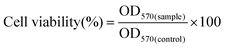Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biofunctionalized surface-modified silver nanoparticles for gene delivery†
Kishor
Sarkar
a,
Sovan Lal
Banerjee
b,
P. P.
Kundu
b,
Giridhar
Madras
 a and
Kaushik
Chatterjee
*c
a and
Kaushik
Chatterjee
*c
aDepartment of Chemical Engineering, Indian Institute of Science, Bangalore-560012, India
bDepartment of Polymer Science and Technology, University of Calcutta, 92 A. P. C. Road, Kolkata-700009, India
cDepartment of Materials Engineering, Indian Institute of Science, Bangalore-560012, India. E-mail: kchatterjee@materials.iisc.ernet.in; Tel: +91-80-2293-3408
First published on 1st June 2015
Abstract
Silver nanoparticles (AgNPs) find use in different biomedical applications including wound healing and cancer. We propose that the efficacy of the nanoparticles can be further augmented by using these particles for gene delivery applications. The objective of this work was to engineer biofunctionalized stable AgNPs with good DNA binding ability for efficient transfection and minimal toxicity. Herein, we report on the one-pot facile green synthesis of polyethylene glycol (PEG) stabilized chitosan-g-polyacrylamide modified AgNPs. The size of the PEG stabilized AgNPs was 38 ± 4 nm with a tighter size distribution compared to the unstabilized nanoparticles which showed bimodal distribution of particle sizes of 68 ± 5 nm and 7 ± 4 nm. To enhance the efficiency of gene transfection, the Arg–Gly–Asp–Ser (RGDS) peptide was immobilized on the silver nanoparticles. The transfection efficiency of AgNPs increased significantly after immobilization of the RGDS peptide reaching up to 42 ± 4% and 30 ± 3% in HeLa and A549 cells, respectively, and significantly higher than 34 ± 3% and 23 ± 2%, respectively, with the use of polyethyleneimine (25 kDa). These nanoparticles were found to induce minimal cellular toxicity. Differences in cellular uptake mechanisms with RGDS immobilization resulting in improved efficiency are elucidated. This study presents biofunctionalized AgNPs for potential use as efficient nonviral carriers for gene delivery with minimal cytotoxicity toward augmenting the therapeutic efficacy of AgNPs used in different biomedical products.
1. Introduction
Silver nanoparticles (AgNPs) have gained immense popularity in recent years for a variety of biotechnological biomedical applications owing to their unique antimicrobial, optical and electronic properties.1 The antibacterial properties of AgNPs have been utilized in different applications including consumer personal care products, water purification, and in medical devices. With the emergence of antibiotic resistant bacterial strains, the interest in AgNPs as an antimicrobial agent has increased significantly.2 The optical properties of AgNPs are better than those of gold nanoparticles (AuNPs) with higher plasmon excitation efficiency which may significantly aid in applications such as biosensing and bioimaging. AgNPs in wound healing products not only impart antimicrobial properties but have also been shown to facilitate the healing process through their anti-inflammatory activity3 and differential effects on keratinocytes and fibroblasts.4 More recently, the use of AgNPs for treatment of cancer has been proposed due to their ability to induce apoptosis in cancer cells.5With ease of production, ability to tailor particle shape and size, and flexibility in surface modification and bioconjugation, AgNPs will continue to find use in different biomedical applications. However, the toxicity associated with the use of nanoparticles in general and AgNPs in particular is a concern. There is no clear consensus on the toxicity of AgNPs in the literature. Whereas many studies have reported cytotoxicity,6–8 others have observed minimal toxicity up to high particle concentration.9 It appears that the concentration, size and surface modification, etc., determine the toxicology of AgNPs. However, the clinical success of wound care products containing AgNPs suggests that AgNPs at low concentration may be used in vivo with minimal toxicity.
We propose that the biomedical efficacy of the AgNPs in applications such as cancer therapy and wound healing could be further augmented by utilizing these nanoparticles to deliver known therapeutic genes. Thus, it is envisaged that such a strategy could yield better therapies that synergistically leverage the various advantages offered by AgNPs and in addition utilize them for gene delivery. However, AgNPs lack the ability to bind double stranded DNA (dsDNA). Therefore, the specific objective of this study was to prepare surface functionalized AgNPs to bind dsDNA and demonstrate their ability to deliver genes efficiently following cellular uptake. Unlike AuNPs, AgNPs have not been studied for gene delivery applications. A few recent studies have reported on the preparation of DNA stabilized AgNPs.10,11 Transfection efficiency increased significantly after metallization of plasmid DNA (pDNA) with minimal toxicity.10,11 Typically, AgNPs are synthesized by chemical reduction of silver ions using toxic reducing agents such as sodium borohydride, hydrazine, formaldehyde, N,N-dimethylformamide, and elemental hydrogen in the presence of organic solvents, or non-biodegradable stabilizing agents, which are potentially hazardous for use in biomedical applications.12–14 One-pot facile green routes to synthesize metallic nanoparticles avoiding the use of toxic chemicals during synthesis are preferred for nanomedicine.
Chitosan is a cationic linear polysaccharide consisting of both acetylated and deacetylated (1 → 4)-2-amino-2-deoxy-β-D-glucan repeating units.15 Chitosan can be used as a nonviral carrier for gene delivery because it is cationic in nature, biocompatible, biodegradable and low toxic.16–18 However, the poor water solubility of chitosan due to its strong intramolecular hydrogen bonding is the main drawback.19 Due to its low pKa value (pKa 6.5), chitosan is only soluble at pH < 6.5.20 However, its water solubility can be improved through functionalization of its primary amine and hydroxyl groups. Different types of water soluble chitosan derivatives such as PEGylated (polyethylene glycol) chitosan,21 carboxylated chitosan,22 succinylated chitosan,23 and dendronized chitosan24 have been synthesized.
Targeted gene delivery is essential to improve transfection efficiency. Various targeting ligands such as folic acid, HIV-1 twin arginine translocation (TAT) peptides, mannose and Arg–Gly–Asp (RGD) peptides have been conjugated to nonviral vectors to improve the transfection efficiency through receptor mediated internalization of the plasma membrane followed by nucleus targeting.25–30 Arg–Gly–Asp–Ser (RGDS) motifs in extracellular matrix proteins are recognized to facilitate ligation of integrins to facilitate cell adhesion.31 Among the targeting ligands, the RGDS motif has been found to have high affinity towards cancer cells due to the presence of abundant αv integrins on cancer cells.32 Similarly, the αv integrins are up-regulated in migrating keratinocytes and endothelial cells during wound healing.33,34
In this work, we report on the one-pot facile green synthesis of AgNPs through reduction of silver nitrate solution by water soluble chitosan-g-polyacrylamide (CTS-g-PAAm) copolymers in the presence of PEG to stabilize the AgNPs. As it has been previously utilized for gene delivery, chitosan was used to modify the surface of the AgNPs to efficiently bind DNA for gene delivery. For use in gene delivery, the formation of AgNP/pDNA complexes was characterized. In vitro cytotoxicity and transfection efficiency of AgNPs were studied in HeLa and A549 cells. To improve the transfection efficiency, these PEG stabilized CTS-g-PAAm AgNPs were modified using the RGDS peptide and the resulting changes in cellular uptake mechanisms were evaluated.
