable limits for different foods have been set up by regulatory authorities in various countries, such as for example 20 ng mL−1 in case of the US Food and Drug Administration (FDA).31 Therefore, the accurate quantification of AFB1 is necessary to meet food safety requirements.
In this study, a bioluminescence-based LFIA (BL-LFIA) for the detection of AFB1 using a Nanobody-Nluc fusion protein was developed, and its practical applicability demonstrated in an oat milk matrix. The small molecular size and high water solubility of the fusion protein enabled the highly sensitive detection of AFB1 with simple LFIA device manipulation. For comparison purposes and to demonstrate the potential of bioluminescence in the enhancement of sensitivity, a fluorescence-based LFIA using a Nanobody-GFP fusion protein was also fabricated. Due to the bright bioluminescence emission, the developed LFIA device does not require any special external equipment for signal readout, but data for quantitative analysis can be collected with a standard digital camera.
Materials and methods
Materials
Aflatoxin B1 (AFB1), AFB1-BSA conjugate, zearalenone (ZEN) and ochratoxin A (OTA) were purchased from Sigma-Aldrich (St Louis, MO, USA). The AFB-1 targeting Nanobody-Nluc and Nanobody-GFP fusion proteins prepared as previously described were obtained from the Department of Entomology and Nematology and UCD Comprehensive Cancer Center, University of California Davis (Davis, CA, USA). Bovine serum albumin (BSA), Tween-20 and ethanol were purchased from FUJIFILM Wako Pure Chemical Industries (Osaka, Japan). Methanol and phosphate buffered saline (PBS) were purchased from Nacalai Tesque (Kyoto, Japan). The Nano-Glo® Luciferase Assay System (N1110) was acquired from Promega Corporation (Tokyo, Japan). For the preparation of all solutions, ultrapure water (18.2 MΩ cm) from a PURELAB flex water purification system (ELGA, Veolia Water, Marlow, U.K.) was used.Nitrocellulose membrane (NC) cards (HF90MC100) and glass fiber membranes (GFDX203000) were purchased from Millipore (Billerica, MA, USA). A different type of nitrocellulose membrane (CN140) was acquired from Sartorius (Göttingen, Germany). Backing cards used for assembly of LFIAs (GL-56338) were purchased from Lohmann (Neuwied, Germany). Cellulose fiber membrane (CF7) used for absorbent pads was purchased from GE healthcare (Pittsburgh, PA, USA).
Instruments and software
Materials used for LFIA device fabrication (sample pads, conjugate pads, nitrocellulose membranes and absorbent pads) were cut using a Silhouette CAMEO 3 (Lindon, UT, USA) cutting device and a compact strip cutter SCM-100DX (Fuji Shoko Machinery, Saitama, Japan). An office inkjet printer Canon PIXUS TS203 (Canon, Tokyo, Japan) was slightly modified and used to deposit the AFB1-BSA conjugate on the test line. Details regarding printer modification are given in Fig. S1 of the ESI.†The fluorescence of AFB1 measured on a ChemiDoc Touch MP imaging system (Bio-Rad, Hercules, CA, USA) with 10 s exposure time in oriole gel mode (302 nm excitation, 590 ± 55 nm emission) was used to monitor the inkjet deposition of the AFB1-BSA conjugate. The same instrument was also applied to acquire the bioluminescence or fluorescence signal on test strips either with 1.0 s of exposure time in chemiluminescent blot mode (647 nm shortpass filter) or with 0.2 s of exposure time in fluorescein blot mode (460–490 nm excitation, 532 ± 14 nm emission). In addition, a LUMIX GF9 compact digital camera (Panasonic, Osaka, Japan) was also used to acquire the bioluminescence signal on test strips. Both the Image Lab 6.1 software (Bio-Rad, Hercules, CA, USA) and ImageJ (NIH, Bethesda, MD, USA) were used to quantify the measured signals. Bioluminescence emitted from test lines was corrected by subtracting the background signal intensity adjacent to both sides of the test line (Fig. S2†). Experimental data was fitted to Hill's equation
| y = [(A − B)/(1 − (x/C)D)] + B | (1) |
Fabrication of bioluminescence-based LFIAs and assay procedures
