Open Access Article
Open Access ArticleCompartmentalisation of enzymes for cascade reactions through biomimetic layer-by-layer mineralization†
Gousia
Begum
a,
W. Brandon
Goodwin
b,
Ben M.
deGlee
b,
Kenneth H.
Sandhage
b and
Nils
Kröger
*ac
aB CUBE Center for Molecular Bioengineering, Dresden Institute of Technology (TU Dresden), Dresden, Germany. E-mail: kroeger@bcube-dresden.de
bSchool of Materials Science and Engineering, Georgia Institute of Technology, Atlanta, GA, USA
cDepartment of Chemistry and Food Chemistry, Dresden Institute of Technology (TU Dresden), Dresden, Germany
First published on 26th May 2015
Abstract
Cellular metabolic pathways are paradigms for the rapid and waste-free conversion of molecules into useful products through multiple enzyme-catalyzed steps (cascade reactions). Attempts to establish efficient cascade reactions for technological applications have focused on mimicking nature's high degree of organization by controlling the positioning of enzymes through immobilization in tailor-made compartments. The present work utilized peptide-mediated layer-by-layer mineralization as a facile and generic method for the compartmentalisation of multi-enzyme systems in nanoscale silica layers. It is demonstrated that, in a multilayer system, the overall rate of the reaction cascade was primarily affected by the placement of the enzyme catalyzing the first step, with the placement of the enzyme possessing the lowest catalytic efficiency also being an important factor. As the rate-limiting enzymes were positioned closer to the external silica surface, the overall rate of cascade reactions increased. Furthermore, distributing the enzymes into different adjacent silica compartments yielded higher overall cascade reaction rates compared to placement of the enzymes into the same silica layer. The synthetic methods and kinetic analyses presented here provide guidance for improving the performance of immobilized multi-enzyme systems for a wide range of technological applications.
1. Introduction
Enzymes find numerous technological applications including chemical sensing, syntheses of bioactive compounds, medical diagnostics, and biomass degradation for fuel production.1 To increase their reusability, enzymes can be immobilized in vitro on a wide variety of solid matrices, which typically results in increased temperature and pH stability of the enzymes. The conversion of a substrate molecule into a desired product may require a linear cascade of reactions via several intermediates, in a manner analogous to cellular metabolic pathways (e.g., glycolysis, citric acid cycle, β-oxidation of fatty acids). High efficiency product formation in cascade reactions is achieved in vivo through the formation of multi-enzyme complexes. The close proximity of the enzymes catalyzing the reaction cascade allows for short diffusion distances for the intermediates, thereby drastically increasing the overall reaction rate. To mimic such natural multi-enzyme complexes, gene-technology based fusion of multiple enzymes or attachment of multiple enzymes to selective DNA and protein scaffolds has been successfully performed.2–4As alternatives to these rather elaborate approaches, less work-intensive methods for co-localization of enzyme cascades have been developed using encapsulation methods. So far, the encapsulated enzyme cascades have been comprised of either two or three enzymes. Betancor et al. co-encapsulated nitrobenzene nitroreductase and glucose-6-phosphate dehydrogenase in silica nanoparticles, which in the presence of glucose-6-phosphate enabled the continuous conversion of nitrobenzene to hydroxylaminobenzene with recycling of NADPH.5 In this prior work, no attempts were made to control the relative placement of the two enzymes. Placement of cascade enzymes into different compartments has been achieved through self-assembly of amphiphilic polymers6–9 or through layer-by-layer assembly of polymer capsules.10–12 Shi and co-workers assembled, through layer-by-layer mineralization and subsequent selective removal of mineral layers, microcapsules comprised of a protamine–titania shell and an inner layer made of protamine and oxidised alginate. Within the microcapsules, the enzyme formaldehyde dehydrogenase was placed in the core, and the enzyme formate dehydrogenase was positioned in the space between the outer mineral shell and the inner organic layer. Upon the addition of NADH, the two enzymes catalysed a two-step reaction cascade that converted CO2 to formaldehyde via formic acid as an intermediate.13
These works demonstrated the feasibility of catalysing cascade reactions by two or three enzymes that were incorporated into different nano- or microcompartments. Furthermore, they established some of the factors (e.g., spacing between compartments, rate-limiting enzymes, enzyme stability) that influence the overall activity of the reaction cascade. However, systematic kinetic analysis of compartmentalised multi-enzyme systems is still in its infancy, and general design principles that govern the overall activity of the cascade reactions have remained poorly developed.
The aim of the present work was to establish a facile and highly reproducible method to allow for the detailed analysis of the correlation between the relative arrangement of the cascade enzymes within different compartments and the rate of the cascade reaction. To achieve this, peptide-mediated layer-by-layer mineralization was utilized as a means for compartmentalisation of enzymes in nanoscale layers of silica under mild conditions with high preservation of enzyme activity.14 Previously, glucose oxidase was immobilized through layer-by-layer mineralization, which required covalent cross-linking of the polycationic peptide protamine to glucose oxidase.14 In the present work, a new protein tag was developed, consisting of six consecutive arginine residues (hexaarginine, R6), which confers silica-binding capability and can be genetically fused to any enzyme of interest. An artificial enzyme cascade consisting of three hexaarginine-tagged enzymes (i.e., cellulase CelB-R6, glucokinase GKin-R6, and phosphoglucose dehydrogenase G6PDH-R6)15–17 was immobilized in five layers of silica that were coated onto diatom silica microparticles. By systematically varying the compartmentalisation of the enzymes into different silica layers, detailed insight into the structure–activity correlation of the immobilized enzyme cascade was obtained.