2. Experimental
2.1 Materials
Chitosan was purchased from Himedia, India. Its molecular weight (MW) was 365 kDa, determined by gel permeation chromatography using a Waters 2414 RI detector and a PC2 separation module (Waters, USA) against PEG standard calibration curves. The degree of deacetylation (DDA) determined by the potentiometric method was 80%.15 PEG (5000 Da), acrylamide (AAm), ammonium persulphate (APS) and silver nitrate (AgNO3) were purchased from Merck, India. RGDS peptides, ethidium bromide (EtBr), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and branched PEI (25 kDa) were obtained from Sigma-Aldrich, USA. Dulbecco's modified Eagle's medium (DMEM), penicillin–streptomycin, trypsin, and fetal bovine serum (FBS) were purchased from Gibco, Life Technologies. pDNA encoding green fluorescence (LifeAct-TagGFP2) under the control of the cytomegalovirus promoter/enhancer was propagated in Escherichia coli (E. coli). pDNA was isolated using a midiprep pDNA isolation Kit (Qiagen, USA) according to the manufacturer's instructions and stored at −20 °C for future use. All other reagents were used as received.2.2 Synthesis of CTS-g-PAAm copolymers
The CTS-g-PAAm copolymer was synthesized following a previous report with slight modifications.23 Briefly, 1.0 g of chitosan was dissolved in 60 mL of 1% acetic acid solution in a two neck round bottom flask followed by nitrogen purging for 30 min. 0.01 g of APS dissolved in 5 mL of water was added to the chitosan solution and the reaction mixture was continuously stirred for 10 min at 60 °C. 2.0 g of acrylamide monomer dissolved in water was added drop-by-drop to the above reaction mixture and the reaction was carried out for additional 1 h at 60 °C under a nitrogen atmosphere. Finally, the reaction mixture was cooled to room temperature followed by the addition of ethanol to stop the reaction. The final product was obtained by filtration followed by Soxhlet extraction with ethanol for 48 h to remove the polyacrylamide homopolymer.2.3 Preparation and characterization of AgNPs
CTS-g-PAAm coated AgNPs (hereafter designated as CTS-g-PAAm@AgNPs) were prepared by the one-pot green synthesis method without using any reducing agent where the copolymer acts as a self reducing agent (Fig. 1). To synthesize the nanoparticles, 0.5 g of CTS-g-PAAm copolymer dissolved in 15 mL of double distilled water and 10 mL of 0.01 M silver nitrate solution were taken in a three necked round bottomed flask and the whole mixture was stirred at 60 °C under a nitrogen atmosphere for 2 h. In another batch of reaction, CTS-g-PAAm@AgNPs were prepared as above in the presence of PEG to stabilize the nanoparticles (hereafter designated as PEG/CTS-g-PAAm@AgNPs) (Fig. 1). Thereafter, the RGDS peptide at different contents with respect to the copolymer (1%, 3%, 6% and 10%) was mixed with nanoparticles under constant stirring for 12 h to prepare RGDS peptide decorated CTS-g-PAAm@AgNPs (Fig. 1). The free peptides were removed by dialysis.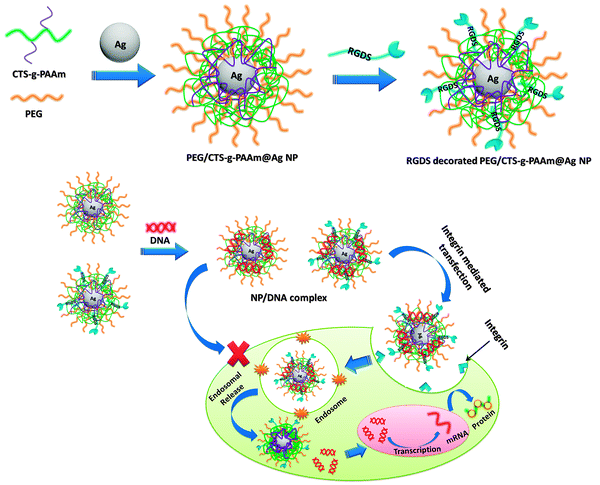 | ||
| Fig. 1 Schematic representation of the preparation of biofunctionalized AgNPs and the resultant receptor mediated cellular uptake and transfection. | ||
The formation of the polymer stabilized AgNPs was confirmed by measuring the UV absorbance at different time intervals during the reaction using a UV-vis spectrophotometer (Optizen Pop Bio, Mecasys). The hydrodynamic diameter of nanoparticles was measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instrument, UK) at 37 °C. A transmission electron microscope (TEM, JEOL JSM-7600 F) was used to measure the size and characterize the morphology of the AgNPs.
2.4 Preparation and characterization of AgNP/pDNA complexes
Polymer stabilized AgNP/pDNA complexes were prepared according to a previous report.35 AgNPs were diluted to 1 mg mL−1 concentration using MES buffer (pH 5.5) and filtered using a 0.22 μm filter (Millipore) before complex formation. pDNA was separately diluted in 25 mM of sodium sulfate solution at a concentration of 100 μg mL−1. The complexes were prepared by mixing equal volumes of nanoparticles and pDNA solution at different ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6
1, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8
1, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10
1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; AgNP/pDNA weight ratio) followed by vortex mixing for 15–30 s and incubated at room temperature for 30 min.
1; AgNP/pDNA weight ratio) followed by vortex mixing for 15–30 s and incubated at room temperature for 30 min.
The hydrodynamic diameter of AgNP/pDNA particles at different ratios was measured by DLS at a scattering angle (θ) of 90°. The zeta potential of the complexes was also determined using the same instrument. The morphology of the RGDS decorated polymer stabilized AgNP (10 wt% RGDS content)/pDNA complex at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was observed using TEM.
1 was observed using TEM.
2.5 Agarose gel electrophoresis assay
The binding ability of nanoparticles with pDNA was confirmed by agarose gel electrophoresis assay. Freshly prepared AgNP/pDNA complexes at different weight ratios were loaded in 0.8% agarose gel, prepared using TAE buffer containing EtBr (10 μg mL−1) as a DNA visualizer. The gel was run at 100 V for 45 min and subsequently the gel was imaged using a gel documentation system (Syngene, USA).2.6 Ethidium bromide assay
EtBr assay was carried out by following a previous report.17 Briefly, pDNA was stained with EtBr at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (EtBr
10 (EtBr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA molar ratio). Polymer stabilized AgNP/pDNA complexes at different weight ratios were added to this EtBr–DNA solution. After incubation at room temperature for 30 min, the fluorescence intensity of the solution was recorded using a Synergy HT multimode microplate reader (BioTek Instrument) with excitation and emission wavelengths of 510 and 605 nm, respectively. The fluorescence intensity of EtBr in the absence of DNA was considered as blank and the relative fluorescence intensities of EtBr–DNA solution in the presence of the nanoparticles were measured after subtracting the blank values. The effect of RGDS peptide concentration on DNA complexation ability of the polymer stabilized AgNPs was also assessed.
DNA molar ratio). Polymer stabilized AgNP/pDNA complexes at different weight ratios were added to this EtBr–DNA solution. After incubation at room temperature for 30 min, the fluorescence intensity of the solution was recorded using a Synergy HT multimode microplate reader (BioTek Instrument) with excitation and emission wavelengths of 510 and 605 nm, respectively. The fluorescence intensity of EtBr in the absence of DNA was considered as blank and the relative fluorescence intensities of EtBr–DNA solution in the presence of the nanoparticles were measured after subtracting the blank values. The effect of RGDS peptide concentration on DNA complexation ability of the polymer stabilized AgNPs was also assessed.