Two types of LFIA test strips consisting of a glass fiber sample pad, a nitrocellulose membrane, and a cellulose fiber absorbent pad manually assembled onto backing cards, either with or without a glass fiber conjugate pad, were used in this work. The dimensions and arrangement of all components are shown in Fig. S3.† For the test line, 0.01 w/v% AFB1-BSA conjugate solution in PBS (pH 7.4) was printed multiple times on the nitrocellulose membrane using the inkjet printer. For the printing process, the nitrocellulose membrane (25 mm × 300 mm) attached to the backing card was temporarily fixed to an A4-sized sheet of paper using a double-sided adhesive. The number of printing cycles varied between experiments. During optimization experiments using devices without conjugate pad, the number of printing cycles of AFB1-BSA conjugate was 10, 15, 20 or 30 cycles. For devices used in final proof-of-concept experiments, 20 printing cycles were applied. A waiting time of 2 minutes was applied between printing cycles, during which the printed area was exposed to a flow of cold air from a hair dryer. After completion of test line printing, nitrocellulose membranes were dried in an oven at 37 °C for 2 hours, before cutting into strips of 5 mm width using the compact strip cutter. Glass fiber sample pad (20 mm × 300 mm by CAMEO cutting device) and conjugate pad (8 mm × 300 mm by CAMEO cutting device), as well as cellulose fiber absorbent pads (20 mm wide strips) were cut into 5 mm width using the compact strip cutter. Finally, all parts were manually assembled with overlaps as indicated in Fig. S3.†All experiments with LFIA devices were performed by placing the bare test strips (no plastic casing) into the well of a 96-well microtiter plate. To guarantee sufficient contact between the nitrocellulose membrane and the absorbent pad, a 3D-printed plastic sleeve was used as shown in Fig. S4.†
For optimization experiments using test strips without a conjugate pad, 50 μL of Nanobody-Nluc of various concentrations in PBS (pH 7.4) containing 0.2% Tween-20 and 2 mg mL−1 BSA (referred to as running buffer) and 50 μL of AFB1 solution of various concentrations in PBS containing 20% methanol were added to a 96-well microtiter plate and incubated at room temperature for 5 minutes. The LFIA devices were then inserted into the wells and the liquid allowed to wick for 15 minutes. The devices were then inserted into another well with 100 μL of washing buffer (PBS containing 0.1% Tween-20 and 1 mg mL−1 BSA) for 20 minutes. After removal of devices from the microtiter plate, 20 μL of furimazine solution (360 μM in Nano-Glo® assay buffer) was dropped directly onto the nitrocellulose membrane. The generated bioluminescence was captured by the ChemiDoc system, and the background-corrected signal value was calculated by the Image Lab software.
The LFIA test strips used in application proof-of-concept experiments consisted of a sample pad, a conjugate pad, a nitrocellulose membrane (CN140), and an absorbent pad. Conjugate pads (8 mm × 5 mm) were pretreated with 25 μL of 0.1% BSA aqueous solution and dried at 37 °C for 2 h. Then, 25 μL of 10 μg mL−1 Nanobody-Nluc solution (PBS containing 1 w/v% sucrose, 0.1 w/v% BSA) was applied on the pretreated conjugate pad and dried at 37 °C for 2 h, following the assembly of devices. 50 μL per well of AFB1 solution and an equal volume of PBS containing 0.2% methanol were added to a 96-well microtiter plate, and the test strips were placed for 15 min. After 20 min of washing as described above, devices were removed and 20 μL of furimazine solution was added directly to the nitrocellulose membrane. The bioluminescence was captured 30 seconds after furimazine application with a digital camera (camera settings: ISO 1600, F 2.8, exposure time 15 s) in the dark condition. The background-corrected test line value was calculated by the Image J software.
For selectivity tests, solutions of 100 ng mL−1 AFB1, ZEN or OTA (in PBS containing 20% methanol) were prepared and analyzed accordingly with the developed bioluminescence-based LFIAs (BL-LFIAs).
For the analysis of AFB1 in a real-world sample matrix, oat milk purchased from a local supermarket was used after filtration through a syringe filter (0.45 μm; Millex®HA, Millipore) to remove solid matter. A series of concentrations of AFB1 (0, 0.01, 0.1, 0.5, 1, 2.5, 5, 10, 25, 50, 100, 1000 ng mL−1) were spiked into this oat milk. Then, 25 μL of the AFB1-spiked samples were diluted with 25 μL of 2× PBS containing 20% methanol, and the analysis performed according to the procedure explained above.
