New structural alerts for Ames mutagenicity discovered using emerging pattern mining techniques
Laurence
Coquin
a,
Steven J.
Canipa
a,
William C.
Drewe
a,
Lilia
Fisk
a,
Valerie J.
Gillet
b,
Mukesh
Patel
a,
Jeffrey
Plante
a,
Richard J.
Sherhod
ab and
Jonathan D.
Vessey
*a
aLhasa Limited, Granary Wharf House, 2 Canal Wharf, Holbeck, Leeds, LS11 5PS, UK. E-mail: jonathan.vessey@lhasalimited.org
bInformation School, University of Sheffield, Regent Court, 211 Portobello Street, Sheffield S1 4DP, UK
First published on 5th November 2014
Abstract
Emerging pattern mining techniques have been applied to datasets of Ames mutagens. The discovered patterns give rise to clusters of compounds from large and biased datasets which are used to develop new structural alerts for mutagenicity in the Derek Nexus expert system.
Introduction
Knowledge based expert systems can be used to predict the potential toxicity of novel chemicals and typically do so by means of identifying toxicophoric chemical substructures known as structural alerts.1–5 The creation of a structural alert is commonly done by human experts who can investigate literature and private source data for supporting mechanistic information. Knowledge base development can, therefore, be a very time consuming process so methods of aiding human experts to identify a structural alert are of interest.Emerging pattern (EP) mining is a data mining technique to distinguish combinations of binary descriptors that are more common in one class (such as toxic compounds) than in another (such as non-toxic compounds).6 EP mining techniques have been used to investigate a variety of biological targets and toxicity endpoints.7–9 In this paper we show how the techniques have been applied to investigate areas of chemical space containing mutagens identified by the Ames test and how they have been used to discover relevant clusters of compounds. These clusters were further investigated and subsequently used to develop new structural alerts in the knowledge base of the Derek Nexus expert system.3
Background
Previously, we have described how emerging pattern technology can be used to mine chemical data sets. The first study10 concentrated on jumping emerging patterns (JEPs) – that is, patterns of descriptors which occur exclusively in compounds of one class; in this case the class is Ames-positive compounds.JEPs are by their nature very intolerant of noisy data: in practice this results in the production of many overlapping JEPs describing similar chemical space. In contrast, the second study11 focussed on emerging patterns, which are patterns of descriptors which are more common in one class over another – for example more common in active rather than inactive compounds – and which are much more noise tolerant.
Both EPs and JEPs can be mined from binary fingerprints of compounds in a dataset. In this study the descriptors tried were binary fingerprints generated from the freely available RDKit tools12 and simple structural fragments generated in-house by a procedure described below.
It is important to distinguish the aims of this paper – new knowledge discovery – from other data mining attempts, particularly a QSAR approach to predictive model building. The motivation for this study was to expedite development of expert predictions by identifying clusters of compounds suitable for the attention of experienced scientists to enhance a knowledge base. It was expected that clusters of compounds which share easily interpretable features would become apparent in the analysis of Ames mutagenicity because that endpoint is relatively well understood in terms of molecular initiating events some of which can be attributed to electrophilic functional groups which themselves are relatively easy to describe.
The datasets used in this study were a curated version of the Hansen data set13 and a curated CFSAN dataset14 from which compounds also contained in the Hansen data set had been deleted.
Pre-computed sets of fragments – such as those available in commercial packages such as Dragon15 or Leadscope Enterprise16 – were found to be of limited value in this study because (a) too many closely related descriptors were available and, conversely, (b) some fragments known to be closely allied to Ames mutagenicity, for example N-nitro groups, were not contained in off-the-shelf descriptor sets. In that any information in a dataset relating structure to activity must come from within the dataset, it was decided to generate a fragment dictionary from the dataset itself and that the dictionary should consist of functional groups which could be related by human experts to mutagenicity on a mechanistic basis.
Results and discussion
Method
The reader is referred to Fig. 1 for the steps in the approach reported in this paper. | ||
| Fig. 1 Flowchart of the approach reported in this paper. Step numbers are explained in more detail in the text. | ||
Step 1: the structures in the datasets were curated as reported previously11 after which each molecule in the training set was represented in its fully hydrogen expressed format. Properties such as number of neighbouring atoms and whether or not the atom had aromatic bonds were added to each atom.
The EP mining process requires that data associated with the compounds in the training set are expressed as a binary fingerprint. Steps 2–6 detail two different fingerprinting methods used in this study.
Step 2: functional group fragments were generated from the curated structures by removing all the carbon–carbon single bonds, carbon–carbon aromatic bonds and the carbon–hydrogen bonds. Of the resulting fragments, those with more than one atom represent discrete functional groups within the molecule and were considered for inclusion in the emerging patterns analysis.
The atoms in the fragments retained their information about the number of neighbours and aromaticity so that groups such as N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O would not match N+(
O would not match N+(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O− or that aromatic cN(H)(H) would not match aliphatic CN(H)(H); the chemical moieties here are represented in SMILES format.17
O)O− or that aromatic cN(H)(H) would not match aliphatic CN(H)(H); the chemical moieties here are represented in SMILES format.17
No further filtering of the fragments, for example by fragment size or by finding subset–superset relationships, was found to be necessary. The method generated 1296 fragments from the Hansen dataset, 1288 of which had 20 atoms or fewer, the exceptions being fragments derived from polypeptide structures.
Step 3: ring fragments were generated by a similar method: this involved, for each molecule in the training set, removing all bonds other than those in rings, exo-double bonds and ring positions substituted by heteroatoms. Again fragments with more than one atom present were retained. This generated 2382 fragments from the Hansen dataset, 2312 of which had 30 atoms or fewer; again larger fragments were those derived from polypeptide structures.
Step 4: the combined fragments generated by both methods were represented as canonicalised SMARTS18 patterns which allowed duplicate fragments to be identified and eliminated. This produced a dictionary of 3678 different fragments from the Hansen dataset.