2.7 In vitro cytotoxicity assay
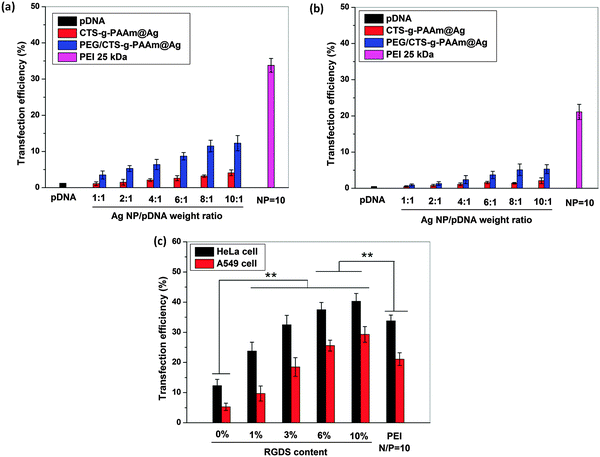
In vitro cytotoxicity of the different AgNP/pDNA complexes in HeLa and A549 cells was assessed by MTT assay. Cells were cultured in DMEM supplemented with 10% FBS. 5 × 103 cells per well were seeded in a 96-well plate for 24 h. Thereafter, the cells were treated with the polymer stabilized AgNP/pDNA complexes at different weight ratios and compared with the PEI–pDNA complex at a N/P ratio (nitrogen to phosphate ratio) of 10 as a positive control. The effect of RGDS peptide concentration on the cell toxicity of polymer stabilized AgNPs was also observed. After 24 h of treatment, 20 μL of MTT solution (5 mg mL−1) in 80 μL of DMEM culture medium was added to each well and incubated for 4 h in a CO2 incubator. The medium was replaced with 100 μL of DMSO to dissolve the formazan crystals. The absorbance was measured at 570 nm using a microplate reader. The cell viability(%) was determined by the following equation:where OD570(sample) and OD570(control) represent the absorbance values from the wells treated with medium containing nanoparticles and fresh growth medium only, respectively. All data are reported based on three measurements.
2.8 In vitro transfection studies
Cells were seeded in a 24 well plate at a density of 5 × 104 cells per well in 0.5 mL of complete culture medium and cultured until 70–80% confluency was reached. CTS-g-PAAm@AgNP/pDNA, PEG/CTS-g-PAAm@AgNP/pDNA and RGDS-decorated PEG/CTS-g-PAAm@AgNP/pDNA complexes at different weight ratios were freshly prepared, as described above, prior to the transfection. Polymer functionalized AgNP/pDNA complexes at different weight ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6
1, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8
1, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10
1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added into individual wells. 1 μg of DNA was used for each well. The transfection media were replaced by fresh complete medium after 6 h. Naked pDNA and PEI–pDNA (N/P = 10) were used as negative and positive controls, respectively. The green fluorescence protein (GFP) expression of the transfected cells was observed after 48 h of transfection using a fluorescence microscope (Olympus IX53, Japan). The transfection efficiency was further quantified by fluorescence-activated cell sorting (FACS, Becton-Dickinson) analysis. 104 events were counted for each sample.
1) were added into individual wells. 1 μg of DNA was used for each well. The transfection media were replaced by fresh complete medium after 6 h. Naked pDNA and PEI–pDNA (N/P = 10) were used as negative and positive controls, respectively. The green fluorescence protein (GFP) expression of the transfected cells was observed after 48 h of transfection using a fluorescence microscope (Olympus IX53, Japan). The transfection efficiency was further quantified by fluorescence-activated cell sorting (FACS, Becton-Dickinson) analysis. 104 events were counted for each sample.
2.9 Intracellular kinetics
The cellular uptake pathways of complexes were characterized by studying transfection at low temperature or in the presence of endocytic inhibitors. To block energy-dependent endocytosis, the cellular uptake study was performed at 4 °C for a 6 h period. Alternatively, cells were pre-incubated with endocytic inhibitors including genistein (100 mg mL−1), amiloride (10 mg mL−1), and chlorpromazine (10 mg mL−1) for 1 h prior to the addition of polymer stabilized AgNP/pDNA complexes. PEG/CTS-g-PAAm@AgNP/pDNA and RGDS decorated (10 wt% RGDS) PEG/CTS-g-PAAm@AgNP/pDNA complexes at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 containing 1 μg of DNA were transfected in both HeLa and A549 cells. The effect of exogenous RGDS peptides (100 nmol l−1) on transfection efficiency of RGDS peptide decorated nanoparticles was also observed. The cellular uptake level was determined by flow cytometry 24 h post transfection, as described above.
1 containing 1 μg of DNA were transfected in both HeLa and A549 cells. The effect of exogenous RGDS peptides (100 nmol l−1) on transfection efficiency of RGDS peptide decorated nanoparticles was also observed. The cellular uptake level was determined by flow cytometry 24 h post transfection, as described above.
2.10 Cellular uptake and intracellular distribution
In order to track the cellular uptake of DNA inside cells, pDNA was first labeled with Cy3 according to the protocol of a Label IT® Tracker™ intracellular nucleic acid localization kit (Mirus Bio, USA). HeLa cells were seeded on cover-glass slides with 12.5 mm diameter in 24 well plates at a density of 1 × 105 cells per well and cultured for 24 h. Cells were incubated with CTS-g-PAAm@AgNP/pDNA, PEG/CTS-g-PAAm@AgNP/pDNA and RGDS decorated (10 wt% RGDS) PEG/CTS-g-PAAm@AgNP/pDNA complexes at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 containing 1 μg of Cy3-labeled pDNA for 6 h. The PEI/pDNA complex (N/P ratio 10
1 containing 1 μg of Cy3-labeled pDNA for 6 h. The PEI/pDNA complex (N/P ratio 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used as positive control. The cells were washed with PBS and fixed with 3.7% (w/v) formaldehyde for 30 min and stained with diamidino phenylindole (DAPI) for 10 min. The intracellular distribution of complexes was imaged by confocal laser scanning microscopy (CLSM; Leica TCS Sp5).
1) was used as positive control. The cells were washed with PBS and fixed with 3.7% (w/v) formaldehyde for 30 min and stained with diamidino phenylindole (DAPI) for 10 min. The intracellular distribution of complexes was imaged by confocal laser scanning microscopy (CLSM; Leica TCS Sp5).