Fluorescence-based LFIA
The experimental procedure for the fluorescence readout-based LFIA using the Nanobody-GFP fusion protein was identical to the above-mentioned, using test strips based on CN140 nitrocellulose membrane without conjugate pad with 20 printing cycles of AFB1-BSA conjugate. 50 μL of a 5.4 μg mL−1 solution of Nanobody-GFP in running buffer was used instead of Nanobody-Nluc, with all other experimental conditions being identical to the bioluminescence-based assay. The fluorescence signal was captured by the ChemiDoc system, and the background-corrected signal value was calculated by the Image Lab software.Results and discussion
Assay principle
Due to the small molecular weight nature of the targeted analyte AFB1, a competitive LFIA format has been adapted, wherein the Nanobody-Nluc fusion protein has been applied as the detection entity to provide selectivity for this mycotoxin, while at the same time acting as a bioluminescence signal transducer. During capillary force driven sample flow along the LFIA strip, AFB1 present in the sample liquid and the AFB1-BSA capture reagent deposited on the test line compete for the binding sites of the nanobody, resulting in an AFB1 target concentration-dependent immunocomplex formation at the test line (Fig. S5†). Highest levels of Nanobody-Nluc will be bound at the test line in the absence of analyte, leading to the signal turn-off behavior in presence of analyte well known for competitive immunoassays.Selection of nitrocellulose membrane
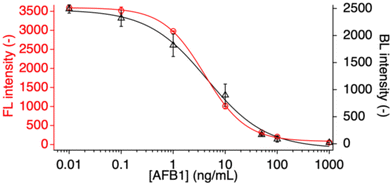
The nitrocellulose membrane is an essential component in LFIAs that directly affects the analytical sensitivity and reproducibility. In general, membranes with slower liquid flow speed result in higher assay sensitivity, due to the longer contact time between the analyte and the capture reagent immobilized on the test line. However, slower flow speeds are generally also a cause for increased non-specific adsorption of proteins including labeled signaling antibodies to the membrane, resulting in enhanced background signals. Moreover, loss of analyte through adsorption without binding at the test line becomes also problematic. Therefore, two types of nitrocellulose were evaluated using devices without a conjugate pad as shown in Fig. S3a.† At first, the influence of the flow speed on the limit of detection (LOD) was investigated. In this study, the IC90, the target analyte concentration at which the signal intensity corresponds to 90% of its maximum value, was used as the limit of detection (LOD).22 In this work, the value was extracted from the corresponding fitting curve according to the Hill equation (eqn (1)) using the IgorPro software. In the same series of experiments, the influence of the timing of bioluminescence signal readout was also evaluated. For this purpose, the time between application of the furimazine substrate and the recording of bioluminescence was varied. As expected, and shown in Fig. 1, devices fabricated with the slower flow speed nitrocellulose membrane (CN140) resulted in a significantly higher dynamic signal range compared to the membrane with faster flow (HF90), indicating that the absolute amount of Nluc-labeled detection nanobody trapped at the test line is higher than when using the membrane with faster liquid flow. No background signal was observed with either nitrocellulose membrane, indicating efficient flow of Nanobody-Nluc through the membrane without non-specific adsorption (data not shown). With increasing time intervals between furimazine substrate application and bioluminescence recording, overall signal intensities are decreasing. The overall lowest LOD as well as the best reproducibility indicated by the smallest mean relative standard deviation over the investigated concentration range of 0.01–1000 ng mL−1 AFB1 were obtained with the device based on the CN140 nitrocellulose with the bioluminescence emission measurements performed immediately after application of furimazine (Table S1†). Thus, this experimental condition was selected for all further optimization experiments.Fabrication of test line by inkjet printing