Step 5: the fragment dictionary was then matched against all of the molecules in the training set with the presence or absence of the fragment in the molecule being recorded producing the fingerprint for each compound based on the generated fragments. It would have been possible at this stage to remove any entries in the dictionary that fell below the threshold for occurrence in the EP mining step, but in practice this was not necessary.
Step 6: fingerprints from the RDKit KNIME19 node were also generated; these were only used for the JEP study.
Step 7: EPs were mined from the full Hansen dataset. The EP mining used the previously described method:11 the minimum threshold on support in both active compounds and inactive compounds was set at 1% and the curve frontier parameter to control noise was set at 1.3. Under these conditions the discovery of the EPs took ca. 10 minutes and was not, therefore significant in the time taken to develop the alerts. A total of 604 EPs were generated and organised into 181 hierarchies of structurally related support sets.
Step 8: it was anticipated that the EPs might represent the chemical signatures of structural alerts. Thus, when developing new structural alerts for a knowledge base prediction system, the clusters of interest will be those supported by the highest number of false negatives and lowest number of true negatives. In this study the false negatives (FNs) and true negatives (TNs) were classified as those compounds for which Derek Nexus did not contain an alert and were found experimentally to be active and inactive respectively. Of the 181 ‘root’ EPs in these hierarchies, the three that were supported by the highest number of false negatives and lowest number of true negatives were selected as the most promising candidates for new structural alerts.
Step 9: as an alternative to mining the full data set, JEPs of descriptors of FNs were obtained from a set of FNs and TNs. The method of JEP mining has been described previously.9 As the dataset of FNs and TNs was somewhat smaller than the full dataset, the time taken to discover the JEPs was also a matter of minutes. JEPs were mined from both the fragment fingerprints generated in Steps 2–5 and the RDKit fingerprints generated in Step 8.
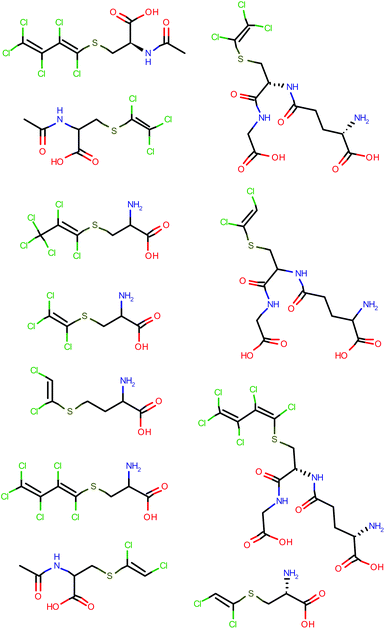
Step 10: where the support set of compounds for a JEP was large enough to merit further assessment (typically 4 or more compounds), they were analysed visually to identify SMARTS patterns which best summarised the support set. This was done without reference to the descriptors which made up the JEP as the supporting sets were typically small (10 compounds or fewer) whereas the SMARTS patterns covered more chemical space. For example, the 11 compounds in Fig. 2 form the support set for the JEP {CSCCl, C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)OH} discovered from the simple fragments fingerprint from the Hansen training set; the set was summarised with the SMARTS pattern ClC
O)OH} discovered from the simple fragments fingerprint from the Hansen training set; the set was summarised with the SMARTS pattern ClC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CS. The SMARTS patterns were matched against the training set to generate clusters which were candidates for further investigation.
CS. The SMARTS patterns were matched against the training set to generate clusters which were candidates for further investigation.
Step 11: each cluster sdf file was imported into an Excel sheet using JChem for Excel.20 The chemical name and CAS number were retrieved for each compound by browsing Chemspider,21 CHEBI,22 ChemID plus23 or other chemistry databases. Toxicological data were retrieved if possible for each active compound in the cluster which did not already activate an alert in Derek Nexus by browsing TOXNET,24 the NTP toxicity studies database25 or querying Vitic Nexus26 by CAS number. Wider searches involving querying TOXNET (through ChemID plus) and Vitic Nexus using substructure searches were also performed to ensure that all relevant or related compounds were found in the data searches. All data were checked against the source publications.
Step 12: finally, when possible, the mechanistic rationale of activity was investigated and assessed using literature found from the PubMed database.27
Study with EPs
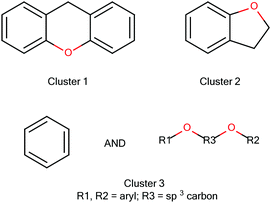
Owing to its inherent noise tolerance, EP mining is well suited to the initial exploratory analysis of previously unseen datasets. A relatively small number of EPs are typically generated, with relatively large support sets; therefore, a knowledge base scientist will be presented with a manageable number of clusters and a set of easily interpretable common features requiring less post-processing modification than needed for JEP analysis.The structural fragments comprising the three selected EPs are shown in Fig. 3. The EPs for Cluster 1 and Cluster 2 are single fragments, while the EP for Cluster 3 is composed of a benzene ring and a dimethoxy group between two aromatic carbon atoms.
Where an EP is defined by a single fragment, the technique effectively produces the same result as a common substructure analysis, however one of the advantages of the EP mining techniques is that the user does not assume this to be the case before performing the analysis and indeed Cluster 3 could not have been found from a common substructure analysis.
Study with JEPs
As shown in Table 1, the simple fragments produced many fewer JEPs than did the RDKit fingerprints, whereas the JEPs from the RDKit fingerprint were more supported.| Training set | Fingerprint | Number of minimal JEPs | Greatest support | Number of JEPs assessed further i.e. support ≥4 |
|---|---|---|---|---|
| Hansen | RDKit | 2485 | 13 | 195 |
| Hansen | Simple fragments | 308 | 11 | 31 |
| CFSAN | RDKit | 4444 | 23 | 209 |
| CFSAN | Simple fragments | 149 | 4 | 4 |
Each SMARTS pattern was evaluated against the training set from which the JEPs had been derived (internal validation) and one other dataset – either of Hansen or CFSAN (whichever had not been used to derive the JEPs; i.e. external validation). For example, the SMARTS pattern ClC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CS found 14 structures in the Hansen data set of which all were FNs and 1 in the CFSAN data set, which again was a FN.