2. Experimental
2.1 Materials
MilliQ purified H2O (resistivity: 18.2 MΩ cm) was used throughout. Diaphorase from Clostridium kluyveri (3–20 U mg−1), Adenosine 5′-diphosphate sodium salt, cellobiose (≥98%), glucose-6-phosphate, protamine sulfate (from salmon), tetramethoxysilane (TMOS), 4-nitrophenyl-β-D-glucopyranoside (≥98%), and tris-(hydroxymethyl-) aminomethane (Tris) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Nicotinamide adenine dinucleotide phosphate disodium salt (NADP+), and kanamycin were purchased from Calbiochem (Merck Millipore, USA). Glucose was purchased from Alfa Aesar (MA, USA). Nitro-blue tetrazolium chloride (NBT) and dNTPs were purchased from Thermo Scientific (Pierce Biotechnology, lL, USA). Dibasic and monobasic sodium phosphate were purchased from Merck (Merck Millipore, USA). Hydrochloric acid was purchased from VWR (West Chester, PA). Polydimethoxysilane (PDMS) base and curing agent were purchased from Dow Corning (Midland, MI, USA). Ni2+–NTA resin was purchased from Thermo Scientific (Rockwell, MD, USA).The diatom silica microparticles were cleaned diatomaceous earth (DE), which is primarily composed of frustules of Aulacoseira sp. The frustules have a hollow cylindrical shape with a height of 10–20 μm and a diameter of 5–10 μm. Their end faces contain a circular hole (3–6 μm diameter) with a protruding outer rim. DE possesses pores ranging from 2–50 nm and the specific surface area is 0.79 m2 g−1. For details of the cleaning procedure for diatomaceous earth see ref. 18, and for details of the surface area measurements see below (2.10. BET measurements and determination of enzyme densities).
2.2 Cloning, expression, and purification of the enzymes
The enzymes studied in the present work are encoded by genes celB (cellulase), glkA (ADP-dependent glucokinase), and gsdA (glucose-6-phosphate dehydrogenase) from the thermophilic archaea Pyrococcus furiosus and Aquifex aeolicus. To obtain large amounts of the tagged enzymes, the proteins were introduced into expression vector pET24 and heterologously expressed in E. coli.The coding regions of celB and glkA were amplified by PCR from P. furiosus genomic DNA (courtesy Dr Robert Kelly, NC State University, USA) using sense primer 5′-GATT GGATCC AAG TTC CCA AAA AAC TTC ATG TT-3′ and antisense primer 5′-AGCT CTCGAG CTA ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CTT TCT TGT AAC AAA TTT GAG GT-3′ for CelB, and sense primer 5′-GATT GGATCC CCC ACT TGG GAG GAG CTT T-3′ and antisense primer 5′-AGCT CTCGAG CTA
CTT TCT TGT AAC AAA TTT GAG GT-3′ for CelB, and sense primer 5′-GATT GGATCC CCC ACT TGG GAG GAG CTT T-3′ and antisense primer 5′-AGCT CTCGAG CTA ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) GAG AGT GAA TGA AAA CTC ACC-3′ for glkA. The primers introduced BamHI (bold) and XhoI (bold italics) restriction sites and a hexa-arginine tag (underlined).
GAG AGT GAA TGA AAA CTC ACC-3′ for glkA. The primers introduced BamHI (bold) and XhoI (bold italics) restriction sites and a hexa-arginine tag (underlined).
The coding region of gsdA was amplified by PCR from A. aeolicus genomic DNA (courtesy Dr Harald Huber, University Regensburg, Germany) using sense primer 5′-GATT GGATCC AGT TCG GGA GAC ACA GAG TT-3′ and antisense primer 5′-AGCT CTCGAG TCA ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) AAG GCA GGA AAT GAA ATC CTC-3′ which introduced BamHI (bold) and XhoI (bold italics) restriction sites and a hexa-arginine tag (underlined).
AAG GCA GGA AAT GAA ATC CTC-3′ which introduced BamHI (bold) and XhoI (bold italics) restriction sites and a hexa-arginine tag (underlined).
For PCR, 300 ng of genomic DNA was mixed with 10 μL Taq DNA polymerase high fidelity buffer (Fermentas, USA), 1.5 μL 2 mM dNTPs, 0.25 μL 100 μM of each sense and antisense primer, 0.5 μL of phusion enzyme (Fermentas, USA), and adjusted to 50 μL by adding H2O. The reaction mixture was then subjected to 20 cycles of amplification (94 °C for 20 s, 60 °C for 20 s and 72 °C for 90 s), and the resulting PCR products were isolated using the GeneJET PCR purification kit (Fermentas, USA). The PCR product and pET24 vector were digested with BamHI/XhoI (Fermentas, USA) for 2 hours at room temperature, and then subjected to agarose gel electrophoresis and gel elution. The plasmid was extracted using the GeneJET gel extraction kit (Thermoscientific, USA) and introduced into Escherichia coli DH5α.
For isolation of the enzymes, E. coli clones harboring the expression vectors were inoculated into LB medium containing 30 μg mL−1 kanamycin. The culture was grown overnight at 37 °C and diluted 50-fold with fresh kanamycin containing LB medium, and further cultivated until the cell density reached an absorbance of 0.6 at 600 nm. Expression of the enzyme coding genes was induced by adding isopropyl-β-D-1-thiogalacto-pyranoside at 1 mM final concentration, and the cultures were incubated at 37 °C for 3 h with constant shaking. Cells were harvested by centrifugation at 6000g for 20 min, washed once with 1 w/w% NaCl, resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 1 M NaCl, 10 mM imidazole), and lysozyme was added at a final concentration of 1 mg mL−1. The cell suspension was kept on ice for 30 min, sonicated for 10 min (Misonix sonicator 3000), centrifuged at 4 °C for 20 min at 6000g, and the supernatant was used for protein purification using a Ni2+–NTA column. The column was prepared by allowing 1 mL of Ni2+–NTA resin to settle in a column, washing the resin with 5 mL H2O, and then equilibrated with 5 mL lysis buffer. The equilibrated resin was then transferred to the supernatant of the E. coli extract and incubated at 4 °C overnight with rotation. The suspension was then poured into an empty column and washed with 5 mL purification buffer (50 mM Tris-HCl pH 8.0, 1 M NaCl) containing 20 mM imidazole. Stepwise elution of proteins was then performed with 1.5 mL each of purification buffer containing 50 mM, 100 mM, 200 mM, and 1000 mM imidazole. In each case, the 200 mM and 1000 mM imidazole fractions contained the majority of the enzymatic activity and were pooled. The enzymes were dialyzed against three changes of 50 mM sodium acetate pH 5.1 (cellulase), or 50 mM Tris-HCl pH 7.5 (kinase), or 50 mM Tris-HCl pH 7.0 (dehydrogenase), and stored at 4 °C in their respective buffers.