In the case of lateral flow immunoassays, the ratio of the concentration of capture reagent immobilized at the test line and the labeled detection reagent added to the sample or pre-deposited on the conjugate pad has a significant impact on the overall sensitivity of the analysis. First, the amount of AFB1-BSA conjugate used as capture reagent on the test line was examined. In this study, test lines were fabricated using an inkjet printer, and the amount of reagent on the test line can be easily adjusted by changing the number of printing cycles during the deposition process. As previously experienced in our group, the maximum protein concentration reproducibly and long-term printable by an office type inkjet printer was found to be around 0.01 w/v%, due to viscosity limitations at higher protein levels in the ink. Thus, in the current study, AFB1-BSA conjugate solutions at that concentration were printed 10, 15, 20, and 30 times to evaluate the suitable volume of capture reagent. The observed intensity of the intrinsic blue fluorescence of AFB1 on the test line increased with the number of printing cycles (Fig. S6a†), indicating that the reagent could be successfully printed up to 30 times. Accordingly, the bioluminescence emission intensity also increased with the amount of deposited AFB1-BSA conjugate when a blank sample solution was added to the printed device (Fig. S6b†). It is expected that the bioluminescence signal intensity will continue increasing with more printing cycles until the protein adsorption capacity of the nitrocellulose membrane is exceeded. However, passing the brittle nitrocellulose repeatedly through the printer increases the risk of physical damage to the membrane. In addition, higher numbers of repetitions enhance the possibility of misalignment between prints. In fact, as observed in Fig. S6b,† the size of the error bar for the bioluminescence signal increased significantly with 30 printing cycles. Therefore, 20 prints were selected as the most appropriate for the deposition of the test line reagent. This corresponds to approximately 0.1 μg of AFB1-BSA conjugate, as estimated from a calibration of inkjet-deposited volumes in the printing process.32 Finally, the suitable concentration of detection reagent was examined. AFB1 response curves were recorded using running buffer containing Nanobody-Nluc at 1.0, 2.5, 5.0, and 10 μg mL−1, which corresponds to molar amounts of approximately 1.0, 2.5, 5.0, and 10 times the amount of AFB1-BSA conjugate at the test line (Fig. S7†). The lowest LOD was obtained when the Nanobody-Nluc concentration was set to 5.0 μg mL−1 (Table S2†) and this condition was used for further experiments.Comparison to fluorescence-based LFIA
The performance of the developed bioluminescence-based LFIA was compared with a fluorescence signaling version of the same assay. For this purpose, the AFB1-targeting Nanobody-Nluc was replaced with Nanobody-GFP, a green fluorescent protein (GFP)-fused nanobody targeting the same toxin.22 Since the AFB1-targeting nanobodies are identical, both Nanobody-GFP and Nanobody-Nluc share the same affinity for AFB1 and other properties. Hence, the fluorescence-based LFIA using Nanobody-GFP was performed under the same experimental conditions as the bioluminescence-based LFIA (BL-LFIA) without further optimization. Calculated from the concentration response curves shown in Fig. 2, the LOD (IC90) was found to be 0.57 ng mL−1, which is of a similar order to the BL-LFIA (0.26 ng mL−1). While the signal of fluorescence-based LFIAs can be influenced by background fluorescence and excitation light reflection from the nitrocellulose membrane, the signal variability for the BL-LFIA is caused by factors originating from the enzymatic reaction, such as timing, mixing and diffusion of substrate applied to the LFIA strips. In general, bioluminescence, which is an enzymatic reaction, is considered to have a higher signal-to-noise ratio than fluorescence, which requires excitation light. Overall, the analytical performance of both LFIAs compared in the current study was similar, but the BL-LFIA features the advantage of not requiring an excitation light source.Digital camera-based signal detection
One advantage of bioluminescence other than its high sensitivity is that no special external equipment is required for signal detection. While a dedicated imaging system has been applied for the basic characterization of LFIA devices, subsequent bioluminescence signal detection was achieved with a digital camera and signal intensities were extracted using the Image J software. For a simpler and more user-friendly assay procedure, conjugate pads were introduced into the devices (Fig. S3(b)†), eliminating the requirement to add the Nanobody-Nluc to the sample solution. The detection limit for AFB1 of this device was estimated as 1.12 ng mL−1 (Fig. 3a), which is about four times higher than that of the device without the conjugate pad as mentioned above. Although the sensitivity was reduced by limited camera performance (cooled CCD detector vs. consumer digital camera) and reagent adsorption on the conjugate pad, the LOD is still well below the AFB1 concentration required to be detected (e.g. the maximum allowable residual concentration in the U.S. is 20 ng mL−1) despite the simpler system.Assay specificity