CS found 14 structures in the Hansen data set of which all were FNs and 1 in the CFSAN data set, which again was a FN.
Distinct SMARTS patterns which produced clusters from the internal or external validation sets which were enriched in FNs relative to the validation dataset as a whole are shown in Table 2. In some cases substructures were suggested by more than one set of JEPs, e.g. from both RDKit and functional group fragments of compounds in the Hansen data set or from compounds in both Hansen and CFSAN datasets; in these cases the substructures are only recorded once.
| CFSAN (training) | Hansen (test) | |||||
|---|---|---|---|---|---|---|
| TN | FN | Ratio | TN | FN | Ratio | |
| All data | 1486 | 335 | 0.22 | 2216 | 787 | 0.36 |
| SMARTS summarising JEPs from RDKit fingerprints | ||||||
| c1@C(O)@C(O)@[#6]@[#6]c1 | 0 | 14 | ∞ | 17 | 41 | 2.4 |
| [#6]N([CH2][CH3])[CH2;R0][#6] | 21 | 27 | 1.29 | 24 | 21 | 0.88 |
[#6]C([#6])![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1C C1C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC( CC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) [N+]([#6])[#6])C [N+]([#6])[#6])C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1 C1 |
3 | 15 | 5 | 4 | 5 | 1.25 |
| SMARTS summarising JEPs from functional group fingerprints | ||||||
| c12ccccc1ccnc2 | 3 | 6 | 2 | 6 | 12 | 2 |
c12ccccc1COC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
10 | 6 | 0.6 | 7 | 2 | 0.28 |
| c1cc[o+]cc1 | 1 | 5 | 5 | 2 | 2 | 1 |
| CFSAN (test) | Hansen (training) | |||||
| SMARTS summarising JEPs from RDKit fingerprints | ||||||
c1c(c)c(c)cc(@C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)@[#6])c1 O)@[#6])c1 |
1 | 2 | 2 | 7 | 19 | 2.71 |
| SMARTS summarising JEPs from functional group fingerprints | ||||||
ClC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/i_char_e001.gif) CS CS
|
0 | 1 | ∞ | 0 | 14 | ∞ |
c1cccc2[#6](![[double bond, length as m-dash]](https://www.rsc.org/images/entities/i_char_e001.gif) O)c3ccccc3[#8,#16]c12 O)c3ccccc3[#8,#16]c12
|
0 | 2 | ∞ | 1 | 15 | 15 |
| c1cccc2cc3ccccc3nc12 | 1 | 4 | 4 | 2 | 11 | 5.5 |
| C1OOC1 | 0 | 0 | — | 3 | 14 | 4.5 |
C1OC1C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
1 | 8 | 8 | 7 | 13 | 1.86 |
c12ccccc1CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N2 N2 |
0 | 0 | — | 1 | 5 | 5 |
| c1ccnn1 | 2 | 0 | 0 | 7 | 9 | 1.28 |
| a12aaaaa1a3aaaaa3n2 | 7 | 8 | 1.12 | 8 | 30 | 3.75 |
NC([CH2;R0]S)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O O)O |
4 | 3 | 0.75 | 11 | 11 | 1 |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N[#7] N[#7] |
8 | 4 | 0.5 | 9 | 11 | 1.22 |
The FN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TN ratios in Table 2 show how the emerging pattern technique is more useful than others, such as a common substructure approach, in cases where there is significant bias in the training data: patterns of descriptors are generated and investigated automatically until either the signal contained in the support set becomes interesting to the user, or until it becomes clear that no further investigation of a combination of features will provide a pattern that fulfils the user's requirements. In the case of this investigation, clusters can be found and investigated where there is still a preponderance of TNs.
TN ratios in Table 2 show how the emerging pattern technique is more useful than others, such as a common substructure approach, in cases where there is significant bias in the training data: patterns of descriptors are generated and investigated automatically until either the signal contained in the support set becomes interesting to the user, or until it becomes clear that no further investigation of a combination of features will provide a pattern that fulfils the user's requirements. In the case of this investigation, clusters can be found and investigated where there is still a preponderance of TNs.
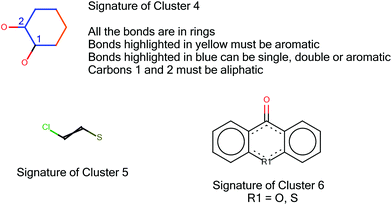
In Table 2 interesting clusters have been italicised and these were taken forward for investigation for new structural alerts; the clusters’ signatures are shown in Fig. 4. As the signature of Cluster 6 is similar to that of Cluster 1, Cluster 6 was not analysed further.