2.3 Enzyme activity assays
The cellulase, kinase, and dehydrogenase investigated in this study are thermophilic enzymes with activity optima around 75 °C. Therefore, reactions were stopped by removing the enzyme solution from the 75 °C incubator and rapid cooling down to room temperature.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min at room temperature. The absorbance of 100 μL of the supernatant was recorded at 410 nm on a Synergy 2 (Biotek, Winooski, VT, USA) plate reader.
000g for 5 min at room temperature. The absorbance of 100 μL of the supernatant was recorded at 410 nm on a Synergy 2 (Biotek, Winooski, VT, USA) plate reader.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min at room temperature. To the supernatant, 4 μL of 0.6 mg mL−1 glucose-6-phosphate dehydrogenase, 3.2 μL of 15.2 mM NBT, and 8 μL of 11 mg mL−1 diaphorase were added, and the reaction mixture was incubated for 10 min at 37 °C. The reaction was terminated by adding HCl to a final concentration of 60 mM and the absorbance was recorded at 550 nm on a Synergy 2 plate reader (Biotek, Winooski, VT, USA).
000g for 5 min at room temperature. To the supernatant, 4 μL of 0.6 mg mL−1 glucose-6-phosphate dehydrogenase, 3.2 μL of 15.2 mM NBT, and 8 μL of 11 mg mL−1 diaphorase were added, and the reaction mixture was incubated for 10 min at 37 °C. The reaction was terminated by adding HCl to a final concentration of 60 mM and the absorbance was recorded at 550 nm on a Synergy 2 plate reader (Biotek, Winooski, VT, USA).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min at room temperature. The concentration of NADPH was then determined by adding NBT and diaphorase as described in ‘Kinase assay’.
000g for 5 min at room temperature. The concentration of NADPH was then determined by adding NBT and diaphorase as described in ‘Kinase assay’.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min at room temperature. The concentration of NADPH was then determined by adding NBT and diaphorase as described in ‘Kinase assay’. As a reference, the activity of the enzyme cascade in solution was determined as follows. A mixture of three enzymes consisting of 75 μL of 0.3 mg mL−1 CelB-R6, 110 μL of 0.4 mg mL−1 ADPGK-R6, 125 μL of 0.6 mg mL−1 G6PDH-R6, 40 μL of 0.5 M sodium phosphate buffer pH 7.0, and 50 μL of H2O was prepared. From this mixture, a 50 μL aliquot was withdrawn and mixed with 24 μL of 0.5 M sodium phosphate buffer pH 7.0, 12.5 μL of 4 M NaCl, 10 μL of 20 mM ADP, 2 μL of 100 mM NADP+, and 2 μL of 0.1 M MgCl2, and the reaction was started with 2 μL 50 mM cellobiose. After incubation for 10 min at 75 °C under constant shaking at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany), the solution was centrifuged at 16
000g for 5 min at room temperature. The concentration of NADPH was then determined by adding NBT and diaphorase as described in ‘Kinase assay’. As a reference, the activity of the enzyme cascade in solution was determined as follows. A mixture of three enzymes consisting of 75 μL of 0.3 mg mL−1 CelB-R6, 110 μL of 0.4 mg mL−1 ADPGK-R6, 125 μL of 0.6 mg mL−1 G6PDH-R6, 40 μL of 0.5 M sodium phosphate buffer pH 7.0, and 50 μL of H2O was prepared. From this mixture, a 50 μL aliquot was withdrawn and mixed with 24 μL of 0.5 M sodium phosphate buffer pH 7.0, 12.5 μL of 4 M NaCl, 10 μL of 20 mM ADP, 2 μL of 100 mM NADP+, and 2 μL of 0.1 M MgCl2, and the reaction was started with 2 μL 50 mM cellobiose. After incubation for 10 min at 75 °C under constant shaking at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany), the solution was centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min at room temperature. The concentration of NADPH was then determined by adding NBT and diaphorase as described in ‘Kinase assay’.
000g for 5 min at room temperature. The concentration of NADPH was then determined by adding NBT and diaphorase as described in ‘Kinase assay’.
2.4 Determining Km and Vmax
The KM and Vmax values were determined for the free enzymes in the solution and for the diatom silica immobilized enzymes (note: saturated amounts of the enzymes were encapsulated in silica layer 2). The substrates were pNPG for CelB-R6, glucose for ADPGK-R6, and glucose-6-phosphate for G6PDH-R6. The activity assays were performed at 75 °C with varied substrate concentration (0.2–4.0 mM pNPG, 0.2–1.2 mM glucose, 0.02–2 mM glucose-6-phosphate). The data were analyzed by a Lineweaver–Burke plot, and KM and Vmax were obtained from the negative x-intercept and reciprocal y-intercept, respectively.2.5 Determining the binding capacity of diatom silica for the enzymes
Diatom silica (2 mg) was washed with 50 mM Tris-HCl pH 7.0 by three centrifugation (2 min at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) and resuspension (30 s vortexing) cycles. The washed diatom silica was resuspended in 50 mM sodium phosphate pH 7.0, and incubated with different amounts of an enzyme (10, 20 and 50 μg of CelB-R6; 10, 20, 50, 100 and 150 μg of ADPGK-R6; 10, 20, 50, 100 and 150 μg of G6PDH-R6) in a total volume of 200 μL (for CelB-R6) or 400 μL (for ADPGK-R6 and G6PDH-R6). The suspension was shaken at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany) for 30 min at 25 °C. The activities of the enzymes in all supernatants and on the silica was determined as described under ‘Enzyme activity assays’.
