Zone analysis by two-dimensional electrophoresis with accelerator mass spectrometry of in vivo protein bindings of idiosyncratic hepatotoxicants troglitazone and flutamide bioactivated in chimeric mice with humanized liver†
Hiroshi
Yamazaki
*a,
Shunji
Kuribayashi
b,
Tae
Inoue
c,
Tomohiro
Honda
d,
Chise
Tateno
c,
Ken
Oofusa
e,
Shinichi
Ninomiya
f,
Toshihiko
Ikeda
g,
Takashi
Izumi
d and
Toru
Horie
h
aShowa Pharmaceutical University, Machida, Tokyo 194-8543, Japan. E-mail: hyamazak@ac.shoyaku.ac.jp
bOtsuka Pharmaceutical Factory, Inc., Naruto, Tokushima 772-8601, Japan
cPhoenixBio, Co., Higashi-Hiroshima, Hiroshima 739-0046, Japan
dDaiichi Sanyo, Co., Shinagawa-ku, Tokyo 140-8710, Japan
eIdea Consultants Inc., Suminoe-ku, Osaka 559-8519, Japan
fSekisui Medical Co., Chuo-ku, Tokyo 103-0027, Japan
gYokohama College of Pharmacy, Totsuka-ku, Yokohama 245-0066, Japan
hDrug Discovery and Development Institute, Tsukuba, Ibaragi 305-0036, Japan
First published on 10th September 2014
Abstract
Analyses using electrophoresis with accelerator mass spectrometry revealed that in vivo bioactivated radiolabeled troglitazone and flutamide, both known to be hepatotoxic in humans, bound nonspecifically to a variety of microsomal and cytosolic proteins in livers from chimeric mice with humanized liver. Unlike those of radiolabeled diazepam (rarely hepatotoxic) and previously reported 5-n-butyl-pyrazolo[1,5-a]pyrimidine (limited hepatotoxicity), some troglitazone and flutamide binding proteins were located in the top right area in a zone analysis, representing high covalent binding contents and high target protein concentrations. Among a variety of liver microsomal proteins bound, the binding target proteins of troglitazone and flutamide with the highest covalent binding contents (in terms of pmol equivalent per μg target protein) were 17β-hydroxysteroid dehydrogenase and 3β-hydroxysteroid dehydrogenase, respectively. Troglitazone and flutamide were activated to reactive metabolites and apparently bound to different target proteins in livers from chimeric mice with humanized liver. The highest covalent binding contents for troglitazone were higher than that for flutamide under the present conditions. These results indicate that the drug metabolism mediated by humanized livers (leading to binding in vivo) in combination with a zone analysis of covalent binding contents/target protein concentration data could be a good tool for evaluating the relationship between the nonspecific protein binding behavior of medicines and potential hepatotoxicity in humans. Thus, testing whether protein binding data of new medicines are unbalanced with respect to deviation from an inverse relationship or the presence of data points in the high covalent binding/high protein concentration zone might be an important concept in evaluating hepatotoxic potential.
Introduction
Drug-induced liver toxicity is the single most prominent adverse event causing non-approval of a drug or withdrawal of a drug from the market by regulatory agencies. In evaluating the potential for metabolic activation prior to market approval as well as during the post-marketing period, in vitro and in vivo studies play roles in the safe development and effective use of medicines. The US National Institutes of Health LiverTox database was developed to promote basic and clinical research on drug-induced liver toxicity.1 It has been reported that drug-induced hepatotoxicity may be caused by active intermediates formed by animal and/or human P450 enzymes from the common analgesic acetaminophen2 or from an idiosyncratic reaction to troglitazone, which was withdrawn from the market.3 Species differences between experimental animals and humans in the roles of cytochrome P450 enzymes in drug metabolism are crucial factors in the evaluation of drug toxicity.4 Chimeric mice with humanized liver or with human P450 genes are used as animal models to investigate human metabolites during drug development.5–7 There is an industrywide need for predictive tools and an established knowledge base for quantifying the risks in humans.The purpose of this study was to characterize in vivo idiosyncratic hepatotoxicants troglitazone3 and flutamide8 in comparison with diazepam (rarely hepatotoxic1) and previously reported 5-n-butyl-pyrazolo[1,5-a]pyrimidine, a proximate metabolite of previous drug candidate OT-7100 (limited hepatotoxic effects in humans9), in terms of their nonspecific protein bindings to a variety of microsomal and cytosolic proteins in livers. We report herein a zone analysis for imbalance between potency and nonspecificity in protein binding using electrophoresis with accelerator mass spectrometry for metabolically activated idiosyncratic hepatotoxicants that bind nonspecifically to a variety of microsomal and cytosolic proteins in chimeric mice with humanized liver.