2.11 Statistical analysis
All the data are presented as mean ± standard deviation. Statistical differences were calculated using a one-way ANOVA and differences were considered significant for p values less than 0.05.3. Results and discussion
3.1 Synthesis and characterization of silver nanoparticles
Water soluble chitosan was prepared by grafting polyacrylamide with chitosan through free radical polymerization using ammonium persulphate (APS) as an initiator (Fig. 1). As shown in the inset of Fig. 2, the color of CTS-g-PAAm@AgNP and PEG/CTS-g-PAAm@AgNP solution changed from colorless to wine red to deep red with the progress of reaction time, which indicates the formation of AgNPs.36 The synthesis of AgNPs was further confirmed by measuring the surface plasmon resonance (SPR) bands of respective CTS-g-PAAm@AgNP and PEG/CTS-g-PAAm@AgNP solutions at different time intervals using UV-visible spectroscopy (Fig. 2). The characteristic SPR bands of silver were obtained for CTS-g-PAAm@AgNPs at 425 nm which confirm the formation of AgNPs.37 The peak was shifted to 417 nm in PEG/CTS-g-PAAm@AgNPs. For the PEG/CTS-g-PAAm@AgNP system, the characteristic SPR band appeared within 5 min of start of the reaction compared to that of the CTS-g-PAAm@AgNP system wherein the peak appeared after 15 min. Recently, Ahmad et al. prepared AgNPs using chitosan dissolved in aqueous acidic solution and the SPR band appeared after 1 h.38 In acidic pH, the lone pair of electrons on both nitrogen and oxygen atoms is less available for the reduction of silver ions, resulting in longer time to form AgNPs. In contrast, water soluble chitosan formed AgNPs rapidly due to the absence of such an impediment and hence this route is more economical. The intensity of the SPR band was much higher for the PEG modified particle after 2 h of reaction indicating that the rate of reduction by CTS-g-PAAm in conjugation with PEG was faster compared to CTS-g-PAAm alone. In the PEG/CTS-g-PAAm@AgNP system, both the PEG molecules and the CTS-g-PAAm copolymer participated in the reduction process whereas only the CTS-g-PAAm copolymer reduced the silver ion in the CTS-g-PAAm@AgNP system. The color intensity of the solution of PEG/CTS-g-PAAm@AgNP was much higher than that of CTS-g-PAAm@AgNP solution (Fig. 2).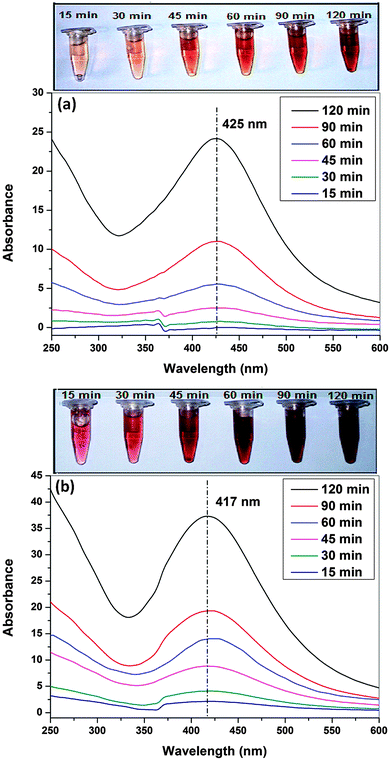 | ||
| Fig. 2 Ultraviolet-visible absorption spectra of CTS-g-PAAm@AgNPs (a) and PEG/CTS-g-PAAm@AgNPs (b) at different reaction times (15, 30, 45, 60, 90 and 120 min). | ||
The particle size and the morphology of the AgNPs are shown in Fig. S1 (ESI†). From DLS (Fig. S1a–d, ESI†), it was found that the PEG/CTS-g-PAAm system yielded nanoparticles with a unimodal size distribution of 38 ± 4 nm (Fig. S1c, ESI†). In contrast, the CTS-g-PAAm copolymer alone yielded particles with bimodal distribution of sizes 68 ± 5 nm and 7 ± 4 nm with a PDI (polydispersity index) value of 0.43 (Fig. S1a, ESI†). It was also found that the CTS-g-PAAm@AgNPs were more stable with unimodal distribution and showed a low PDI value (0.28) in the presence of PEG even after 30 days. After 30 days, the particle size of PEG/CTS-g-PAAm@AgNPs remained nearly unchanged at 40 ± 2 nm (Fig. S1d, ESI†) whereas CTS-g-PAAm@AgNPs aggregated to form bigger particles of size 140 ± 25 nm (Fig. S1b, ESI†). The particle size of AgNPs determined by DLS was slightly larger compared to TEM images as DLS measures hydrodynamic diameter in contrast to TEM which images the dried nanoparticles. Previously, PEG has been used to enhance the blood circulation stability of nonviral carriers which is an important consideration for successful gene therapy.39–41 From the TEM images, it was also found that the AgNPs were spherical in shape for both systems but the particle size of CTS-g-PAAm@AgNPs was larger than PEG stabilized CTS-g-PAAm@AgNPs corroborating the findings from DLS. TEM images also showed that CTS-g-PAAm@AgNPs aggregated after 30 days whereas PEG stabilized CTS-g-PAAm@AgNPs remained unchanged in size and shape.
3.2 Preparation of AgNP/pDNA complexes
The DNA binding capability of AgNPs was confirmed by agarose gel electrophoresis and EtBr assay as shown in Fig. S2 (ESI†). From Fig. S2b (ESI†), it was observed that PEG/CTS-g-PAAm@AgNPs were able to bind all DNA at the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio (polymer AgNP
1 weight ratio (polymer AgNP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA weight ratio) whereas some unbound DNA remained in the lane of CTS-g-PAAm@Ag/pDNA complexes at the same weight ratio (Fig. S2a, ESI†). AgNPs were more stable with smaller particle size in the presence of PEG molecules even after 30 days and as a result PEG stabilized AgNPs showed better DNA complexation capability due to the availability of more surface area compared to that of aggregated CTS-g-PAAm@AgNPs. The binding capacity of AgNPs with pDNA was further evaluated using the EtBr assay. In this assay, EtBr intercalates in the specific sites of DNA and gives a strong fluorescence signal. Any strong cationic compound such as the carrier developed herein may replace EtBr from the DNA–EtBr complex as it is a reversible interaction. As a consequence, the fluorescence intensity is lowered due to the displacement of EtBr by the incoming cationic compound. The relative decrease in the intensity with the decrease in fluorescence from the DNA–EtBr complex upon addition of the carrier is taken as a measure of its DNA binding capability. Fig. S2c (ESI†) shows that the maximum fluorescence intensity was obtained from the DNA–EtBr complex alone but the fluorescence intensity gradually decreased upon addition of the polymer coated AgNPs to the solution containing the DNA–EtBr complex. The fluorescence intensity significantly decreased with PEG/CTS-g-PAAm@AgNPs, and attained a constant value (∼10%) with further increase of weight ratio beyond 6
DNA weight ratio) whereas some unbound DNA remained in the lane of CTS-g-PAAm@Ag/pDNA complexes at the same weight ratio (Fig. S2a, ESI†). AgNPs were more stable with smaller particle size in the presence of PEG molecules even after 30 days and as a result PEG stabilized AgNPs showed better DNA complexation capability due to the availability of more surface area compared to that of aggregated CTS-g-PAAm@AgNPs. The binding capacity of AgNPs with pDNA was further evaluated using the EtBr assay. In this assay, EtBr intercalates in the specific sites of DNA and gives a strong fluorescence signal. Any strong cationic compound such as the carrier developed herein may replace EtBr from the DNA–EtBr complex as it is a reversible interaction. As a consequence, the fluorescence intensity is lowered due to the displacement of EtBr by the incoming cationic compound. The relative decrease in the intensity with the decrease in fluorescence from the DNA–EtBr complex upon addition of the carrier is taken as a measure of its DNA binding capability. Fig. S2c (ESI†) shows that the maximum fluorescence intensity was obtained from the DNA–EtBr complex alone but the fluorescence intensity gradually decreased upon addition of the polymer coated AgNPs to the solution containing the DNA–EtBr complex. The fluorescence intensity significantly decreased with PEG/CTS-g-PAAm@AgNPs, and attained a constant value (∼10%) with further increase of weight ratio beyond 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio. However, CTS-g-PAAm@AgNPs were not as effective in replacing EtBr from the DNA–EtBr complex reaching a relative intensity value of ∼20% even at a higher weight ratio of 10
1 weight ratio. However, CTS-g-PAAm@AgNPs were not as effective in replacing EtBr from the DNA–EtBr complex reaching a relative intensity value of ∼20% even at a higher weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Thus, these results suggest that the PEG/CTS-g-PAAm@AgNP exhibits higher DNA binding capacity even at low concentration and corroborate the findings from gel electrophoresis.
1. Thus, these results suggest that the PEG/CTS-g-PAAm@AgNP exhibits higher DNA binding capacity even at low concentration and corroborate the findings from gel electrophoresis.
3.3 Size and zeta potential of AgNP/pDNA complexes
The particle size of the AgNPs was measured after complexation with pDNA. The particle size and the zeta potential of AgNP/pDNA are shown in Fig. 3a and b, respectively. It was found that the size of both nanoparticles increased after complexation with pDNA compared to the uncomplexed nanoparticles. However, the particle size gradually decreased with increase in the weight ratio and the average particle size of PEG/CTS-g-PAAm@AgNP/pDNA complexes at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio was 100–150 nm. The size of CTS-g-PAAm@Ag/pDNA complexes was larger (250–300 nm) at the same weight ratio (Fig. 3a), which may be attributed to the aggregation of CTS-g-PAAm@AgNPs.
1 weight ratio was 100–150 nm. The size of CTS-g-PAAm@Ag/pDNA complexes was larger (250–300 nm) at the same weight ratio (Fig. 3a), which may be attributed to the aggregation of CTS-g-PAAm@AgNPs.