000g) and resuspension (30 s vortexing) cycles. The washed diatom silica was resuspended in 50 mM sodium phosphate pH 7.0, and incubated with different amounts of an enzyme (10, 20 and 50 μg of CelB-R6; 10, 20, 50, 100 and 150 μg of ADPGK-R6; 10, 20, 50, 100 and 150 μg of G6PDH-R6) in a total volume of 200 μL (for CelB-R6) or 400 μL (for ADPGK-R6 and G6PDH-R6). The suspension was shaken at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany) for 30 min at 25 °C. The activities of the enzymes in all supernatants and on the silica was determined as described under ‘Enzyme activity assays’.
2.6 Determining the silica immobilization efficiency for the enzymes
Diatom silica (2 mg) was washed with 50 mM Tris-HCl pH 7.0 by three centrifugation (2 min at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) and resuspension (30 s vortexing) cycle. The washed diatom silica was resuspended in 50 mM sodium phosphate pH 7.0, and incubated with saturating amounts of an enzyme (50.0 μg CelB-R6, 93.6 μg ADPGK-R6, 157.4 μg G6PDH-R6) in a total volume of 200 μL (for CelB-R6) or 400 μL (for ADPGK-R6 and G6PDH-R6). The suspension was shaken at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany) for 30 min at 25 °C, and then washed with 200 μL 50 mM sodium phosphate pH 7.0 by three centrifugation–resuspension cycles. The enzyme bearing diatom was then resuspended in 500 μL freshly prepared protamine sulfate (20 mg mL−1 in 50 mM Tris-HCl pH 7.0), incubated for 15 min at 25 °C under constant shaking at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany), and then washed with 200 μL 50 mM sodium phosphate pH 7.0 by three centrifugation–resuspension cycles. The enzyme and protamine bearing diatom silica particles were resuspended in 1 mL containing 50 mM Tris-HCl pH 7.0 and 40 mM freshly prepared silicic acid (generated from TMOS by hydrolysis in 1 mM HCl). The suspension was shaken at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany) for 30 min at 25 °C, and then washed with 200 μL of 50 mM sodium phosphate pH 7.0 as described above. The pellet was washed once with 200 μL of 50 mM sodium phosphate pH 7.0, and resuspended in 100 μL of the same buffer. The activities of the enzymes in all supernatants were determined as described under ‘Enzyme activity assays’.
000g) and resuspension (30 s vortexing) cycle. The washed diatom silica was resuspended in 50 mM sodium phosphate pH 7.0, and incubated with saturating amounts of an enzyme (50.0 μg CelB-R6, 93.6 μg ADPGK-R6, 157.4 μg G6PDH-R6) in a total volume of 200 μL (for CelB-R6) or 400 μL (for ADPGK-R6 and G6PDH-R6). The suspension was shaken at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany) for 30 min at 25 °C, and then washed with 200 μL 50 mM sodium phosphate pH 7.0 by three centrifugation–resuspension cycles. The enzyme bearing diatom was then resuspended in 500 μL freshly prepared protamine sulfate (20 mg mL−1 in 50 mM Tris-HCl pH 7.0), incubated for 15 min at 25 °C under constant shaking at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany), and then washed with 200 μL 50 mM sodium phosphate pH 7.0 by three centrifugation–resuspension cycles. The enzyme and protamine bearing diatom silica particles were resuspended in 1 mL containing 50 mM Tris-HCl pH 7.0 and 40 mM freshly prepared silicic acid (generated from TMOS by hydrolysis in 1 mM HCl). The suspension was shaken at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany) for 30 min at 25 °C, and then washed with 200 μL of 50 mM sodium phosphate pH 7.0 as described above. The pellet was washed once with 200 μL of 50 mM sodium phosphate pH 7.0, and resuspended in 100 μL of the same buffer. The activities of the enzymes in all supernatants were determined as described under ‘Enzyme activity assays’.