Results and discussion
Four 14C-labeled substrates, namely diazepam (5 mg dose per kg body weight), 5-n-butyl-pyrazolo[1,5-a]pyrimidine (3 mg dose per kg body weight), flutamide (10 mg dose per kg body weight), and troglitazone (150 mg dose per kg body weight), were administered to chimeric mice. Liver microsomes and cytosol fractions were prepared 24 h, 12 h, 1 h, and 1 h after the treatments with 14C-troglitazone, 14C-flutamide, 14C-5-n-butyl-pyrazolo[1,5-a]pyrimidine, and 14C-diazepam, respectively. Many protein bands were present in the sodium dodecyl sulfate–polyacrylamide gel electrophoretogram after Coomassie blue staining; ten 1 mm wide gel strips in the 35–100 kDa range in the liver microsome analysis were used to focus on diazepam and flutamide. After measurements of protein contents and radioactivities of the 10 analyte strips, covalent bindings per mg protein were calculated, as shown in Fig. 1. Per milligram of total microsomal protein, approximately 1–6 pmol and 2–15 pmol equivalents of negative control diazepam and positive control flutamide, respectively, bound to a variety of liver microsomal proteins. As judged from the immunoblotting and molecular weights on the gel in combination with the radioactivity from the radiolabeled substrates, metabolically activated substrates bound to human P450 1A2 and 3A4 (strips 7 and 8, respectively), which were the primary enzymes involved in substrate activation,11,12 and to other many microsomal proteins in humanized livers. | ||
| Fig. 1 Electrophoresis for microsomal proteins, including P450 enzymes, bound to 14C-labeled diazepam (A), 14C-labeled 5-n-butyl-pyrazolo[1,5-a]pyrimidine, a primary metabolite of OT-7100 (B), and 14C-labeled flutamide (C) in chimeric mice with humanized liver. Gel slices 1 mm wide were cut out by hand between 35 and 100 kDa. Data for 5-n-butyl-pyrazolo[1,5-a]pyrimidine were taken from Yamazaki et al.9 for comparison. | ||
As can be seen in Fig. 1, the overall protein binding levels reflect the hepatotoxicity of the three substrates, i.e., flutamide > OT-7100 > diazepam. Unfortunately, troglitazone could not be included in this part of the analysis because of sample limitations.
After administration of the four radiolabeled substrates to chimeric mice with humanized liver, covalent binding profiles of liver microsomal and cytosolic fractions were further investigated by two-dimensional electrophoresis. Protein samples (100 μg) were separated by isoelectric point (pI 3–10) and molecular weight (10–225 kDa). The target protein concentrations and covalent binding contents of the resulting analyte spots in the gels were determined. Two-dimensional electrophoresis with accelerator mass spectrometry analyses revealed that troglitazone, flutamide, 5-n-butyl-pyrazolo[1,5-a]pyrimidine, and diazepam bonded covalently to a variety of liver microsomal (Fig. 2A) and/or cytosolic (Fig. 2B) proteins. The analyte spots with the 50 highest covalent binding contents were included in this analysis. The highest binding level of troglitazone was observed with 17β-hydroxysteroid dehydrogenase (0.42 pmol equivalent per μg target protein) in microsomal proteins (target protein concentration 0.51 μg protein per mg microsomal protein, as shown in Fig. 2A) and with glutathione S-transferase M2-2 (0.27 pmol equivalent per μg target protein) in cytosolic proteins (target protein concentration 0.14 μg protein per mg cytosolic protein, as shown in Fig. 2B). These values for binding levels and target protein concentrations are considered to be high in the zone analysis.