To test the specificity of the developed BL-LFIA, cross-reactivity to zearalenone (ZEN) and ochratoxin A (OTA), mycotoxins often found in real samples, was evaluated. As expected, significant decrease of bioluminescence signals was only observed in the presence of AFB1, while the signal intensities were similar to that of the blank when other mycotoxins were applied (Fig. 3b), confirming the high specificity of the device.LFIA application in oat milk sample matrix
Sample matrix effects are an important factor to be investigated in LFIAs, because proteins, ions, and various other components in the sample matrix often affect the sample liquid flow, the antigen–antibody interactions and the bioluminescence reaction. In this study, the response behavior of the LFIA to AFB1 spiked into filtered and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 diluted oat milk was investigated. Oat milk has become a popular daily food that can be contaminated with AFB1, and there is a demand for mycotoxin monitoring during beverage production.33 The IC50 and LOD values determined for the BL-LFIA in this sample matrix were 35.40 ng mL−1 and 4.09 ng mL−1, respectively (Fig. 4), demonstrating the practical applicability of the device. Note that the sensitivity in the matrix was corrected by a factor of two to account for the dilution of the sample with buffer.
1 diluted oat milk was investigated. Oat milk has become a popular daily food that can be contaminated with AFB1, and there is a demand for mycotoxin monitoring during beverage production.33 The IC50 and LOD values determined for the BL-LFIA in this sample matrix were 35.40 ng mL−1 and 4.09 ng mL−1, respectively (Fig. 4), demonstrating the practical applicability of the device. Note that the sensitivity in the matrix was corrected by a factor of two to account for the dilution of the sample with buffer.
Conclusions
In this work, a low molecular weight nanobody-nanoluciferase fusion protein targeting AFB1 has been applied to a lateral flow immunoassay with bioluminescence signal readout using a standard digital camera. To the best of our knowledge, this represents the first report of integrating a nanobody with bioluminescence signal transduction into an LFIA device. It has been found that the Nanobody-Nluc protein pre-deposited on glass fiber conjugate pads reliably resolubilizes into the sample solution wicking through the test strip, resulting in a clear bioluminescence signal at the test line with low background in surrounding areas. The assay time between immersion of the BL-LFIA test strip into the sample solution and the recording of the bioluminescence emission is 35 minutes, requiring only a single washing step and the application of the bioluminescent substrate directly to the strip. The limit of detection of 4 ng mL−1 achieved in a filtered and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 diluted oat milk matrix without any further sample processing indicates the potential applicability of the BL-LFIA for practical on-site monitoring of the mycotoxin. While the bioluminescence readout-based LFIA reported in this work might not necessarily outperform the fluorescence readout-based counterpart, there might be situations, where the absence of an excitation light source and stray light reflected from the LFIA strip encountered in the fluorescence-based approach might be advantageous, whereas in other cases, the requirement of an excitation light source might outweigh the added complexity of the bioluminescence-based approach requiring a washing and substrate addition step. However, this work has demonstrated that a bioluminescence-based LFIA relying on a nanobody-luciferase fusion protein is a viable alternative, expanding the toolbox of simple analytical assays performable outside of sophisticated laboratory environments.
1 diluted oat milk matrix without any further sample processing indicates the potential applicability of the BL-LFIA for practical on-site monitoring of the mycotoxin. While the bioluminescence readout-based LFIA reported in this work might not necessarily outperform the fluorescence readout-based counterpart, there might be situations, where the absence of an excitation light source and stray light reflected from the LFIA strip encountered in the fluorescence-based approach might be advantageous, whereas in other cases, the requirement of an excitation light source might outweigh the added complexity of the bioluminescence-based approach requiring a washing and substrate addition step. However, this work has demonstrated that a bioluminescence-based LFIA relying on a nanobody-luciferase fusion protein is a viable alternative, expanding the toolbox of simple analytical assays performable outside of sophisticated laboratory environments.