2.7 Layer-by-layer immobilization of the enzymes on diatom silica
Diatom silica (2 mg) resuspended in a final volume of 300 μL of 50 mM sodium phosphate pH 7.0 were incubated with saturating amounts of the enzymes (50.0 μg CelB-R6, 93.6 μg ADPGK-R6, 157.4 μg G6PDH-R6) at the indicated stages during the layer-by-layer mineralization process. The suspension was incubated for 15 min at 25 °C under constant shaking at 1400 rpm in a thermomixer (Eppendorf, Hamburg, Germany), and then washed with 200 μL of 50 mM sodium phosphate pH 7.0 by three centrifugation–resuspension cycles. Subsequently, coating with protamine and deposition of silica layers was performed as described under ‘Determining the silica immobilization efficiency for the enzymes’. This cyclic process was performed up to five times.2.8 Zeta potential measurements
Determination of the enzymes' zeta potentials was performed with a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, United Kingdom). The enzymes were dissolved at a concentrations of 0.075 mg mL−1 for cellulase, 0.05 mg mL−1 for kinase, and 0.15 mg mL−1 for dehydrogenase in 100 mM sodium phosphate pH 7.0. Data were averaged over 10 measurement runs for three independent samples.2.9 AFM measurements
AFM measurements of the thicknesses of enzyme–silica films on silicon wafer substrates were conducted with a Veeco Dimension 3100 Scanning Probe Microscope (Veeco, Plainview, NY, USA) using a 10 nm radius general purpose probe (n-Silicon Pointprobe, Nanosensors, Inc., Neuchatel, Switzerland). The thickness of a given film was obtained by reference to a region of the substrate from which the film was removed (by scraping with a razor blade) prior to conducting the thickness measurements. Samples were placed on the AFM stage and tapping-mode measurements were executed over a 5 μm × 5 μm area that spanned the boundary between coated and uncoated (film removed) regions. The Nanoscope Analysis v1.20 program was used for data analysis. The film-free surface of each scan was plane-leveled and a Bearing Analysis was performed on the entire sample area. For each type of sample, a total of twenty-six measurements were obtained by analyzing two to three different locations on three independently-prepared sister samples.2.10 BET measurements and determination of enzyme densities
Nitrogen adsorption (BET) analysis of diatom silica coated with five nanoscale layers of protamine–silica was conducted using an Autosorp-1 Surface Area and Pore Size Analyzer (Quantachrome Instruments, Boynton Beach, FL, USA). The coated diatom silica sample (∼1 g) was washed extensively with H2O, flash-frozen in liquid nitrogen, lyophilized for 24 h, and degassed in the analyzer at room temperature. The sample was backfilled with nitrogen gas, weighed, and isothermal gas adsorption/desorption was performed in a liquid nitrogen bath (∼77 K) with nitrogen gas as the adsorbate. A total of 40 adsorption and 40 desorption data points were collected and BJH analysis was performed on the isothermal desorption data to ascertain the pore volume in the meso- and macroporous ranges. The analysis yielded a specific surface area of 0.49 m2 g−1 for the protamine–silica coated diatom silica.To determine the mass of an enzyme per volume of one layer of nanoscale silica coating, the amount of enzyme mass per mass of diatom silica retained after silica coating (22 mg CelB-R6, 42.3 mg for GKin-R6, 51.0 mg for G6PDH-R6), the thickness of a protamine–enzyme–silica layer (9.8 nm for CelB-R6, 6.3 nm for GKin-R6, 7.0 nm for G6PDH-R6), and the specific surface area of the protamine silica coated diatom silica was used (0.49 m2 g−1).
3. Results and discussion
3.1 Establishment of the enzyme cascade
For the purpose of this study, an artificial, three-step enzyme cascade was designed that consisted of a cellulase (encoded by gene celB from Pyrococcus furiosus16), an ADP-dependent glucokinase (encoded by glkA from P. furiosus17), and a glucose-6-phosphate dehydrogenase (encoded by gsdA from Aquifex aeolicus18). Together, the three enzymes catalyze the conversion of cellobiose to 6-phosphogluconolactone via the intermediates glucose and glucose-6-phosphate (Scheme 1). | ||
| Scheme 1 Reactions catalyzed by the enzyme cascade. CelB = cellulase, GKin = ADP-dependent glucokinase, G6PDH = glucose-6-phosphate dehydrogenase. | ||
The enzyme cascade was designed to allow for straightforward determination of both the overall catalytic activity and the individual activity of each enzyme (see Experimental section for details). The three enzymes were separately expressed in E. coli as fusion proteins (coined CelB-R6, GKin-R6, G6PDH-R6) with each bearing a hexahistidine-tag (His6) on the N-terminus and six arginine residues (R6-tag) on the C-terminus. The His-tag allowed for one-step purification of the enzymes by immobilized metal ion affinity chromatography, while the R6-tag was introduced to enable immobilization of the enzymes (see below). Catalytically active preparations of the enzymes were obtained with high yield and purity (Table S1 and Fig. S1, ESI†). When the enzyme substrates cellobiose, ADP, and NADP+ were added to a mixture of the three recombinant enzymes rapid production of NADPH was observed (Fig. 1). No NADPH was produced when one of the substrates or one of the enzymes was omitted (Fig. 1). In the absence of cellobiose, NADPH production was restored by adding glucose (Fig. 1). These results demonstrated that the designed enzyme cascade operated in solution as intended (see Scheme 1).
 | ||
| Fig. 1 Activity of the enzyme cascade in solution. The reactions were performed in the presence of ADP, NADP+, and the enzymes and substrates indicated under the horizontal axis. | ||
3.2 Immobilization of the enzymes on diatom silica
To investigate the possibility of immobilizing the enzyme cascade on diatom silica through layer-by-layer mineralization, the attachment of the recombinant enzymes onto diatom silica and the stability of this interaction during mineralization were investigated. Due to its high mesoporosity, diatom silica is an advantageous enzyme support material, particularly in flow-through applications.19–21 Previously, it was demonstrated that polycationic peptides, including the arginine-rich peptide protamine, exhibit strong binding to silica surfaces and silica forming activity.22–26 Therefore, each of the three enzymes was engineered to contain six arginine residues to promote binding to silica. When CelB-R6 was incubated with a suspension of diatom silica at pH 7.0 (note: the diatom silica was coated beforehand with a freshly deposited layer of silica using the layer-by-layer mineralization method) followed by extensive washing of the silica with the same buffer, 75 ± 4% of the cellulase activity was attached to the diatom silica, with no cellulase activity in the supernatants after incubation and washing. In contrast, in the same experiment with Hexahistidine-tagged CelB that lacked the R6-tag only 45 ± 2% of the cellulase activity was attached to the diatom silica, whereas 50 ± 0.5% of the activity remained in the supernatants. This demonstrated that the R6-tag strongly enhanced the amount of enzyme activity that could be attached to silica. The same result was observed for the other two enzymes, GKin-R6 and G6PDH-R6. However, all three diatom silica-bound enzymes exhibited reduced activities of 75 ± 4% for CelB-R6, 90 ± 3% for GKin-R6, and 80 ± 2% for G6PDH-R6 relative to the activities of the enzymes in solution. This indicated either that upon binding to the silica surface ∼10–25% of the enzyme molecules became inactivated, or that the diatom silica exerted a partially inhibiting effect on all enzyme molecules. The maximum capacities of diatom silica for adsorption of the enzymes were (in mg enzyme per g silica) 25 ± 2 mg g−1 for CelB-R6, 45 ± 2 mg g−1 for GKin-R6, and 75 ± 3 mg g−1 for G6PDH-R6.The encapsulation of individual surface-adsorbed enzymes in nanoscale layers of silica, using the layer-by-layer mineralization method,14 was then investigated. The method consists of two steps: (i) coating the enzyme-bearing diatom silica with protamine (note: protamine exhibits silica-forming activity),27 and then (ii) incubating the enzyme- and protamine-loaded particles with silicic acid. During the procedure, substantial amounts of G6PDH-R6 of (32 ± 2%), and smaller amounts of CelB-R6 (13 ± 3%) and of GKin-R6 (6 ± 2%) were desorbed from the diatom silica (Fig. 2). Almost all of the G6PDH-R6 desorption occurred during the protamine coating step (Fig. 2), which suggested that the electrostatic interaction with the negatively charged silica surface was weaker for G6PDH-R6 than for the other two enzymes. The zeta potentials of the three enzymes are negative and almost identical (Table S2, ESI†). Thus, positively charged patches on the enzymes' surfaces, rather than their net negative charges may be mainly responsible for silica binding. Therefore, it can be hypothesized that the R6-tag of G6PDH-R6, compared to the other two recombinant enzymes, may be less exposed on the protein surface, thus resulting in weaker silica affinity.