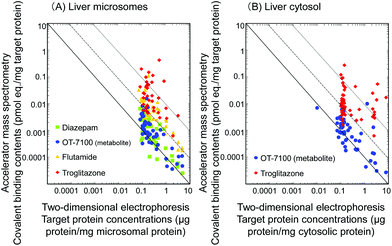 | ||
| Fig. 2 Covalent binding profiles of liver microsomal (A) and cytosolic (B) protein fractions separated by two-dimensional electrophoresis. The fractions were obtained from chimeric mice with at least 70% humanized liver after administration of radiolabeled substrates. Loaded protein samples (100 μg) underwent isoelectric focusing (pI 3–10) and were then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (10–225 kDa). In vivo protein bindings with metabolically activated 14C-substrates were analyzed by accelerator mass spectrometry. Lines are drawn through convenient axis intersections to indicate an inverse relationship. Data for 5-n-butyl-pyrazolo[1,5-a]pyrimidine, an OT-7100 metabolite, were taken from Yamazaki et al.9 for comparison. Note that the data points in the top right zone (i.e., those with high covalent binding contents and high target protein concentrations) of the left-hand graph (A) are predominantly those of troglitazone and flutamide, both of which are known to be hepatotoxic. | ||
Flutamide also bound covalently to a variety of liver microsomal proteins, the highest covalent binding target being 3β-hydroxysteroid dehydrogenase (0.11 pmol equivalent per μg target protein, target protein concentration 0.18 μg protein per mg microsomal protein, as shown in Fig. 2A); however, the highest covalent binding contents for flutamide were lower than that for troglitazone. It is worth noting that the three target proteins quoted above with the highest covalent binding levels were different. The identities of the other analyte spot proteins are shown in Tables 1 and 2, including the names for reanalyzed binding proteins for 5-n-butyl-pyrazolo[1,5-a]pyrimidine. The top right zone of Fig. 2A contains activated drugs bound to abundant target protein molecules, indicating a nonspecific binding manner.
| Protein | Name |
|---|---|
| a The peptides were analyzed by MASCOT database software (Matrix Science, Tokyo, Japan). | |
| 1 | 3-Ketoacyl-CoA-thiolase |
| 2 | NADPH-dependent carbonyl reductase |
| 3 | Plasma retinol-binding protein [precursor] |
| 4 | Branched-chain-amino-acid aminotransferase |
| 5 | 3-Hydroxyanthranilic acid dioxygenase |
| 6 | Fatty acid desaturase 1 |
| 7 | 7-Dehydrocholesterol reductase |
| 8 | Selenium binding liver protein |
| 9 | Retinol dehydrogenase type III, |
| 10 | UDP-glucuronosyltransferase |
| 11 | Endoplasmic reticulum protein |
| 12 | NADPH quinone oxide reductase 1 |
| 13 | Acetaminophen binding protein |
| 14 | 17β-Hydroxysteroid dehydrogenase |
| 15 | Steroid dehydrogenase homolog |
| 16 | Flavin-containing monooxygenase |
| 17 | Prolyl-4-hydroxylase |
| 18 | Retinol dehydrogenase type I |
| 19 | Superoxide dismutase |
| 20 | Sterol-4-α-carboxylate 3-dehydrogenase |
| 21 | Dihydrolipoamide acetyltransferase component |
| 22 | Putative UST1-like organic anion transporter |
| 23 | Tropomyosin 5 |
| 24 | 2,4-CoA reductase |
| 25 | Betaine-homocysteine S-methyltransferase |
| 26 | Carbonyl reductase, NADPH |
| 27 | Methionine adenosyltransferase |
| 28 | 3β-Hydroxysteroid dehydrogenase type III |