Investigation of new alerts from EPs and JEPs
Having identified several clusters of compounds with defined and easily recognisable commonality which showed a preponderance for activity, they were then assessed as new chemical classes for the development of structural alerts.Clusters from EP mining
Cluster 1: Cluster 1 contained 30 compounds with 13 FNs; Fig. 5 and Table 3 summarise the data found for 17 compounds of this class. Table 4 summarises the metrics of Cluster 1 as a whole. Although grouped together in a single cluster from the EP analysis, two different mechanisms of action of compounds in the cluster are observed and thus two new structural alerts could be made. Firstly, the mutagenic activity of the 1,3-dihydroxyxanthones is likely to involve a non-covalent DNA intercalation28 with a 1,3-dihydroxyxanthone metabolite as evidenced by several experiments including ethidium bromide displacement and changes to DNA viscosity and transition temperature.29 Secondly, the thioxanthones and analogues have been shown to undergo activation at the C4 position30 leading to a benzylic cation which is likely to be the active mutagen, which binds or intercalates and then alkylates DNA causing frameshift and other mutations.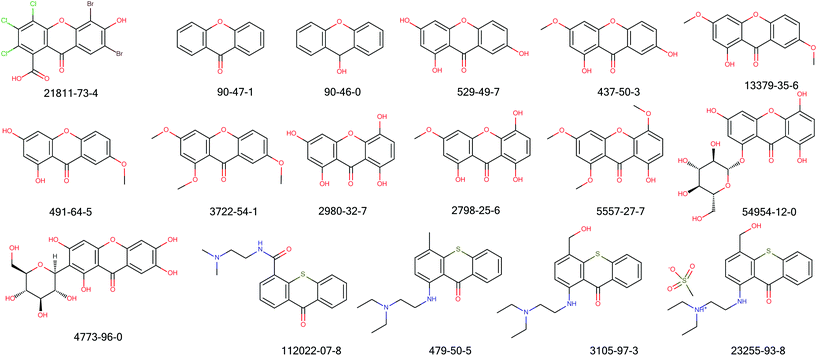 | ||
| Fig. 5 Structures of compounds whose toxicity data are reported in Table 3. | ||
| CAS number | Strains | Overall call | Ref. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TA100 | TA97 | TA98 | TA1537 | |||||||
| −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | −S9 | +S9 | |||
| a Activity seen versus control but not determined to be significantly strong enough to be a clear positive. b If β-glucosidase is present in the S9 activation medium, the compounds are positive. c Also negative in TA 1538. d Also positive in TA 1538. | ||||||||||
| 21811-73-4 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 31 | ||
| 90-47-1 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 32 | ||
| 90-46-0 | Neg | Neg | Pos | Pos | Pos | Pos | Pos | 32 | ||
| 529-49-7 | Neg | Pos | Neg | Pos | Neg | Neg | Pos | 32 | ||
| 437-50-3 | Neg | Pos | Neg | Pos | Neg | Neg | Pos | 32,33 | ||
| 13379-35-6 | Neg | Pos | Neg | Pos | Neg | Neg | Pos | 32 | ||
| 491-64-5 | Neg | Pos | Neg | Pos | Neg | Neg | Pos | 32,33 | ||
| 3722-54-1 | Neg | Pos | Neg | Pos | Neg | Neg | Pos | 32 | ||
| 2980-32-7 | Neg | Neg | Neg | Pos | Neg | Neg | Pos | 32 | ||
| 2798-25-6 | Neg | Neg | Pos | Pos | Neg | Neg | Pos | 32 | ||
| 5557-27-7 | Neg | Neg | Equa | Pos | Neg | Neg | Pos | 32 | ||
| 54954-12-0 | Neg | Negb | Neg | Negb | Neg | Neg | Neg | 32 | ||
| 4773-96-0 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 32 | ||
| 112022-07-8 | Pos | Pos | 34 | |||||||
| 479-50-5 | Negc | Posc | Pos | 35 | ||||||
| 3105-97-3 | Pos | Pos | Pos | 35 | ||||||
| 23255-93-8 | Posd | Posd | Pos | 35 | ||||||
| Experimental toxicity | Predicted toxicity | Total | |
|---|---|---|---|
| + | − | ||
| + | 10 | 13 | 23 |
| − | 6 | 1 | 7 |
| Total | 16 | 14 | 30 |
Cluster 2: Cluster 2 contained 62 compounds with 19 FNs. Table 5 summarises the metrics of Cluster 2. Most of the FNs are benzofuran dioxetane derivatives with a core shown in Fig. 6.
| Experimental toxicity | Predicted toxicity | Total | |
|---|---|---|---|
| + | − | ||
| + | 25 | 19 | 44 |
| − | 8 | 10 | 18 |
| Total | 33 | 29 | 62 |
Fig. 7 and Table 6 summarise the data found for a series of benzofuran dioxetane compounds. Mechanistically, this class of compounds is thought to interact with DNA via alkylating properties, the ultimate mutagen is proposed to be the epoxide formed by deoxygenation.36 A new structural alert for mutagenicity of aryl fused furan 2,3-dioxetanes was constructed.
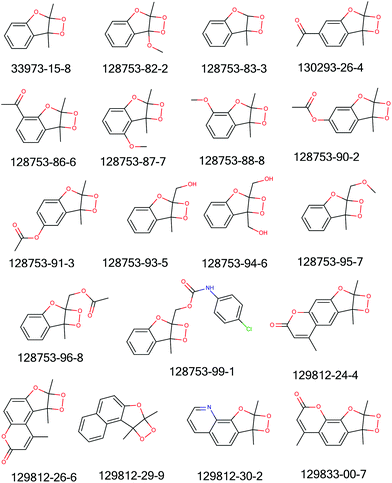 | ||
| Fig. 7 Structures of compounds whose toxicity data are reported in Table 6. | ||
| CAS number | Strains | Ref. |
|---|---|---|
| TA100 − S9 | ||
| a Highest dose tested not stated. b Tested to 100 μg per plate. | ||
| 33973-15-8 | Pos | 44,45 |
| 128753-82-2 | Pos | 44,45 |
| 128753-83-3 | Pos | 44,45 |
| 130293-26-4 | Pos | 44,45 |
| 128753-86-6 | Pos | 44 |
| 128753-87-7 | Pos | 44 |
| 128753-88-8 | Pos | 44 |
| 128753-90-2 | Pos | 44 |
| 128753-91-3 | Pos | 44,45 |
| 128753-93-5 | Nega | 44 |
| 128753-94-6 | Nega | 44 |
| 128753-95-7 | Pos | 44,45 |
| 128753-96-8 | Pos | 44,45 |
| 128753-99-1 | Pos | 44 |
| 129812-24-4 | Pos | 45,46 |
| 129812-26-6 | Pos | 46 |
| 129812-29-9 | Negb | 45,46 |
| 129812-30-2 | Negb | 46 |
| 129833-00-7 | Pos | 46 |
Cluster 3: Cluster 3 contained 55 compounds with 19 FNs. Table 7 summarises the metrics of Cluster 3. This cluster was too general and picked up a part of bigger molecules containing a polyaromatic hydrocarbon skeleton, PAH, (a class that is already covered in the Derek Nexus knowledge base) which seems not to be responsible for any mutagenicity. This investigation did not lead to the development of a new mutagenicity alert.