Author contributions
Shun Takahashi: conceptualization, methodology, investigation, data curation, formal analysis, writing – original draft. Yuki Hiruta: validation, writing – review & editing, supervision. Daniel Citterio: conceptualization, methodology, validation, writing – review & editing, supervision, project administration, funding acquisition.Data availability
Data created and analysed in this study are included in the article and the ESI file.† Raw data supporting the findings of this study are available from the corresponding author upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support through Keio University internal funding. They gratefully acknowledge Dr Natalia Vasylieva and Prof. Dr Bruce D. Hammock from the Department of Entomology and Nematology and UCD Comprehensive Cancer Center at the University of California Davis (USA) for the donation of the AFB-1 targeting Nanobody-Nluc and Nanobody-GFP fusion proteins.References
- C. Parolo, A. Sena-Torralba, J. F. Bergua, E. Calucho, C. Fuentes-Chust, L. Hu, L. Rivas, R. Álvarez-Diduk, E. P. Nguyen, S. Cinti, D. Quesada-González and A. Merkoçi, Nat. Protoc., 2020, 15, 3788–3816 CrossRef CAS PubMed.
- G. A. Posthuma-Trumpie, J. Korf and A. van Amerongen, Anal. Bioanal. Chem., 2009, 393, 569–582 CrossRef CAS PubMed.
- Y.-J. Xie, Y. Yang, W.-J. Kong, S.-H. Yang and M.-H. Yang, Chin. J. Anal. Chem., 2015, 43, 618–628 CAS.
- C. Ruppert, N. Phogat, S. Laufer, M. Kohl and H.-P. Deigner, Microchim. Acta, 2019, 186, 119 Search PubMed.
- N. Jiang, R. Ahmed, M. Damayantharan, B. Unal, H. Butt and A. K. Yetisen, Adv. Healthc. Mater., 2019, 8, e1900244 CrossRef PubMed.
- P. Tripathi, N. Upadhyay and S. Nara, Crit. Rev. Food Sci. Nutr., 2018, 58, 1715–1734 CrossRef CAS PubMed.
- J. H. Soh, H.-M. Chan and J. Y. Ying, Nano Today, 2020, 30, 100831 CrossRef CAS.
- B. B. Dzantiev, N. A. Byzova, A. E. Urusov and A. V. Zherdev, TrAC, Trends Anal. Chem., 2014, 55, 81–93 Search PubMed.
- M. Xiao, Q. Fu, H. Shen, Y. Chen, W. Xiao, D. Yan, X. Tang, Z. Zhong and Y. Tang, Talanta, 2018, 178, 644–649 Search PubMed.
- Q. Fu, Y. Tang, C. Shi, X. Zhang, J. Xiang and X. Liu, Biosens. Bioelectron., 2013, 49, 399–402 Search PubMed.
- M. Mirasoli, A. Buragina, L. S. Dolci, P. Simoni, L. Anfossi, G. Giraudi and A. Roda, Biosens. Bioelectron., 2012, 32, 283–287 CrossRef CAS PubMed.
- Y. Ding, X. Hua, H. Chen, F. Liu, G. González-Sapien and M. Wang, Anal. Chem., 2018, 90, 2230–2237 CrossRef CAS PubMed.
- M. P. Hall, J. Unch, B. F. Binkowski, M. P. Valley, B. L. Butler, M. G. Wood, P. Otto, K. Zimmerman, G. Vidugiris, T. Machleidt, M. B. Robers, H. A. Benink, C. T. Eggers, M. R. Slater, P. L. Meisenheimer, D. H. Klaubert, F. Fan, L. P. Encell and K. V. Wood, ACS Chem. Biol., 2012, 7, 1848–1857 CrossRef CAS PubMed.
- K. Tenda, B. Van Gerven, R. Arts, Y. Hiruta, M. Merkx and D. Citterio, Angew. Chem., Int. Ed., 2018, 57, 15369–15373 CrossRef CAS PubMed.
- H. Oyama, Y. Kiguchi, I. Morita, T. Miyashita, A. Ichimura, H. Miyaoka, A. Izumi, S. Terasawa, N. Osumi, H. Tanaka, T. Niwa and N. Kobayashi, Anal. Chim. Acta, 2021, 1161, 238180 CrossRef CAS PubMed.
- F. Wang, Z. F. Li, Y. Y. Yang, D. B. Wan, N. Vasylieva, Y. Q. Zhang, J. Cai, H. Wang, Y. D. Shen, Z. L. Xu and B. D. Hammock, Anal. Chem., 2020, 92, 11935–11942 CrossRef CAS PubMed.