| 29 | Acyl-CoA synthetase short-chain family member 2 |
| 30 | Mitochondrial P450 |
| 31 | Long-chain-fatty-acid-CoA ligase, liver isozyme |
| 32 | Glutathione peroxidase |
| 33 | Epoxide hydrolase |
| 34 | Keap-1 |
| 35 | Oxidoreductase |
| 36 | NADH-cytochrome b5 reductase |
| 37 | Fructose-bisphosphate aldolase B |
| 38 | Urate oxidase |
| 39 | Catechol O-methyltransferase, membrane-bound form |
| 40 | NAD(P) transhydrogenase |
| 41 | Aldose reductase |
| 42 | Aldehyde dehydrogenase |
| 43 | NADPH-cytochrome P450 reductase |
| 44 | Fatty aldehyde dehydrogenase |
| 45 | Glutathione S-transferase |
| 46 | Uricase |
| 47 | ATP-synthetase |
| 48 | Carbonic anhydrase |
| 49 | Protein disulfide isomerase |
| 50 | Serum albumin |
| Protein | Name |
|---|---|
| a The peptides were analyzed using the MASCOT database software (Matrix Science, Tokyo, Japan). ND, not determined because of sample limitations. | |
| 1 | Catalase |
| 2 | Pyrophosphate phosphohydrolase |
| 3 | Protein DJ-1 |
| 4 | UDP-N-acetylglucosamine-peptide N-acetylglucosaminyl-transferase 110 kDa subunit |
| 5 | Golgi-associated protein, GCP360 |
| 6 | Glutathione S-transferase Ya2 |
| 7 | Thiopurine S-methyl transferase |
| 8 | Virus Tat binding protein, TBP-7 |
| 9 | Virus Tat binding protein, TBP-1 |
| 10 | Life tech mouse embryo 8 5dpc 10664019 |
| 11 | Hypothetical protein clone pT-Adv JuaX22 |
| 12 | Non-neural enolase |
| 13 | Phosphoglycerate kinase 1 |
| 14 | Tumor protein p53 |
| 15 | Transformed 3T3 cell double minute 2 |
| 16 | Glutathione S-transferase, M2-2 |
| 17 | Vesicle-associated calmodulin-binding protein |
| 18 | Solute carrier family 22 member-7, OAT2 |
| 19 | Glutathione S-transferase, M2-3 |
| 20 | Senescence marker protein-30 |
| 21 | Heat shock protein 10 |
| 22 | Glutathione S-transferase, M1-1 |
| 23 | Mitochondrial P450 |
| 24 | Phenylpyruvate tautomerase |
| 25 | Glutaredoxin |
| 26 | Kelch-like ECH-associated protein 1, KEAP 1 |
| 27 | Proteasome activator subunit 1 |
| 28 | NADPH quinone oxide reductase 1 |
| 29 | Hydroxyprostaglandin dehydrogenase |
| 30 | Hemooxygenase-1 |
| 31 | Phosphatidylethanolamine-binding protein |
| 32 | ES/130,180 kDa Ribosome receptor |
| 33 | pI 6.1 Esterase |
| 34 | Nuclear factor (erythroid-derived 2)-like 2, NRF 2 |
| 35 | Oxalosuccinate decarboxylase |
| 36 | Ribonuclease UK114 |
| 37 | Glutathione synthetase |
| 38 | S-Adenosylmethionine synthetase isoform type-1 |
| 39 | Tropomyosin g |
| 40 | Tyrosine-ester sulfotransferase |
| 41 | Guanidinoacetate N-methyltransferase |
| 42 | Solute carrier family 10, NTCP |
| 43 | Glutathione peroxidase |
| 44 | Solute carrier family 22, member-1, OCT-1 |
| 45 | Heat shock protein 90 |
| 46 | Eukaryotic translation elongation factor 1 a1 |
| 47 | Hydroxysteroid dehydrogenase |
| 48 | D-Dopachrome tautomerase |
| 49 | Triosephosphate isomerase |
| 50 | 26S Proteasome regulatory subunit 6B |
| 51 | Glutathione S-transferase A1 |
| 52 | Carbonic anhydrase |
| 53 | Guanine aminohydrolase |
| ND | Lamin A (protein no. 21 for troglitazone) |
| ND | Preadipocyte growth factor (protein no. 24 for troglitazone) |