The overall zeta potential of polymer/pDNA complexes is known to affect cellular uptake of the complexes through electrostatic interaction between the overall net positive charge of the polymer/DNA complex and the negatively charged cell surface. The zeta potential of various AgNP/pDNA complexes was measured at different weight ratios, as shown in Fig. 3b. From Fig. 3b, it was found that the zeta potential of both AgNP/pDNA complexes was negative below 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio which indicates incomplete DNA complexation. The zeta potential of both complexes increased with increase in weight ratios and became constant above 8
1 weight ratio which indicates incomplete DNA complexation. The zeta potential of both complexes increased with increase in weight ratios and became constant above 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio. The PEG/CTS-g-PAAm@AgNP/pDNA complex showed the highest zeta potential of 30.5 ± 2.9 mV at a weight ratio of 10
1 weight ratio. The PEG/CTS-g-PAAm@AgNP/pDNA complex showed the highest zeta potential of 30.5 ± 2.9 mV at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 whilst the CTS-g-PAAm@Ag/pDNA complex showed only 15.4 ± 2.1 mV at the same weight ratio.
1 whilst the CTS-g-PAAm@Ag/pDNA complex showed only 15.4 ± 2.1 mV at the same weight ratio.
3.4 Effect of RGDS immobilization on AgNP/pDNA complexes
The RGDS peptide at different concentrations such as 1%, 3%, 6% and 10% was immobilized on PEG/CTS-g-PAAm@AgNPs to improve the uptake and resultant transfection efficiency (Fig. 1). The binding ability of RGDS peptide decorated PEG/CTS-g-PAAm@AgNPs with pDNA at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was evaluated using the EtBr assay for the carrier modified with different RGDS concentrations (1%, 3%, 6% and 10%) as shown in Fig. 3c. It was found that there were no marked differences in DNA binding before and after immobilization of the RGDS peptide on PEG/CTS-g-PAAm@AgNPs even at higher RGDS content. Thus, RGDS modification of the polymer coated AgNPs did not compromise the DNA binding ability of the carrier. The particle size and the zeta potential of PEG/CTS-g-PAAm@AgNP/pDNA complexes (Fig. 3d) also remained unchanged with a PDI value of 0.31 even at high RGDS peptide content. The TEM image (Fig. 3e) of RGDS decorated PEG/CTS-g-PAAm@AgNP/pDNA complexes at 10% peptide content shows that the complex was spherical in shape and its size increased after DNA complexation compared to the uncomplexed PEG/CTS-g-PAAm@AgNPs.
1 was evaluated using the EtBr assay for the carrier modified with different RGDS concentrations (1%, 3%, 6% and 10%) as shown in Fig. 3c. It was found that there were no marked differences in DNA binding before and after immobilization of the RGDS peptide on PEG/CTS-g-PAAm@AgNPs even at higher RGDS content. Thus, RGDS modification of the polymer coated AgNPs did not compromise the DNA binding ability of the carrier. The particle size and the zeta potential of PEG/CTS-g-PAAm@AgNP/pDNA complexes (Fig. 3d) also remained unchanged with a PDI value of 0.31 even at high RGDS peptide content. The TEM image (Fig. 3e) of RGDS decorated PEG/CTS-g-PAAm@AgNP/pDNA complexes at 10% peptide content shows that the complex was spherical in shape and its size increased after DNA complexation compared to the uncomplexed PEG/CTS-g-PAAm@AgNPs.
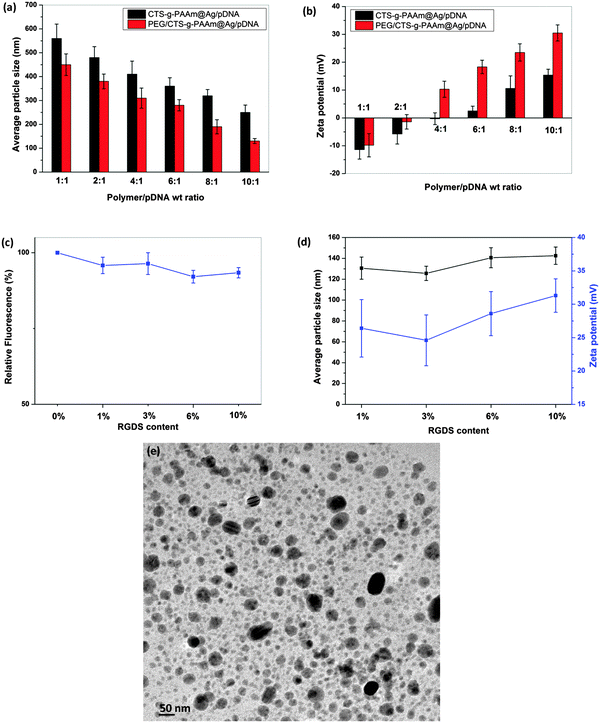
3.5 In vitro cytotoxicity
The cytotoxicity of AgNP/pDNA complexes at different weight ratios was evaluated in HeLa (Fig. 4a) and A549 cells (Fig. 4b) using MTT assay. These are some common cell lines widely used in this field to characterize efficiency and cytotoxicity of novel carriers for gene transfection. Cells without exposure to the nanoparticles were used as the control and the resultant viability was taken to be 100%. From Fig. 4a and b, it can be seen that both cell lines exhibited higher than 80% viability in the presence of PEG/CTS-g-PAAm@AgNP/pDNA complexes even at a high weight ratio. In previous studies, it was reported that small AgNPs are more toxic compared to larger particles.42,43 In another study, Chen et al.44 observed that the acute oral toxicity in mice was strongly dependent on the particle size distribution and the smallest particles showed the highest toxicity. In this work, the particle size of the AgNPs was 100–150 nm, which is in the optimal size range as proposed by previous studies and thus likely resulted in low toxicity. However, the CTS-g-PAAm@AgNP/pDNA complex showed marginally higher toxicity compared to that of the PEG/CTS-g-PAAm@AgNP/pDNA complex at all weight ratios although the toxicity of both AgNP/pDNA complexes increased with increase in carrier/DNA weight ratios. The higher toxicity of CTS-g-PAAm@AgNP/pDNA complexes may be attributed to the increased aggregation of CTS-g-PAAm@AgNPs on the cellular membrane. The PEI/pDNA complex at a N/P ratio of 10 was more toxic compared to both AgNP/pDNA complexes wherein the cell viability was less than 60%. Strong electrostatic interaction between the positive charge of primary amine groups of PEI and the negatively charged cell membrane may disrupt the cell membrane and thereby its functions by aggregating on the cell surface.45The cytotoxicity of RGDS peptide decorated PEG/CTS-g-PAAm@AgNP/pDNA complexes at an AgNP/pDNA weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with different peptide concentrations such as 1%, 3%, 6% and 10% was also evaluated by MTT assay on both cell lines as shown in Fig. 4c. It was found that there was no significant change in toxicity of the PEG/CTS-g-PAAm@AgNP/pDNA complex before and after RGDS peptide immobilization.
1 with different peptide concentrations such as 1%, 3%, 6% and 10% was also evaluated by MTT assay on both cell lines as shown in Fig. 4c. It was found that there was no significant change in toxicity of the PEG/CTS-g-PAAm@AgNP/pDNA complex before and after RGDS peptide immobilization.