The average thickness of silica layers produced through layer-by-layer mineralization has previously been found to be ∼5 nm.14 However, the influence of relatively large amounts of encapsulated enzyme molecules on the thickness of the silica layers has not previously been reported. To investigate this, saturating amounts of the enzymes were first adsorbed on a silicon wafer and five cycles of layer-by-layer mineralization (with saturating amounts of the enzyme in each layer) were conducted. The thicknesses of the films relative to an unmodified region on the wafer were evaluated by AFM (Fig. S2, ESI,†Fig. 3). These analyses revealed an average thickness of 9.8 ± 0.9 nm for a CelB-R6–silica layer, 6.3 ± 0.4 nm for a GKin-R6–silica layer, and 7.0 ± 0.5 nm for a G6PDH-R6–silica layer. The greater thickness of the CelB-R6–silica layer compared to the other two enzyme–silica layers was consistent with the larger size of the native cellulase (which is a homotetramer) relative to the other two enzymes (which are homodimers). The monomeric polypeptide chains of the three enzymes are similar to each other with molecular masses between 47–58 kDa corresponding to folded globular particles of ∼4 nm in diameter.16–18 The thicknesses of the enzyme–silica films suggest that (i) the enzyme molecules may be preferentially bound to the silica surface as monolayers (i.e., height of ∼4 nm for Gkin-R6 and G6PDH-R6, height of ∼8 nm for CelB-R6), and (ii) each layer in the enzyme–silica film may be less mineralized (∼2–3 nm silica) than the silica-only layers (∼5 nm silica). The density of an enzyme within a nanoscale silica layer was 4.5 g cm3 for CelB-R6, 13.7 g cm3 for GKin-R6, and 14.9 g cm3 for G6PDH-R6.
3.3 Kinetic analyses of the immobilized enzyme cascade
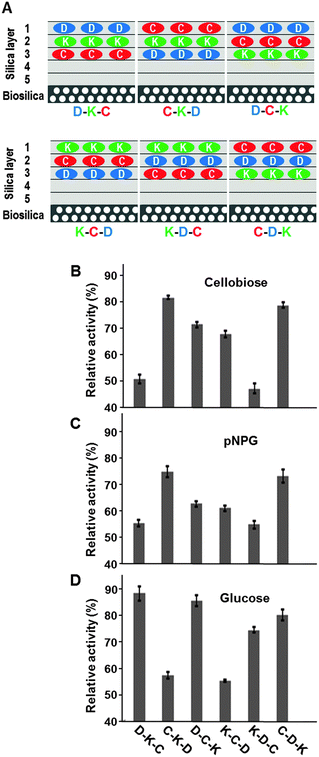
The previous experiments established a method for functional immobilization of the individual recombinant enzymes on diatom silica microparticles covered with a ∼2–3 nm layer of silica. The same approach was then examined for immobilizing a functional enzyme cascade including all three recombinant enzymes. Five layers of silica were deposited on diatom silica with enzyme molecules included in the 3rd, 2nd, and 1st layers (Fig. 4A; note: the order of synthesis of the silica layers is the inverse of the numbering). A saturating amount of only one type of enzyme was incorporated in each layer. This synthesis strategy gave rise to six different relative arrangements of the enzyme layers (referred to as ‘configurations’ in the following), and six different diatom silica-based microparticles representing these configurations were synthesized (Fig. 4A).When the enzyme-loaded diatom silica microparticles were incubated with cellobiose, ADP, and NADP+ as substrates, NADPH production was detected for all six enzyme configurations, which demonstrated that the enzyme cascade was functional in each case. Interestingly, the relative activities among the different configurations varied substantially ranging between 50–80% of the activity of the enzyme cascade in solution (Fig. 4B). Configurations in which the cellulase was placed in silica layer 3 (D–K–C and K–D–C) exhibited the lowest catalytic activities (∼55%), whereas the relative activities of the configurations containing cellulase in the outermost silica layer (i.e., silica layer 1; C–K–D and C–D–K) reached about 80% activity (Fig. 4B). The relative enzyme cascade activities of the two configurations containing the cellulase in silica layer 2 (D–C–K and K–C–D) exhibited intermediate relative activities around 70% (Fig. 4B). In the six different cascade configurations, the relative activities of the cellulase only (Fig. 4C) were consistent with the relative activities of the entire cascade (Fig. 4B). In contrast, the combined activities of kinase and dehydrogenase were not consistent with the activities of the entire cascade (Fig. 4D). As will be outlined in the following paragraph, these results can be rationalized by assuming that the cellulase-catalyzed reaction is the rate-limiting step of the enzyme cascade.