In Fig. 3, fractioned microsomal and cytosolic proteins are listed in order of decreasing covalent binding contents with respect to diazepam (microsomal) and 5-n-butyl-pyrazolo[1,5-a]pyrimidine (cytosolic). Dark colors indicate higher covalent binding contents for all four substrates. Fractioned proteins highly bound to the bioactivated substrates (darker colors in Fig. 3) differed between idiosyncratic hepatotoxicants troglitazone3 and flutamide.8 In fact, the distribution of dark colors in Fig. 3A becomes more imbalanced in the order troglitazone > flutamide > 5-n-butyl-pyrazolo[1,5-a]pyrimidine > diazepam, the same order as for their hepatotoxicities.
 | ||
| Fig. 3 Profiles of liver microsomal (A) and cytosolic (B) protein fractions separated by two-dimensional electrophoresis. The fractions were obtained from chimeric mice with at least 70% humanized liver after administration of radiolabeled substrates. Fractioned microsomal (A) and cytosolic (B) proteins are listed in order of decreasing covalent binding contents for radiolabeled diazepam and 5-n-butyl-pyrazolo[1,5-a]pyrimidine, respectively. Colors in five dark to light shades indicate from high to low covalent binding contents for each substrate [so the color shades in the left-hand columns of (A) and (B) are highly ordered by definition]. D, diazepam; O, 5-n-butyl-pyrazolo[1,5-a]pyrimidine, a primary metabolite of OT-7100; F, flutamide; and T, troglitazone. The numbers indicate individual target proteins, the names of which are shown in Tables 1 and 2. Binding data for 5-n-butyl-pyrazolo[1,5-a]pyrimidine were taken from Yamazaki et al.9 for comparison. For cytosolic proteins, diazepam and flutamide were not included because of sample limitations. NA, not available. | ||
The estimated covalent binding levels (pmol drug eq. per μg target protein) apparently indicated an inverse relationship with the target protein concentrations in the case of the negative control, diazepam (a straight line, Fig. 2A). The selective and balanced data pattern for diazepam may reflect the low hepatotoxicity of diazepam. A little imbalance between covalent binding content and target protein concentration was seen in the case of 5-n-butyl-pyrazolo[1,5-a]pyrimidine, which might be involved as the primary metabolite in the limited hepatotoxic effects of OT-7100 in humans undergoing high-dose, long-term treatments.10 Although data for several binding analyte spots for severely hepatotoxic flutamide overlapped those for diazepam and 5-n-butyl-pyrazolo[1,5-a]pyrimidine, most were in the high covalent binding/high protein concentration zone (above the dashed line, Fig. 2A). Furthermore, analyte spots for troglitazone deviated more from an inverse relationship than those for flutamide did. Both troglitazone and flutamide were activated to bind proteins with an imbalance between covalent binding content and target protein concentration, indicating that nonspecific binding to liver proteins can lead to hepatotoxicity. A zone analysis like that shown in Fig. 2 in terms of intensity and imbalance of covalent binding contents and target protein concentrations could prove to be a useful tool for predicting hepatotoxic effects.