| Experimental toxicity | Predicted toxicity | Total | |
|---|---|---|---|
| + | − | ||
| + | 12 | 19 | 31 |
| − | 4 | 20 | 24 |
| Total | 16 | 39 | 55 |
Clusters from JEP mining
Cluster 4: Cluster 4 was generated from the Hansen data set using the SMARTS pattern c1@C(O)@C(O)@[#6]@[#6]c1; the cluster contained 121 compounds with 41 FNs. Table 8 summarises the metrics of Cluster 4.| Experimental toxicity | Predicted toxicity | Total | |
|---|---|---|---|
| + | − | ||
| + | 50 | 41 | 91 |
| − | 13 | 17 | 30 |
| Total | 63 | 58 | 121 |
As with cluster 3, the cluster was too generic and could not be used directly to derive new structural alerts. However, a look at the FNs in more detail supported the following conclusions. The FNs were reorganised into two subcategories:
Fluoranthene and derivatives have a core as shown in Fig. 8. Fig. 9 and Table 9 show the toxicological data found for 32 fluoranthene derivatives. Under the forward mutation assay conditions in Salmonella typhimurium TM677, the ultimate mutagen is identified as the 2,3-diol-1,10-epoxide fluoranthene.37,38 The implication that this diolepoxide is the ultimate mutagenic form responsible for activity is further supported by evidence suggesting that (i) diastereoisomers of the 2,3-dihydrodiol-1,10b-epoxide of benzo[ghi]fluoranthene were demonstrated to react with DNA in vitro,39,40 and (ii) fluoranthene formed similar DNA adducts in vitro in the presence of metabolic activation, which were identified as being formed through the diolepoxide metabolites.41 Based on this research an alert covering the mutagenicity of fluoranthenes and their 2,3-diol derivatives was developed.
 | ||
| Fig. 9 Structures of compounds whose toxicity data are reported in Table 9. | ||
| CAS number | Strains | Ref. |
|---|---|---|
| TA100 + S9 | ||
| 83606-71-7 | Pos | 57 |
| 205-82-3 | Pos | 57 |
| 207-08-9 | Pos | 57 |
| 76479-15-7 | Pos | 57 |
| 5385-75-1 | Pos | 58 |
| 74340-04-8 | Pos | 58 |
| 74339-98-3 | Pos | 58 |
| 74339-99-4 | Pos | 58 |
| 15299-08-8 | Neg | 58 |
| 60032-80-6 | Neg | 58 |
| 93285-74-6 | Neg | 58 |
| 205-99-2 | Pos | 59 |
| 95741-48-3 | Pos | 59 |
| 95741-50-7 | Pos | 59 |
| 95741-52-9 | Neg | 59 |
| 95741-49-4 | Weakly pos | 59 |
| 95741-46-1 | Weakly pos | 59 |
| 95741-47-2 | Neg | 59 |
| 95741-51-8 | Pos | 59 |
| 95741-53-0 | Pos | 59 |
| 116208-67-4 | Pos | 60 |
| 113600-17-2 | Pos | 60 |
| 113600-15-0 | Pos | 60 |
| 112575-92-5 | Weakly pos | 60 |
| 112575-91-4 | Weakly pos | 60 |
| 76479-15-7 | Pos | 60 |
| 206-44-0 | Pos | 61 |
| 33543-31-6 | Pos | 60 |
| 1706-01-0 | Pos | 61 |
| 83606-70-6 | Pos | 61 |
| 83606-71-7 | Pos | 61 |
| 82911-12-4 | Pos | 61 |
Pah dihydrodiol derivatives have a core as shown in Fig. 10. These compounds are formed by CYP450 oxidation and results, after a subsequent oxidation, in the ultimate mutagen of bay containing-PAH, namely the 1,2-diol-3,4-epoxide as shown in Scheme 1. Fig. 11 and Table 10 show the toxicological data found for 23 such compounds. Based on the mechanistic evidence and toxicological data,42 the mutagenicity of the 1,2-dihydrodiol derivatives of bay-PAH could be covered by being included in the scope of an existing structural alert for the mutagenicity of PAH. In contrast, metabolism of K-region epoxides of PAHs to 9,10-dihydro diols are considered to be a detoxification pathway and these diols are reported to be negatives in Ames tests43 (see Scheme 2).
 | ||
| Fig. 10 bay-PAH-1,2-dihydrodiols. At least one of the bonds marked * must be fused to an aromatic ring. | ||
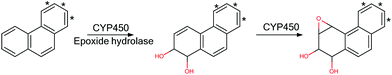 | ||
| Scheme 1 Generation of the mutagenic 1,2-dihydrodiol-3,4-epoxide. At least one of the bonds marked * must be fused to an aromatic ring. | ||
 | ||
| Fig. 11 Structures of compounds whose toxicity data are reported in Table 10. | ||
| CAS number/identifier | Strains | Overall call | Ref. | ||
|---|---|---|---|---|---|
| TA100 + S9 | TA98 + S9 | E. Coli WP2 uVrA + S9 | |||
| 98601-00-4 | Pos | Pos | 50 | ||
| 98600-98-7 | Pos | Pos | 50 | ||
| 98601-01-5 | Weakly pos | Weakly pos | 50 | ||
| 93673-37-1 | Pos | Pos | Pos | 51,52 | |
| 132172-57-7 | Pos | Pos | 53 | ||
| 132172-58-8 | Pos | Pos | Pos | 53 | |
| 72100-19-7 | Pos | Pos | Pos | 53 | |
| 160637-30-9 | Pos | Pos | Pos | 54 | |
| 160543-23-7 | Neg | Neg | Neg | 54 | |
| 160637-29-6 | Pos | Pos | Pos | 54 | |
| 28622-72-2 | Pos | Pos | Pos | 43 | |
| 96383-86-7 | Pos | Neg | Pos | 43 | |
| 87707-06-0 | Neg | Neg | 55 | ||
| 87976-64-5 | Neg | Neg | 55 | ||
| 1 | Pos | Pos | 54 | ||
| 87480-50-0 | Neg | Neg | 54 | ||
| 87436-71-3 | Neg | Neg | 54 | ||
| 87425-69-2 | Neg | Neg | 54 | ||
| 134109-01-6 | Pos | Pos | 54 | ||
| 134109-03-8 | Neg | Neg | 54 | ||
| 134109-02-7 | Neg | Neg | 54 | ||
| 1421-82-5 | Pos | Pos | 56 | ||
| 1421-83-6 | Pos | Pos | 56 | ||
Cluster 5: Cluster 5 was generated from the Hansen data set using the SMARTS pattern ClC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CS, it contained 17 compounds with 13 FNs. Table 11 summarises the metrics of Cluster 5. Although the signature of the cluster represents beta-halo alkenyl thiol derivatives, the cluster led to the identification of a range of mutagenic alpha-halo alkenyl-thiol derivatives, including S-glutathione and S-cysteine conjugates of haloalkenes, and a number of S-benzyl and disulphide derivatives. The mutagenicity of these compounds is believed to involve metabolic or abiotic transformation to the corresponding thiol, which may either lose halide to give a thioketene or tautomerise to a thioacyl halide.47,48 These metabolites are electrophilic and may form DNA adducts via reaction with nucleophilic groups in DNA.49 In the Derek Nexus version 3.0.1 knowledge base, an alert covers the mutagenicity of halogenated alkenes but that alert is based on a different mechanism (epoxidation of the double bond). Therefore, a new alert covering the activity of S-haloalkenyl derivatives, via formation of thioketene or thioacyl halide metabolites, was implemented.