3.6 In vitro transfection efficiency
The transfection efficiency of CTS-g-PAAm@AgNP/pDNA and PEG/CTS-g-PAAm@AgNP/pDNA complexes at different weight ratios ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 was studied in HeLa and A549 cells. pDNA was used as the negative control and the PEI (25 kDa)/pDNA complex at a N/P ratio of 10 served as the positive control.46 As shown in Fig. S3 (ESI†), PEG/CTS-g-PAAm@AgNP/pDNA complexes (Fig. S3b1–b4, ESI†) showed better transfection efficiency compared to CTS-g-PAAm@AgNP/pDNA complexes (Fig. S3a1–a4, ESI†) in HeLa cells. It was found that the fraction of GFP positive cells increased with increase in AgNP/pDNA weight ratio. The transfection efficiency of the PEG/CTS-g-PAAm@AgNP/pDNA complex at a weight ratio of 10
10 was studied in HeLa and A549 cells. pDNA was used as the negative control and the PEI (25 kDa)/pDNA complex at a N/P ratio of 10 served as the positive control.46 As shown in Fig. S3 (ESI†), PEG/CTS-g-PAAm@AgNP/pDNA complexes (Fig. S3b1–b4, ESI†) showed better transfection efficiency compared to CTS-g-PAAm@AgNP/pDNA complexes (Fig. S3a1–a4, ESI†) in HeLa cells. It was found that the fraction of GFP positive cells increased with increase in AgNP/pDNA weight ratio. The transfection efficiency of the PEG/CTS-g-PAAm@AgNP/pDNA complex at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was significantly increased after immobilization of the RGDS peptide (Fig. S3c1–c4, ESI†) and further increased with the increase in peptide content. The maximum fraction of GFP positive cells was obtained at 10% peptide content (Fig. S3c4, ESI†) which was higher than that of PEI at a N/P ratio of 10 (Fig. S3d2, ESI†). The transfection efficiency followed the same trend in A549 cell lines (Fig. S4, ESI†) although the fraction of GFP positive cells at all weight ratios of AgNP/pDNA complexes was lower compared to that of HeLa cells even in the presence of the RGDS peptide. There was no discernable change in the cellular morphology of both cell lines after transfection compared to untransfected cells.
1 was significantly increased after immobilization of the RGDS peptide (Fig. S3c1–c4, ESI†) and further increased with the increase in peptide content. The maximum fraction of GFP positive cells was obtained at 10% peptide content (Fig. S3c4, ESI†) which was higher than that of PEI at a N/P ratio of 10 (Fig. S3d2, ESI†). The transfection efficiency followed the same trend in A549 cell lines (Fig. S4, ESI†) although the fraction of GFP positive cells at all weight ratios of AgNP/pDNA complexes was lower compared to that of HeLa cells even in the presence of the RGDS peptide. There was no discernable change in the cellular morphology of both cell lines after transfection compared to untransfected cells.
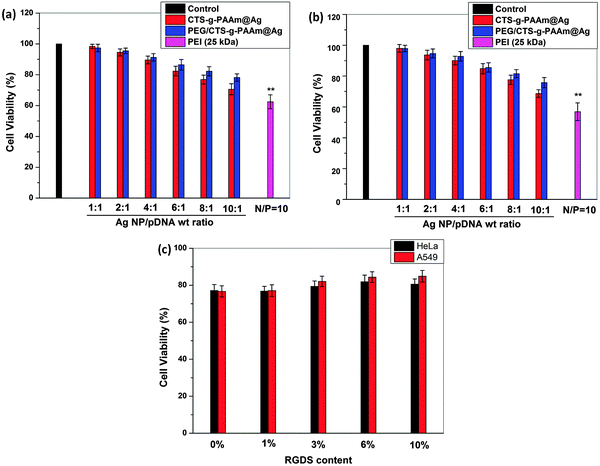
The transfection efficiency of various polymer stabilized AgNP/pDNA complexes at different weight ratios in HeLa and A549 cells was quantified by flow cytometry as shown in Fig. 5. It was observed that the transfection efficiency of both AgNP/pDNA complexes increased gradually with increase in the AgNP/pDNA weight ratio although the transfection efficiency of PEG/CTS-g-PAAm@AgNP/pDNA complexes was higher compared to that of CTS-g-PAAm@AgNP/pDNA complexes at all weight ratios. The PEG/CTS-g-PAAm@AgNP/pDNA complex at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 showed ∼15% transfection efficiency whereas the CTS-g-PAAm@AgNP/pDNA complex showed only ∼5% efficiency at the same weight ratio against HeLa cell lines (Fig. 5a). The transfection efficiency of both kinds of polymer stabilized AgNP/pDNA complexes at the same weight ratio was only ∼6% and ∼2.5%, respectively against A549 cell lines (Fig. 5b). Here, the particle size of AgNPs played an important role in transfection efficiency. It was found PEG/CTS-g-PAAm formed smaller particles compared to CTS-g-PAAm alone and resulted in higher transfection efficiency. Previous studies reported that the particle size of 100–200 nm results in good transfection efficiency.16,35 The transfection efficiency of PEG/CTS-g-PAAm@AgNP/pDNA complexes was significantly increased after immobilization of the RGDS peptide on PEG/CTS-g-PAAm@AgNPs in both cell lines corroborating the results from fluorescence microscopy reported above. Previous reports showed that the conjugation of RGD peptides to nonviral carriers significantly increases the transfection efficiency.28–30 The presence of net positive charges on the carrier/pDNA complex is believed to facilitate nonspecific interaction with negatively charged cell membranes through electrostatic interaction followed by internalization into the cell. Conjugation of the integrin binding sequence RGD peptide improves uptake and reduces the nonspecific cellular internalization resulting in higher transfection efficiency. The transfection efficiency of PEG/CTS-g-PAAm@AgNP/pDNA complexes at a weight ratio of 10
1 showed ∼15% transfection efficiency whereas the CTS-g-PAAm@AgNP/pDNA complex showed only ∼5% efficiency at the same weight ratio against HeLa cell lines (Fig. 5a). The transfection efficiency of both kinds of polymer stabilized AgNP/pDNA complexes at the same weight ratio was only ∼6% and ∼2.5%, respectively against A549 cell lines (Fig. 5b). Here, the particle size of AgNPs played an important role in transfection efficiency. It was found PEG/CTS-g-PAAm formed smaller particles compared to CTS-g-PAAm alone and resulted in higher transfection efficiency. Previous studies reported that the particle size of 100–200 nm results in good transfection efficiency.16,35 The transfection efficiency of PEG/CTS-g-PAAm@AgNP/pDNA complexes was significantly increased after immobilization of the RGDS peptide on PEG/CTS-g-PAAm@AgNPs in both cell lines corroborating the results from fluorescence microscopy reported above. Previous reports showed that the conjugation of RGD peptides to nonviral carriers significantly increases the transfection efficiency.28–30 The presence of net positive charges on the carrier/pDNA complex is believed to facilitate nonspecific interaction with negatively charged cell membranes through electrostatic interaction followed by internalization into the cell. Conjugation of the integrin binding sequence RGD peptide improves uptake and reduces the nonspecific cellular internalization resulting in higher transfection efficiency. The transfection efficiency of PEG/CTS-g-PAAm@AgNP/pDNA complexes at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 reached 43% and 32% in HeLa and A549 cells, respectively, whereas PEI at a N/P ratio of 10 showed only 34% and 23% efficiency, respectively.
1 reached 43% and 32% in HeLa and A549 cells, respectively, whereas PEI at a N/P ratio of 10 showed only 34% and 23% efficiency, respectively.
3.7 Cellular uptake pathways
The cytoplasmic compartment is a major barrier and the gene transfection efficiency of a nonviral vector depends on the intracellular kinetics such as the internalization pathway and the endosomal escape mechanism.47 We studied the intracellular kinetics of undecorated and RGDS decorated PEG/CTS-g-PAAm@AgNPs in the presence of different known endocytotic inhibitors such as chlorpromazine (to inhibit clathrin mediated endocytosis, CME), genistein (to inhibit caveolae mediated endocytosis) and amiloride (to inhibit macropinocytosis). The transfection was also carried out at low temperature (4 °C) to study energy dependence of the uptake and in the presence of soluble RGDS peptides to elucidate the role, if any, of integrin receptors in the uptake leading to transfection.Fig. S5a (ESI†) shows that the transfection efficiency of undecorated PEG/CTS-g-PAAM@AgNP/pDNA complexes was significantly attenuated in the presence of chlorpromazine and remained largely unaffected in the presence of exogenous RGDS in both HeLa and A549 cells indicating that CME was the primary pathway in its uptake. Interestingly, the transfection was significantly reduced by genistein and soluble RGDS peptides for the RGDS modified PEG/CTS-g-PAAM@AgNPs (Fig. S5b, ESI†). This indicates that the cellular uptake was mediated by the caveolae-dependent pathway and involved engagement of integrin receptors. It is also observed that the cellular uptake was energy independent and was not mediated by macropinocytosis. Previous reports reveal that if the cellular uptake of a nonviral carrier occurs through the CME pathway, the complex has to pass through acidic and degradative lysosomal compartments leading to reduced transfection efficiency.48 It is reported that ligand conjugation may endow the vector-specific receptors targeting capability and reduce non-specific contact delivery functions.49 Higher transfection efficiency of RGDS decorated PEG/CTS-g-PAAM@AgNPs can be attributed to the cellular uptake by caveolae-mediated endocytosis and binding to the integrin.