It can be speculated that N-oxidation of flutamide after hydrolysis11 in humanized liver might lead to binding to P450 itself and to many other proteins in a nonselective and extensive manner. Troglitazone was withdrawn from the market and is reportedly activated to a quinone epoxide metabolite by human P450 3A4.3,12 The drug–protein adducts of activated troglitazone are likely to nonselectively and extensively bind any abundant proteins in microsomal or cytosolic proteins shown in Fig. 1 and 2. This would lead to a less inverse relationship as shown in Fig. 2. These lines of evidence suggest that substrates were bioactivated by human P450 enzymes in humanized liver in vivo and could bind to any proteins in both microsomes and cytosol, including the catalyst P450 itself, implying relatively low selectivity in target protein bindings. In the present study, two idiosyncratic hepatotoxic medicines, flutamide and troglitazone, were activated to reactive metabolites and apparently bound to different target proteins. It would be of great interest to test other nonhepatotoxic substrates and hepatotoxic medicines using this zone analysis approach and to accumulate target protein information in future studies.
Experimental
Chimeric mice with human liver cells (PXB-mice®, PhoenixBio Co., Hiroshima, Japan, n = 3)6 were orally treated with 14C-troglitazone (150 mg kg−1 body weight, 1.75 MBq per mouse), 14C-flutamide (the positive [hepatotoxic] control, 10 mg kg−1, 0.866 MBq per mouse), and 14C-diazepam (the negative [rarely hepatotoxic] control, 5.0 mg kg−1, 0.370 MBq per mouse) and intravenously treated with 14C-5-n-butyl-pyrazolo[1,5-a]pyrimidine (3.0 mg kg−1, 0.645 MBq per mouse). In the chimeric mice, more than 70% of liver cells were estimated to have been replaced with human hepatocytes, as judged by measurements of human albumin concentrations in plasma. Liver microsome and cytosol fractions were prepared 24 h, 12 h, 1 h, and 1 h after the treatments with 14C-troglitazone, 14C-flutamide, 14C-5-n-butyl-pyrazolo[1,5-a]pyrimidine,9 and 14C-diazepam, respectively, based on their drug disposition. All animal experiments were performed in compliance with the relevant laws and institutional guidelines. The institutional Animal Care and Use committee of PhoenixBio approved the experiments. This study was performed in accordance with the Guidelines for Proper Conduct of Animal Experiments by the Science Council of Japan (2006). All animals were handled in strict accordance with good animal practice under the supervision of veterinarians. Every effort was made to alleviate animal discomfort and pain by appropriate and routine use of anesthetic and/or analgesic agents.In a one-dimensional analysis, sodium dodecyl sulfate–polyacrylamide gel electrophoretograms in 12% acrylamide gels with liver microsomes (10 μg) from chimeric mice were stained with 0.08% Coomassie Brilliant Blue R350 (GE Healthcare Bio-Science, Tokyo, Japan).9 The radioactivity content of each band on the gel was determined by a BAS-5000 Image Analysis System (Fujifilm, Tokyo, Japan). Ten 1 mm-wide gel slices from separate experiments were cut out by hand from the gel between 35 and 100 kDa after electrophoresis and were subjected to mass spectrometry to detect P450 enzymes.9 After measurements of the protein contents and radioactivities of the analyte strips, covalent binding levels per μg microsomal protein were calculated.