The overall activity of an enzyme cascade is determined by the rate-limiting enzyme. As more silica layers are placed atop an immobilized enzyme, its catalytic activity should decrease, because the activity of a silica-immobilized enzyme has been found to be limited by reactant diffusion.14 Therefore, the cascade configurations in which the rate-limiting enzyme is buried most deeply inside the silica layers (i.e., layer 3 in this study) are expected to exhibit the lowest overall catalytic activities. This is exactly the case for configurations D–K–C (51 ± 1.5% activity) and K–D–C (47 ± 1.7% activity), in which the cellulase is positioned in layer 3. Furthermore, the overall catalytic activity of the cascade should increase as the rate limiting enzyme is positioned closer to the external surface.
Such correlation between enzyme placement and cascade activity was only observed for cellulase with the relative activities increasing from ∼55% to ∼60% and to ∼75% when the cellulase was present in layer 3, layer 2 and layer 1, respectively. In contrast, the combined activities of the kinase and dehydrogenase for the different cascade configurations (Fig. 4D) are not consistent with these two enzymes being rate limiting for the overall activity of the enzyme cascade.
To further validate the rate-limiting role of cellulase, three different types diatom silica particles with the configuration C–K–D (see Fig. 4A) were synthesized. Microparticles of Type 1 contained only 1/3rd of the saturating amount of cellulase, but the other two enzymes were present in saturating amounts. C–K–D microparticles of Type 2 contained only 1/3rd of the kinase, and Type 3 only 1/3rd of the dehydrogenase, with both microparticles containing saturating amounts of the two other enzymes of the cascade.
As expected, the diatom silica microparticles bearing reduced amounts of cellulase (Type 1) exhibited the lowest enzyme cascade activity (Fig. 5), which confirmed the limiting role of the cellulase for the overall reaction rate. Reduction of the amount of the kinase (Type 2) had no significant effect on the overall cascade activity (83 ± 1.5% activity with reduced amount of kinase versus 82 ± 0.8% activity with saturating amount), which demonstrated that the overall speed of the cascade was not limited by this enzyme. However, reduction of the amount of dehydrogenase (Type 3) resulted in a significant decrease of cascade activity (Fig. 5) indicating that the dehydrogenase also plays a significant, although lesser, role than the cellulase for determining the overall enzyme cascade activity. The rate-limiting effect of the dehydrogenase may be a consequence of its low catalytic efficiency (kcat/KM), which is the lowest of the three enzymes of the cascade (see Table S1, ESI†). This demonstrated that the overall reaction rate of the silica-immobilized reaction cascade is not exclusively governed by the enzyme catalyzing the first step, but also depends on the inherent catalytic properties of the enzymes catalyzing the subsequent reaction steps.
In the above experiments the overall rate of the reaction cascade was studied as a function of placement of each of the three enzymes in a different silica layer. Therefore, it remained to be determined how (if at all) the overall rate of the reaction cascade changed when all the three enzymes were placed within the same silica layer. To interrogate this, two different microparticles were synthesized. For L1 particles, a mixture of the cellulase, the kinase, and the dehydrogenase was incorporated in silica layer 1, whereas such enzyme mixture was placed in silica layer 3 for L3 particles (Fig. 6A). In these enzyme mixtures, each enzyme was present at only one-third of the amount required to saturate the silica surface. This approach was taken to promote the binding of the enzymes at approximately the same relative molar ratios as in the microparticles that were investigated above (see Fig. 4A). Microparticles 1/3(D–K–C) and 1/3(C–K–D) were synthesized as references that contained the same amount of each enzyme as particle L1 and L3, but the enzymes were placed in different silica layers (Fig. 6A). Configurations D–K–C and C–K–D were chosen as references because they represent the configurations with the lowest and highest catalytic activities, respectively (see Fig. 4).
Determination of the overall reaction rates of the four different diatom silica microparticles revealed that the L1 microparticles had a significantly lower catalytic activity than the 1/3(C–K–D) microparticles (Fig. 6B). This is remarkable, because diffusion of the final product of the reaction cascade (NADPH) to the surface occurs through ∼15 nm enzyme–silica layer for the 1/3(C–K–D) microparticles but only through ∼3 nm enzyme–silica layer for the L1 microparticles (see Fig. 3 and 6A; note: the diffusion path for the initial substrate, cellobiose, is ∼3 nm for both microparticles). It is also surprising that the activities of L1 and 1/3(D–K–C) microparticles are almost identical, although the diffusion path for the initial substrate is much longer for the 1/3(D–K–C) microparticles (note: the diffusion path for the final product is the same for both microparticles). These data suggest that enzyme–substrate interactions are favored when the enzymes of the cascade are located in adjacent layers rather than within the same silica layer. The comparatively low activity of the L3 microparticles (68 ± 2.0% of the activity of L1 microparticles) can be explained by the long diffusion paths (∼15 nm) for both initial substrate and final product.