CS, it contained 17 compounds with 13 FNs. Table 11 summarises the metrics of Cluster 5. Although the signature of the cluster represents beta-halo alkenyl thiol derivatives, the cluster led to the identification of a range of mutagenic alpha-halo alkenyl-thiol derivatives, including S-glutathione and S-cysteine conjugates of haloalkenes, and a number of S-benzyl and disulphide derivatives. The mutagenicity of these compounds is believed to involve metabolic or abiotic transformation to the corresponding thiol, which may either lose halide to give a thioketene or tautomerise to a thioacyl halide.47,48 These metabolites are electrophilic and may form DNA adducts via reaction with nucleophilic groups in DNA.49 In the Derek Nexus version 3.0.1 knowledge base, an alert covers the mutagenicity of halogenated alkenes but that alert is based on a different mechanism (epoxidation of the double bond). Therefore, a new alert covering the activity of S-haloalkenyl derivatives, via formation of thioketene or thioacyl halide metabolites, was implemented.
| Experimental toxicity | Predicted toxicity | Total | |
|---|---|---|---|
| + | − | ||
| + | 4 | 13 | 17 |
| − | 0 | 0 | 0 |
| Total | 4 | 13 | 17 |
Fig. 12 and Table 12 summarise the data found for this class of compounds.
 | ||
| Fig. 12 Structures of compounds whose toxicity data is reported in Table 12. | ||
| CAS number/identifier | Strains | Overall call | Ref. | ||
|---|---|---|---|---|---|
| TA100 | TA98 | ||||
| −S9 | +S9 | −S9 | |||
| 627-72-5 | Equ | Pos | Weakly pos | Pos | 67,68 |
| 87619-82-7 | Pos | Pos | Pos | Pos | 67,68 |
| 98025-31-1 | Pos | Pos | Pos | 67 | |
| 89784-39-4 | Neg | Pos | Pos | 69 | |
| 111348-61-9 | Pos | Pos | Pos | 70 | |
| 115453-72-0 | Pos | Pos | Pos | 71 | |
| 111959-96-7 | Neg | Pos | Pos | 72 | |
| 91085-62-0 | Neg | Pos | Pos | 72 | |
| 2 | Pos | Pos | Pos | 73 | |
| 3 | Pos | Pos | Pos | 73 | |
| 111574-85-7 | Neg | Pos | Pos | Pos | 73 |
| 4 | Neg | Pos | Pos | Pos | 73 |
| 133831-60-4 | Pos | Pos | 48 | ||
| 117760-95-9 | Pos | Pos | 48 | ||
| 133831-61-5 | Pos | Pos | 48 | ||
| 133831-62-6 | Pos | Pos | 48 | ||
Experimental
The Hansen dataset was obtained and curated as described in a previous publication.11Functional group and heterocycle fragments were generated in KNIME19 using nodes built in-house based on the Ceres62,63 chemical engine. EP mining was done using an in-house Java implementation of the published contrast pattern tree mining algorithm.64 JEP mining was done again in KNIME using an in-house built node implementing published algorithms.6,65,66 Workflows in were built in KNIME version 2.5.2.
Toxicity predictions and TN and FN classifications were made using Derek Nexus version 3.0.1 in Lhasa Knowledge Suite – Nexus 1.5.
Conclusions
EP and JEP mining offer enhanced rates of knowledge discovery in the hands of expert scientists. They allow experts to tackle large and biased datasets from which it is difficult to extract knowledge manually and this has led to EP mining tools being implemented at Lhasa Limited.The success of the approach is significantly impacted by the fragments from which patterns are mined, where commercial sources proved inferior to a custom developed approach.
The alerts discovered in this work have been implemented in the knowledge base of Derek Nexus version 4.0.5.
Acknowledgements
We acknowledge funding from the Technology Strategy Board for a KTP award to RJS.Notes and references
- J. C. Dearden, in In Silico Toxicology, ed. M. Cronin and J. Madden, Royal Society of Chemistry, Cambridge, UK, 2010, pp. 478–507 Search PubMed.
- P. Judson, Knowledge-based Expert Systems in Chemistry: Not Counting on Computers, Royal Society of Chemistry, Cambridge, UK, 2009 Search PubMed.
- Derek Nexus, version 3.0.1, Lhasa Limited, Leeds, UK, 2012 Search PubMed.
- G. Patlewicz, N. Jeliazkova, R. J. Safford, A. P. Worth and B. Aleksiev, SAR QSAR Environ. Res., 2008, 19, 495–524 CrossRef CAS PubMed.