For two-dimensional electrophoresis, microsomal or cytosolic protein samples (100 μg) were applied overnight to Immobiline Drystrip (GE Healthcare Bio-Science) by in-gel rehydration as described previously.9 After the rehydrated gels were dried gently, isoelectric focusing was performed in a Pharmacia Hoefer Multiphor II electrophoresis chamber (GE Healthcare, Buckinghamshire, UK) as the first-dimension analysis, according to the manufacturer's instructions. As the second dimension, sodium dodecyl sulfate–polyacrylamide gel electrophoresis was performed in 9–18% acrylamide gradient gels using an IsoDalt electrophoresis chamber. The second-dimension gels were stained with SYPRO Ruby (Invitrogen, Carlsbad, CA, USA) for accurate quantification of spots using a fluorescent scanner following the manufacturer's protocols. The SYPRO Ruby-stained proteins were quantified using a Molecular Imager FX (Bio-Rad Laboratories, Hercules, CA, USA) and the Image Master Platinum image analysis software® (GE Healthcare) because this fluorescent stain has a wider linear-dynamic range than the silver stain. In the two-dimensional electrophoresis procedure cytochrome P450 isoforms were not evaluated because membrane-bound proteins are generally difficult to resolve by the first pI-dependent separation step in two-dimensional electrophoresis.9
Identification of proteins was performed.9 Because the fluorescent signals of stained gels were weakened during image analysis, the gels were re-stained with silver for accurate picking of spots selected by the image analysis. Protein spots were excised from the dried silver-restained second-dimension gels and rehydrated for 20 min in 100 mM NH4HCO3. The gel spots were then destained for 20 min in a solution of 15 mM potassium ferricyanide and 50 mM thiosulfate, rinsed twice in water, and finally dehydrated in 100% acetonitrile until they turned opaque white. The spots were then dried in a vacuum centrifuge and subsequently rehydrated in a digestion solution consisting of 50 mM NH4HCO3, 5 mM CaCl2, and 0.1 μg μl−1 modified sequence-grade trypsin (Promega, Madison, WI, USA). After overnight incubation at 37 °C, the digestion was terminated in 5% trifluoroacetic acid for 20 min. Peptides were extracted three times (20 min each) with 5% trifluoroacetic acid in 50% acetonitrile, and the extracted peptides were pooled and dried in a vacuum centrifuge. The peptides were purified with ZipTip (Millipore, Billerica, MA, USA) using the manufacturer's protocols and analyzed using the MASCOT database software (Matrix Science, Tokyo, Japan).
Accelerator mass spectrometry analyses were performed with an NEC 1.5SDH-1 0.6-MV Pelletron AMS system (National Electrostatics Corporation, Tokyo, Japan) to determine the 14C/12C content ratio in the microsomal and cytosolic protein samples after dilution, as described previously.13 After measurements of protein contents and radioactivities of the analyte spots, the covalent binding levels were calculated.
Conclusions
In conclusion, the hepatotoxic substrates, flutamide and troglitazone, were activated by human livers to reactive intermediate(s) in vivo in humanized chimeric mice and could relatively nonspecifically bind to biomolecules. A zone analysis for intensity and imbalance in terms of covalent binding contents and target protein concentrations could provide important information on potential hepatotoxic effects. To test whether protein binding analyte spots of new medicines are unbalanced is an important concept, because if the protein binding data deviate greatly from an inverse relationship or if they are in the high covalent binding/high protein concentration zone, this might indicate a tendency to extensive nonspecific protein binding, and suggest the possibility of hepatotoxicity. Thus, the use of chimeric mice with humanized liver to realize in vivo covalent protein bindings of new drug substances in combination with the above-described zone analysis could prove to be a good predictor of drug toxicity in vivo in preclinical human studies.Acknowledgements
The authors thank Drs Tomigahara Yoshitaka, Makiko Shimizu, Norie Murayama, and Shinsaku Naito for their assistance and David Smallbones for his advice on English language. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2013–2017) (H.Y.).Notes and references
- J. H. Hoofnagle, J. Serrano, J. E. Knoben and V. J. Navarro, LiverTox: a website on drug-induced liver injury, Hepatology, 2013, 57, 873–874 CrossRef PubMed.
- L. P. James, P. R. Mayeux and J. A. Hinson, Acetaminophen-induced hepatotoxicity, Drug Metab. Dispos., 2003, 31, 1499–1506 CrossRef CAS PubMed.