- S. Muyldermans, FEBS J., 2021, 288, 2084–2102 CrossRef CAS PubMed.
- C. Hamers-Casterman, T. Atarhouch, S. Muyldermans, G. Robinson, C. Hammers, E. B. Songa, N. Bendahman and R. Hammers, Nature, 1993, 363, 446–448 CrossRef CAS PubMed.
- J. Lin, J. Yu, H. Wang, Y. Xu, F. Li, X. Chen, Y. Liang, J. Tang, L. Wu, Z. Zhou, C. Chen, M. Liu, X. Chun, R. Nian and H. Song, Anal. Bioanal. Chem., 2020, 412, 1723–1728 Search PubMed.
- T. He, Y. Wang, P. Li, Q. Zhang, J. Lei, Z. Zhang, X. Ding, H. Zhou and W. Zhang, Anal. Chem., 2014, 86, 8873–8880 CrossRef CAS PubMed.
- S. Muyldermans, Annu. Rev. Biochem., 2013, 82, 775–797 CrossRef CAS PubMed.
- J. P. Salvador, N. Vasylieva, I. Gonzalez-Garcia, M. Jin, R. Caster, J. B. Siegel and B. D. Hammock, ACS Food Sci. Technol., 2022, 2, 1276–1282 CrossRef CAS.
- D. Li, Y. Cui, C. Morisseau, S. J. Gee, C. S. Bever, X. Liu, J. Wu, B. D. Hammock and Y. Ying, Anal. Chem., 2017, 89, 6248–6256 Search PubMed.
- C. S. Bever, J.-X. Dong, N. Vasylieva, B. Barnych, Y. Cui, Z.-L. Xu, B. D. Hammock and S. J. Gee, Anal. Bioanal. Chem., 2016, 408, 5985–6002 CrossRef CAS PubMed.
- S. Y. Doerflinger, J. Tabatabai, P. Schnitzler, C. Farah, S. Rameil, P. Sander, A. Koromyslova and G. S. Hansman, mSphere, 2016, 1, e00219-16 CrossRef PubMed.
- F. Zhao, Y. Tian, Q. Shen, R. Liu, R. Shi, H. Wang and Z. Yang, Talanta, 2019, 195, 55–61 CrossRef CAS PubMed.
- X. Tang, P. Li, Q. Zhang, Z. Zhang, W. Zhang and J. Jiang, Anal. Chem., 2017, 89, 11520–11528 CrossRef CAS PubMed.
- W. Ren, Z. Li, Y. Xu, D. Wan, B. Barnych, Y. Li, Z. Tu, Q. He, J. Fu and B. D. Hammock, J. Agric. Food Chem., 2019, 67, 5221–5229 CrossRef CAS PubMed.
- S. Marin, A. J. Ramos, G. Cano-Sancho and V. Sanchis, Food Chem. Toxicol., 2013, 60, 218–237 CrossRef CAS PubMed.
- B. R. Rushing and M. I. Selim, Food Chem. Toxicol., 2019, 124, 81–100 CrossRef CAS PubMed.
- Z. Xue, Y. Zhang, W. Yu, J. Zhang, J. Wang, F. Wan, Y. Kim, Y. Liu and X. Kou, Anal. Chim. Acta, 2019, 1069, 1–27 CrossRef CAS PubMed.
- K. Kuwahara, K. Yamada, K. Suzuki and D. Citterio, Analyst, 2018, 143, 1234–1241 RSC.
- E. Miró-Abella, P. Herrero, N. Canela, L. Arola, F. Borrull, R. Ras and N. Fontanals, Food Chem., 2017, 229, 366–372 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Tables showing IC50 values and LODs for various experimental conditions; inkjet printer preparation; bioluminescence signal and background collection method; dimensions and component arrangement for LFIA test strip fabrication; details about 3D-printed plastic sleeve; optimization of amounts of AFB1-BSA conjugate capture reagent; optimization of amounts of Nanobody-Nluc for assays on LFIA devices without conjugate pad. See DOI: https://doi.org/10.1039/d5an00030k |
| This journal is © The Royal Society of Chemistry 2025 |