3.8 Intracellular distribution
To observe the intracellular uptake of pDNA complexed with PEG/CTS-g-PAAm@AgNPs at a weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the presence and the absence of RGDS conjugation, transfection experiments were performed by live cell imaging using Cy3 labeled DNA (red) in HeLa cells (blue nuclei), as shown in Fig. S6 (ESI†). There was no red fluorescence around the nucleus with Cy3 labeled DNA alone, which was used as the negative control (Fig. S6a1, ESI†). However, a few fluorescent red particles were observed for PEG/CTS-g-PAAm@AgNPs (Fig. S6b1, ESI†). The number of red particles was markedly increased for RGDS conjugated PEG/CTS-g-PAAm@AgNPs (Fig. S6c1, ESI†) compared to PEI (25 kDa) used as positive control (Fig. S6d1, ESI†). For the PEG/CTS-g-PAAm@AgNP/pDNA complex, most of the pDNA remained in the cytoplasm after 4 h of transfection resulting in lower transfection efficiency. In contrast, most of the DNA particles were found to have reached the cell nuclei when RGDS decorated PEG/CTS-g-PAAm@AgNP/pDNA complexes (Fig. S6c3, ESI†) were used and even more compared to the PEI/pDNA complexes (Fig. S6d3, ESI†). These trends in colocalization of pDNA and the nucleus closely follow the trends in transfection efficiency of the different carriers studied herein and thus elucidate the underlying mechanism resulting in higher transfection efficiency of the RGDS decorated PEG/CTS-g-PAAm@AgNP/pDNA complexes.
1 in the presence and the absence of RGDS conjugation, transfection experiments were performed by live cell imaging using Cy3 labeled DNA (red) in HeLa cells (blue nuclei), as shown in Fig. S6 (ESI†). There was no red fluorescence around the nucleus with Cy3 labeled DNA alone, which was used as the negative control (Fig. S6a1, ESI†). However, a few fluorescent red particles were observed for PEG/CTS-g-PAAm@AgNPs (Fig. S6b1, ESI†). The number of red particles was markedly increased for RGDS conjugated PEG/CTS-g-PAAm@AgNPs (Fig. S6c1, ESI†) compared to PEI (25 kDa) used as positive control (Fig. S6d1, ESI†). For the PEG/CTS-g-PAAm@AgNP/pDNA complex, most of the pDNA remained in the cytoplasm after 4 h of transfection resulting in lower transfection efficiency. In contrast, most of the DNA particles were found to have reached the cell nuclei when RGDS decorated PEG/CTS-g-PAAm@AgNP/pDNA complexes (Fig. S6c3, ESI†) were used and even more compared to the PEI/pDNA complexes (Fig. S6d3, ESI†). These trends in colocalization of pDNA and the nucleus closely follow the trends in transfection efficiency of the different carriers studied herein and thus elucidate the underlying mechanism resulting in higher transfection efficiency of the RGDS decorated PEG/CTS-g-PAAm@AgNP/pDNA complexes.
The immobilization of RGDS on PEG/CTS-g-PAAm@AgNPs imparts cell targeting capability of recognizing and binding to the integrin receptor and consequently enhancing the cellular uptake followed by higher transfection efficiency. Previously, Chen et al.50 also showed the enhancement of siRNA delivery of liposome after conjugation of the RGD peptide with liposome. In another study, Singh et al.51 showed that the surface functionalization of poly(lactide-co-glycolide) nanoparticles by the RGD peptide enhanced the nanoparticle delivery to the neovascular eye compared to the unmodified nanoparticles.
The use of AgNPs in wound healing and cancer treatment has been proposed in recent years. As proof of principle we demonstrate in this study that AgNPs may be modified for gene therapy with minimal cytotoxicity. Such AgNP-based non-viral carriers could be developed for use in gene therapy wherein the anti-cancer, anti-microbial and anti-inflammatory properties of the particles can be further augmented by targeted delivery of known therapeutic genes for cancer treatment and wound healing.52,53 Further studies are warranted to not only evaluate their efficacy for treatment of such diseases but also to fully characterize potential concerns of cytotoxicity, genotoxicity and elimination from the human body prior to safe clinical use.
4. Conclusions
AgNPs were synthesized by a one-pot, facile green synthesis method using a water soluble CTS-g-PAAm copolymer. The synthesized nanoparticles remained stable up to 30 days after PEG modification. It was found that the DNA complexation capability of polymer modified AgNPs increases with increase in the AgNP/pDNA weight ratio. PEG stabilized nanoparticles showed enhanced DNA complexation. The transfection efficiency of AgNPs was significantly increased compared to that of PEI after RGDS immobilization on AgNPs with minimal toxicity. Therefore, RGDS decorated PEG/CTS-g-PAAm@AgNPs may be used as an efficient nonviral carrier in gene therapy to augment the efficacy of medical products and therapies utilizing AgNPs to treat different diseases.Acknowledgements
The authors acknowledge Prof. Sandhya S. Visweswariah, IISc for providing pDNA. This work was funded by the Department of Science and Technology (DST), India. K.S. was supported by the D. S. Kothari fellowship (BSR/EN/13-14/0005) from the University Grants Commission (UGC), India. K.C. acknowledges the Ramanujan fellowship from DST.References
- A. Ravindran, P. Chandran and S. S. Khan, Colloids Surf., B, 2013, 105, 342–352 CrossRef CAS PubMed.
- J. B. Wright, K. Lam and R. E. Burrell, Am. J. Infect. Control, 1998, 26, 572–577 CrossRef CAS PubMed.
- K. K. Wong, S. O. Cheung, L. Huang, J. Niu, C. Tao, C. M. Ho, C. M. Che and P. K. Tam, ChemMedChem, 2009, 4, 1129–1135 CrossRef CAS PubMed.
- X. Liu, P.-y. Lee, C.-m. Ho, V. C. H. Lui, Y. Chen, C.-m. Che, P. K. H. Tam and K. K. Y. Wong, ChemMedChem, 2010, 5, 468–475 CrossRef CAS PubMed.
- X. Liu, W. He, Z. Fang, A. Kienzle and Q. Feng, J. Biomed. Nanotechnol., 2014, 10, 1277–1285 CrossRef CAS PubMed.
- P. Gopinath, S. K. Gogoi, A. Chattopadhyay and S. S. Ghosh, Nanotechnology, 2008, 19, 075104 CrossRef CAS PubMed.
- Y.-H. Hsin, C.-F. Chen, S. Huang, T.-S. Shih, P.-S. Lai and P. J. Chueh, Toxicol. Lett., 2008, 179, 130–139 CrossRef CAS PubMed.
- E.-J. Park, J. Yi, Y. Kim, K. Choi and K. Park, Toxicol. In Vitro, 2010, 24, 872–878 CrossRef CAS PubMed.
- M. E. Samberg, E. G. Loboa, S. J. Oldenburg and N. A. Monteiro-Riviere, Nanomedicine, 2012, 7, 1197–1209 CrossRef CAS PubMed.