- Genetox Expert Alerts Suite, Leadscope Inc, Columbus, Ohio, 2013 Search PubMed.
- G. Dong and J. Li, in Proceedings of the fifth ACM SIGKDD international conference on Knowledge discovery and data mining - KDD ‘99, ACM Press, New York, New York, USA, 1999, pp. 43–52 Search PubMed.
- V. Namasivayam, Y. Hu, J. Balfer and J. Bajorath, J. Chem. Inf. Model., 2013, 53, 1272–1281 CrossRef CAS PubMed.
- S. Lozano, G. Poezevara, M.-P. Halm-Lemeille, E. Lescot-Fontaine, A. Lepailleur, R. Bissell-Siders, B. Crémilleux, S. Rault, B. Cuissart and R. Bureau, J. Chem. Inf. Model., 2010, 50, 1330–1339 CrossRef CAS PubMed.
- R. Sherhod, V. J. Gillet, P. N. Judson and J. D. Vessey, J. Chem. Inf. Model., 2012, 52, 3074–3087 CrossRef CAS PubMed.
- R. Sherhod, V. Gillet, T. Hanser, P. Judson and J. Vessey, J. Cheminf., 2013, 5, O9 Search PubMed.
- R. Sherhod, P. N. Judson, T. Hanser, J. D. Vessey, S. J. Webb and V. J. Gillet, J. Chem. Inf. Model., 2014, 57, 1864–1879 CrossRef PubMed.
- RDKit, Cheminformatics and Machine Learning Software, 2013, http://www.rdkit.org, (accessed April 2014).
- K. Hansen, S. Mika, T. Schroeter, A. Sutter, A. ter Laak, T. Steger-Hartmann, N. Heinrich and K.-R. Müller, J. Chem. Inf. Model., 2009, 49, 2077–2081 CrossRef CAS PubMed.
- A collection of Ames test data for 8421 compounds derived from the FDA/CFSAN/OFAS knowledge base, Silver Spring, Maryland Search PubMed.
- Dragon version 6, Talete srl, Milan, Italy, 2007 Search PubMed.
- Leadscope Enterprise, Leadscope Inc, Columbus, Ohio, 2012 Search PubMed.
- E. Anderson, G. D. Veith and D. Weininger, SMILES: A line notation and computerized interpreter for chemical structures, U.S. EPA, Environmental Research Laboratory-Duluth. Report No. EPA/600/M-87/021. Duluth, MN, 1987.
- SMARTS - A Language for Describing Molecular Patterns, http://www.daylight.com/dayhtml/doc/theory/theory.smarts.html, (accessed April 2014).
- KNIME, version 2.5.2, KNIME.com AG, Zurich, Switzerland, 2012 Search PubMed.
- JChem for Excel, version 6.2.2, ChemAxon Kft, Budapest, Hungary, 2012 Search PubMed.
- ChemSpider, http://www.chemspider.com/, (accessed April 2014).
- Chemical Entities of Biological Interest (ChEBI), https://www.ebi.ac.uk/chebi/, (accessed April 2014).
- ChemIDplus Advanced, http://chem.sis.nlm.nih.gov/chemidplus/, (accessed April 2014).
- TOXNET, http://toxnet.nlm.nih.gov/, (accessed April 2014).
- National Toxicology Program Database Search Application, http://tools.niehs.nih.gov/ntp_tox/, (accessed April 2014).
- Vitic Nexus, version 2.0.0, Lhasa Limited, Leeds, UK, 2011 Search PubMed.
- PubMed, http://www.ncbi.nlm.nih.gov/pubmed, (accessed April 2014).
- L. R. Ferguson and W. A. Denny, Mutat. Res., 2007, 623, 14–23 CrossRef CAS PubMed.
- R. Shen, P. Wang and N. Tang, J. Fluoresc., 2010, 20, 1287–1297 CrossRef CAS PubMed.
- H. Glatt, FASEB J., 1997, 11, 314–321 CAS.
- H. Aoki, Y. Ogawa, C. Yukawa, M. Nakamura and H. Nakazawa, Food Addit. Contam., 2002, 19, 350–356 CrossRef CAS PubMed.
- T. Matsushima, A. Araki, O. Yagame, M. Muramatsu, K. Koyama, K. Ohsawa, S. Natori and H. Tomimori, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1985, 150, 141–146 CrossRef CAS.
- I. Morimoto, T. Nozaka, F. Watanabe, M. Ishino, Y. Hirose and T. Okitsu, Mutat. Res., Genet. Toxicol., 1983, 116, 103–117 CrossRef CAS.
- W. A. Denny, P. M. Turner, G. J. Atwell, G. W. Rewcastle and L. R. Ferguson, Mutat. Res., 1990, 232, 233–241 CrossRef CAS.
- P. E. Hartman, P. B. Hulbert, E. Bueding and D. D. Taylor, Mutat. Res., 1975, 31, 87–95 CAS.
- W. Adam, L. Hadjiarapoglou, T. Mosandl, C. R. Saha-Moeller and D. Wild, J. Am. Chem. Soc., 1991, 113, 8005–8011 CrossRef CAS.
- J. R. Babson, S. E. Russo-Rodriguez, R. V. Wattley, P. L. Bergstein, W. H. Rastetter, H. L. Liber, B. M. Andon, W. G. Thilly and G. N. Wogan, Toxicol. Appl. Pharmacol., 1986, 85, 355–366 CrossRef CAS.
- W. H. Rastetter, R. B. Nachbar, S. E. Russo-Rodriguez, R. V. Wattley, W. G. Thilly, B. M. Andon, W. L. Jorgensen and F. Ibrahim, J. Org. Chem., 1982, 47, 4873–4878 CrossRef CAS.
- H.-F. Chang, D. M. Huffer, M. P. Chiarelli and B. P. Cho, Chem. Res. Toxicol., 2002, 15, 187–197 CrossRef CAS PubMed.
- H.-F. Chang, D. M. Huffer, M. P. Chiarelli, L. R. Blankenship, S. J. Culp and B. P. Cho, Chem. Res. Toxicol., 2002, 15, 198–208 CrossRef CAS PubMed.