- K. Kassahun, P. G. Pearson, W. Tang, I. McIntosh, K. Leung, C. Elmore, D. Dean, R. Wang, G. Doss and T. A. Baillie, Studies on the metabolism of troglitazone to reactive intermediates in vitro and in vivo. Evidence for novel biotransformation pathways involving quinone methide formation and thiazolidinedione ring scission, Chem. Res. Toxicol., 2001, 14, 62–70 CrossRef CAS PubMed.
- F. J. Gonzalez and A. M. Yu, Cytochrome P450 and xenobiotic receptor humanized mice, Annu. Rev. Pharmacol. Toxicol., 2006, 46, 41–64 CrossRef CAS PubMed.
- M. W. Powley, C. B. Frederick, F. D. Sistare and J. J. DeGeorge, Safety assessment of drug metabolites: implications of regulatory guidance and potential application of genetically engineered mouse models that express human P450s, Chem. Res. Toxicol., 2009, 22, 257–262 CrossRef CAS PubMed.
- C. Tateno, Y. Yoshizane, N. Saito, M. Kataoka, R. Utoh, C. Yamasaki, A. Tachibana, Y. Soeno, K. Asahina, H. Hino, T. Asahara, T. Yokoi, T. Furukawa and K. Yoshizato, Near completely humanized liver in mice shows human-type metabolic responses to drugs, Am. J. Pathol., 2004, 165, 901–912 CrossRef CAS.
- M. Katoh, C. Tateno, K. Yoshizato and T. Yokoi, Chimeric mice with humanized liver, Toxicology, 2008, 246, 9–17 CrossRef CAS PubMed.
- J. L. Gomez, A. Dupont, L. Cusan, M. Tremblay, R. Suburu, M. Lemay and F. Labrie, Incidence of liver toxicity associated with the use of flutamide in prostate cancer patients, Am. J. Med., 1992, 92, 465–470 CrossRef CAS.
- H. Yamazaki, S. Kuribayashi, T. Inoue, C. Tateno, Y. Nishikura, K. Oofusa, D. Harada, S. Naito, T. Horie and S. Ohta, Approach for in vivo protein bindings of 5-n-butyl-pyrazolo[1,5-a]pyrimidine bioactivated in chimeric mice with humanized liver by two-dimensional electrophoresis with accelerator mass spectrometry, Chem. Res. Toxicol., 2010, 23, 152–158 CrossRef CAS PubMed.
- S. Kuribayashi, K. Goto, S. Naito, T. Kamataki and H. Yamazaki, Human cytochrome P450 1A2 involvement in the formation of reactive metabolites from a species-specific hepatotoxic pyrazolopyrimidine derivative, 5-n-butyl-7-(3,4,5-trimethoxybenzoylamino)pyrazolo[1,5-a]pyrimidine, Chem. Res. Toxicol., 2009, 22, 323–331 CrossRef CAS PubMed.
- R. Goda, D. Nagai, Y. Akiyama, K. Nishikawa, I. Ikemoto, Y. Aizawa, K. Nagata and Y. Yamazoe, Detection of a new N-oxidized metabolite of flutamide, N-[4-nitro-3-(trifluoromethyl)phenyl]hydroxylamine, in human liver microsomes and urine of prostate cancer patients, Drug Metab. Dispos., 2006, 34, 828–835 CrossRef CAS PubMed.
- Y. Yamamoto, H. Yamazaki, T. Ikeda, T. Watanabe, H. Iwabuchi, M. Nakajima and T. Yokoi, Formation of a novel quinone epoxide metabolite of troglitazone with cytotoxicity to HepG2 cells, Drug Metab. Dispos., 2002, 30, 155–160 CrossRef CAS.
- T. Miyaoka, Y. Isono, K. Setani, K. Sakai, I. Yamada, Y. Sato, S. Gunji and T. Matsui, Bioanalysis works in the IAA AMS facility: Comparison of AMS analytical method with LSC method in human mass balance study, Nucl. Instrum. Methods Phys. Res., Sect. B, 2007, 259, 779–785 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4tx00068d |
| This journal is © The Royal Society of Chemistry 2015 |