- Y. Tao, E. Ju, J. Ren and X. Qu, Chem. Commun., 2013, 49, 9791–9793 RSC.
- D. V. Berdnikova, H. Ihmels, H. Schonherr, M. Steuber and D. Wesner, Org. Biomol. Chem., 2015, 13, 3766–3770 CAS.
- K. M. Abou El-Nour, A. Eftaiha, A. Al-Warthan and R. A. Ammar, Arabian J. Chem., 2010, 3, 135–140 CrossRef CAS PubMed.
- P. Van Dong, C. H. Ha and J. Kasbohm, Int. Nano Lett., 2012, 2, 1–9 CrossRef.
- T. Quang Huy, N. Van Quy and L. Anh-Tuan, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2013, 4, 033001 CrossRef.
- K. Sarkar, R. Srivastava, U. Chatterji and P. Kundu, J. Appl. Polym. Sci., 2011, 121, 2239–2249 CrossRef CAS PubMed.
- K. Sarkar, A. Chatterjee, G. Chakraborti and P. P. Kundu, Carbohydr. Polym., 2013, 98, 596–606 CrossRef CAS PubMed.
- K. Sarkar, M. Debnath and P. Kundu, Carbohydr. Polym., 2013, 92, 2048–2057 CrossRef CAS PubMed.
- P. P. Kundu and K. Sarkar, in Biopolymers: Biomedical and Environmental Applications, ed. S. Kalia and L. Avérous, John Wiley & Sons, Inc., 2011, pp. 575–603 Search PubMed.
- K. Sarkar and P. P. Kundu, Int. J. Biol. Macromol., 2012, 51, 859–867 CrossRef CAS PubMed.
- K. Tømmeraas, M. Köping-Höggård, K. M. Vårum, B. E. Christensen, P. Artursson and O. Smidsrød, Carbohydr. Res., 2002, 337, 2455–2462 CrossRef.
- J. Du and Y.-L. Hsieh, Cellulose, 2007, 14, 543–552 CrossRef CAS PubMed.
- P. Mukhopadhyay, K. Sarkar, S. Soam and P. Kundu, J. Appl. Polym. Sci., 2013, 129, 835–845 CrossRef CAS PubMed.
- P. Mukhopadhyay, K. Sarkar, S. Bhattacharya, A. Bhattacharyya, R. Mishra and P. P. Kundu, Carbohydr. Polym., 2014, 112, 627–637 CrossRef CAS PubMed.
- P. Mukhopadhyay, K. Sarkar, S. Bhattacharya, R. Mishra and P. Kundu, RSC Adv., 2014, 4, 43890–43902 RSC.
- V. B. Morris, C. K. S. Pillai and C. P. Sharma, Polym. Int., 2011, 60, 1097–1106 CrossRef CAS PubMed.
- H. Y. Nam, J. Kim, S. Kim, J. W. Yockman, S. W. Kim and D. A. Bull, Biomaterials, 2011, 32, 5213–5222 CrossRef CAS PubMed.
- S. Mahor, B. C. Dash, S. O'Connor and A. Pandit, Bioconjugate Chem., 2012, 23, 1138–1148 CrossRef CAS PubMed.
- P. Erbacher, J. Remy and J. Behr, Gene Ther., 1999, 6, 138–145 CrossRef CAS PubMed.
- X.-L. Wang, R. Xu, X. Wu, D. Gillespie, R. Jensen and Z.-R. Lu, Mol. Pharmaceutics, 2009, 6, 738–746 CrossRef CAS PubMed.
- K. Kunath, T. Merdan, O. Hegener, H. Häberlein and T. Kissel, J. Gene Med., 2003, 5, 588–599 CrossRef CAS PubMed.
- R. O. Hynes, Cell, 2002, 110, 673–687 CrossRef CAS.
- U. K. Marelli, F. Rechenmacher, T. R. A. Sobahi, C. Mas-Moruno and H. Kessler, Front. Oncol., 2013, 3, 222 Search PubMed.
- H. Larjava, T. Salo, K. Haapasalmi, R. Kramer and J. Heino, J. Clin. Invest., 1993, 92, 1425 CrossRef CAS PubMed.
- M. Christofidou-Solomidou, M. Bridges, G. F. Murphy, S. M. Albelda and H. M. DeLisser, Am. J. Pathol., 1997, 151, 975 CAS.
- K. Sarkar and P. Kundu, Carbohydr. Polym., 2013, 98, 495–504 CrossRef CAS PubMed.
- S. P. Chaplot and I. D. Rupenthal, J. Pharm. Pharmacol., 2014, 66, 542–556 CrossRef CAS PubMed.
- K. G. Stamplecoskie and J. C. Scaiano, J. Am. Chem. Soc., 2010, 132, 1825–1827 CrossRef CAS PubMed.
- M. B. Ahmad, M. Y. Tay, K. Shameli, M. Z. Hussein and J. J. Lim, Int. J. Mol. Sci., 2011, 12, 4872–4884 CrossRef PubMed.
- T. Merdan, K. Kunath, H. Petersen, U. Bakowsky, K. H. Voigt, J. Kopecek and T. Kissel, Bioconjugate Chem., 2005, 16, 785–792 CrossRef CAS PubMed.
- S. Mishra, P. Webster and M. E. Davis, Eur. J. Cell Biol., 2004, 83, 97–111 CrossRef CAS PubMed.
- M. Neu, O. Germershaus, M. Behe and T. Kissel, J. Controlled Release, 2007, 124, 69–80 CrossRef CAS PubMed.
- C. Carlson, S. M. Hussain, A. M. Schrand, L. K. Braydich-Stolle, K. L. Hess, R. L. Jones and J. J. Schlager, J. Phys. Chem. B, 2008, 112, 13608–13619 CrossRef CAS PubMed.
- G. Oberdörster, E. Oberdörster and J. Oberdörster, Environ. Health Perspect., 2005, 823–839 CrossRef.
- Z. Chen, H. Meng, G. Xing, C. Chen, Y. Zhao, G. Jia, T. Wang, H. Yuan, C. Ye and F. Zhao, Toxicol. Lett., 2006, 163, 109–120 CrossRef CAS PubMed.
- S. M. Moghimi, P. Symonds, J. C. Murray, A. C. Hunter, G. Debska and A. Szewczyk, Mol. Ther., 2005, 11, 990–995 CrossRef CAS PubMed.
- B. Lu, X.-D. Xu, X.-Z. Zhang, S.-X. Cheng and R.-X. Zhuo, Biomacromolecules, 2008, 9, 2594–2600 CrossRef CAS PubMed.
- L. Yin, Z. Song, K. H. Kim, N. Zheng, N. P. Gabrielson and J. Cheng, Adv. Mater., 2013, 25, 3063–3070 CrossRef CAS PubMed.
- J. Rejman, A. Bragonzi and M. Conese, Mol. Ther., 2005, 12, 468–474 CrossRef CAS PubMed.
- A. G. Schätzlein, BioMed Res. Int., 2003, 2003, 149–158 Search PubMed.
- C.-W. Chen, D.-W. Lu, M.-K. Yeh, C.-Y. Shiau and C.-H. Chiang, Int. J. Nanomed., 2011, 6, 2567 CrossRef CAS PubMed.
- S. Singh, H. Grossniklaus, S. Kang, H. Edelhauser, B. Ambati and U. Kompella, Gene Ther., 2009, 16, 645–659 CrossRef CAS PubMed.
- B. Sharma, W. Ma, I. Adjei, J. Panyam, S. Dimitrijevic and V. Labhasetwar, Drug Delivery Transl. Res., 2011, 1, 43–52 CrossRef CAS PubMed.
- S. A. Eming, T. Krieg and J. M. Davidson, Clin. Dermatol., 2007, 25, 79–92 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5tb00614g |
| This journal is © The Royal Society of Chemistry 2015 |