- J. R. Babson, S. E. Russo-Rodriguez, W. H. Rastetter and G. N. Wogan, Carcinogenesis, 1986, 7, 859–865 CrossRef CAS PubMed.
- Y. Ye, C. E. Scharping and G. M. Holder, Carcinogenesis, 1995, 16, 787–793 CrossRef CAS PubMed.
- H. Glatt, A. Seidel, W. Bochnitschek, H. Marquardt, H. Marquardt, R. M. Hodgson, P. L. Grover and F. Oesch, Cancer Res., 1986, 46, 4556–4565 CAS.
- W. Adam, O. Albrecht, E. Feineis, I. Reuther, C. R. Saha-Möller, P. Seufert-Baumbach and D. Wild, Liebigs Ann. Chem., 1991, 1991, 33–40 CrossRef.
- W. Adam, A. Beinhauer, T. Mosandl, C. Saha-Möller, F. Vargas, B. Epe, E. Müller, D. Schiffmann and D. Wild, Environ. Health Perspect., 1990, 88, 89–97 CrossRef CAS.
- W. Adam, H. Hauer, T. Mosandl, C. R. Saha-Möller, W. Wagner and D. Wild, Liebigs Ann. Chem., 1990, 1990, 1227–1236 CrossRef.
- S. Vamvakas, A. A. Elfarra, W. Dekant, D. Henschler and M. W. Anders, Mutat. Res., 1988, 206, 83–90 CAS.
- D.-A. Müller, G. Urban and W. Dekant, Chem.-Biol. Interact., 1991, 77, 159–172 CrossRef.
- M. Müller, G. Birner, M. Sander and W. Dekant, Chem. Res. Toxicol., 1998, 11, 464–470 CrossRef PubMed.
- M. Mushtaq, P. P. Fu, D. W. Miller and S. K. Yang, Cancer Res., 1985, 45, 4006–4014 CAS.
- K. O. Newcomb, R. Sangaiah, A. Gold and L. M. Ball, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1993, 287, 181–190 CrossRef CAS.
- R. Sangaiah, A. Gold, K. O. Newcomb and L. M. Ball, J. Med. Chem., 1991, 34, 546–549 CrossRef CAS.
- S. Kumar, P. L. Kole and H. C. Sikka, Mutat. Res., Genet. Toxicol., 1990, 242, 337–343 CrossRef CAS.
- P. P. Fu, L. S. Von Tungeln, L. E. Unruh, Y.-C. Ni and M. W. Chou, Carcinogenesis, 1991, 12, 371–378 CrossRef CAS PubMed.
- S. Kumar, H. C. Sikka, S. K. Dubey, A. Czech, N. Geddie, C. X. Wang and E. J. LaVoie, Cancer Res., 1989, 49, 20–24 CAS.
- K. L. Platt, M. Schollmeier, H. Frank and F. Oesch, Environ. Health Perspect., 1990, 88, 37–41 CrossRef CAS.
- E. J. LaVoie, S. S. Hecht, S. Amin, V. Bedenko and D. Hoffmann, Cancer Res., 1980, 40, 4528–4532 CAS.
- C. Malaveille, A. Hautefeuille, O. Perin-Roussel, S. Saguem, M. Croisy-Delcey, F. Zajdela and H. Bartsch, Carcinogenesis, 1984, 5, 1263–1266 CrossRef CAS PubMed.
- S. Amin, K. Huie and S. S. Hecht, Carcinogenesis, 1985, 6, 1023–1025 CrossRef CAS PubMed.
- E. H. Weyand, N. Geddie, J. E. Rice, A. Czech, S. Amim and E. J. La Voie, Carcinogenesis, 1988, 9, 1277–1281 CrossRef CAS PubMed.
- E. J. LaVoie, S. S. Hecht, V. Bedenko and D. Hoffmann, Carcinogenesis, 1982, 3, 841–846 CrossRef CAS PubMed.
- T. Hanser, E. Rosser, M. Ulyatt and S. Werner, in 5th Joint Sheffield Conference on Chemoinformatics, Sheffield, UK, 2010.
- T. Hanser, E. Rosser, S. Werner and P. Górny, in 9th International Conference on Chemical Structures, Noordwijkerhout, NL, 2011, p. P-8.
- H. Fan and K. Ramamohanarao, IEEE Trans. Knowl. Data Eng., 2006, 18, 721–737 CrossRef.
- G. Dong and J. Li, Knowl. Inf. Syst., 2004, 8, 178–202 CrossRef PubMed.
- J. Li, G. Dong and K. Ramamohanarao, Knowl. Inf. Syst., 2001, 3, 131–145 CrossRef.
- W. Dekant, S. Vamvakas, K. Berthold, S. Schmidt, D. Wild and D. Henschler, Chem.-Biol. Interact., 1986, 60, 31–45 CrossRef CAS.
- T. Green and J. Odum, Chem.-Biol. Interact., 1985, 54, 15–31 CrossRef CAS.
- D. Reichert and S. Sch
![[g with combining umlaut]](https://www.rsc.org/images/entities/char_0067_0308.gif) tz, Biochem. Pharmacol., 1986, 35, 1271–1275 CrossRef CAS.
tz, Biochem. Pharmacol., 1986, 35, 1271–1275 CrossRef CAS. - S. Vamvakas, W. Dekant, K. Berthold, S. Schmidt, D. Wild and D. Henshler, Biochem. Pharmacol., 1987, 36, 2741–2748 CrossRef CAS.
- S. Vamvakas, K. Berthold, W. Dekant and D. Henschler, Chem.-Biol. Interact., 1988, 65, 59–71 CrossRef CAS.
- S. Vamvakas, W. Dekant and M. W. Anders, Biochem. Pharmacol., 1989, 38, 935–939 CrossRef CAS.
- B. Dreeßen, G. Westphal, J. Bünger, E. Hallier and M. Müller, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2003, 539, 157–166 CrossRef.
| This journal is © The Royal Society of Chemistry 2015 |