4. Conclusions
The present work has demonstrated that layer-by-layer silica immobilization of hexaarginine-tagged enzymes is a generic method for the compartmentalisation of enzymes on a solid support with high preservation of enzyme activity. The enzyme molecules were not covalently attached to the support material (diatom silica) yet they became stably immobilized by silica encapsulation. The 30–50 nm thick enzyme-bearing silica coat is mechanically stabilized by the underlying diatom silica. The sequential nature of the assembly of the multilayer system enabled control over the relative positioning and the incorporated amounts of the cascade enzymes. Detailed kinetic analyses of the individual enzymes and the entire enzyme cascade revealed that the overall reaction rate is largely determined by reactant diffusion and much less by the catalytic efficiency of each enzyme. Therefore, to ensure high overall cascade reaction rate the enzyme catalysing the first reaction step needs to be positioned close to the surface. Furthermore, positioning the cascade enzymes in adjacent silica layers rather than the same silica layer is advantageous, but a clear explanation for this observation is currently lacking. Diffusion of the reactant may be faster between the silica layers compared to lateral reactant diffusion within a protein-packed silica layer. Also the chance for productive enzyme–substrate interactions of a reactant molecule leaving a silica layer might be higher when enzymes are positioned in neighbouring layers rather than only within one layer.The positioning of the enzymes in silica layers in close proximity to each other was intended to enhance the cascade reaction rate by reducing the diffusion path lengths of the intermediate reactants (i.e., glucose and glucose-6-phosphate). However, even the most active configuration of the immobilized enzyme cascade reached no more than 80% of the activity of the enzyme cascade in solution. This indicates that for the cascade configurations that were synthesized here, the benefit of enzyme proximity in the silica layers did not outweigh the decreased reactant diffusion speed within the solid material. It may be possible to enhance the activity of the immobilized enzyme cascade by reducing the mineral content of the silica through (i) changing the mineralization conditions (e.g., pH and time of mineralization, concentration of mineral precursor), or (ii) introducing mineral dopants that can be gently removed post mineralization (e.g., dissolution of CaCO3 nanoparticles using citrate or EDTA at pH 7).
Future work should explore the detailed diffusion mechanisms(s) contributing to the overall reaction rate associated with compartmentalized enzyme cascades. Furthermore, the catalytic performance of particles that exhibit a repetitive enzyme configuration (i.e., that contain multiple layers bearing the same enzyme) has not yet been studied. Using the synthetic methods described here, such materials and analyses are readily accessible.
Acknowledgements
The work of GB and NK was supported by the Deutsche Forschungsgemeinschaft within priority program SPP 1569 ‘Generation of multifunctional inorganic materials by molecular bionics’ (grant no. KR 1853/3-2 to NK), by the European Union (EFRE), and the Free State of Saxony (SMWK). The work of WBG and KHS was supported by the U.S. Air Force Office of Scientific Research via Award No. FA9550-10-1-0555. We are indebted to Dr Harald Huber (University Regensburg, Germany) and Dr Robert Kelly (NC State University, USA) for genomic DNA from A. aeolicus and P. furiosus, respectively. We are grateful to Philip D. Brooke (Georgia Institute of Technology, USA) for help with BET analysis.Notes and references
- A. S. Bommarius and B. Riebel, Biocatalysis, Wiley-VCH Verlag GmbH and Co. KGaA, 2004 Search PubMed.
- F. Jia, B. Narasimhan and S. Mallapragada, Biotechnol. Bioeng., 2014, 111, 209 CrossRef CAS PubMed.
- S. Schoffelen and J. C. M. van Hest, Soft Matter, 2012, 8, 1736 RSC.
- S. Schoffelen and J. C. M. van Hest, Curr. Opin. Chem. Biol., 2013, 23, 613 CAS.
- L. Betancor, C. Berne, H. R. Luckarift and J. C. Spain, Chem. Commun., 2006, 3640 RSC.
- D. M. Vriezema, P. M. L. Garcia, N. S. Oltra, N. S. Hatzakis, S. M. Kuiper, R. J. M. Nolte, A. E. Rowan and J. C. M. van Hest, Angew. Chem., Int. Ed., 2007, 46, 7378 CrossRef CAS PubMed.
- G. Delaittre, I. C. Reynhout, J. J. L. M. Cornelissen and R. J. M. Nolte, Chem. – Eur. J., 2009, 15, 12600 CrossRef CAS PubMed.
- S. F. M. van Dongen, M. Nallani, J. L. L. M. Cornelissen, R. J. M. Nolte and J. C. M. van Hest, Chem. – Eur. J., 2009, 15, 1107 CrossRef CAS PubMed.
- L. Zhang, J. F. Shi, Z. Y. Jiang, Y. J. Jiang, S. Z. Qiao, J. A. Li, R. Wang, R. J. Meng, Y. Y. Zhu and Y. Zheng, Green Chem., 2011, 13, 300 RSC.
- O. Kreft, M. Prevot, H. Möhwald and G. B. Sukhorukov, Angew. Chem., Int. Ed., 2007, 46, 5605 CrossRef CAS PubMed.
- H. Bäumler and R. Georgieva, Biomacromolecules, 2010, 11, 1480 CrossRef PubMed.
- Y. Jiang, Q. Sun, L. Zhang and Z. Jiang, J. Mater. Chem., 2009, 19, 9068 RSC.
- J. Shi, L. Zhang and Z. Jiang, ACS Appl. Mater. Interfaces, 2011, 3, 881 CAS.
- N. R. Haase, S. Shian, K. H. Sandhage and N. Kröger, Adv. Funct. Mater., 2011, 21, 4243 CrossRef CAS.
- Y. Fang, Q. Wu, M. B. Dickerson, Y. Cai, S. Shian, J. D. Berrigan, N. Poulsen, N. Kröger and K. H. Sandhage, Chem. Mater., 2009, 21, 5704 CrossRef CAS.
- S. W. M. Kengen, E. J. Luesink, A. J. M. Stams and A. J. B. Zehnders, Eur. J. Biochem., 1993, 213, 305 CrossRef CAS.
- S. Koga, I. Yoshioka, H. Sakuraba, M. Takahashi, S. Sakasegawa, S. Shimizu and T. Ohshinia, J. Biochem., 2000, 128, 1079 CrossRef CAS.
- R. B. Iyer, J. Wang and L. G. Bachas, Extremophiles, 2002, 6, 283 CrossRef CAS PubMed.
- N. Poulsen, C. Berne, J. Spain and N. Kröger, Angew. Chem., Int. Ed., 2007, 46, 1843 CrossRef CAS PubMed.
- V. C. Sheppard, A. Scheffel, N. Poulsen and N. Kröger, Appl. Environ. Microbiol., 2012, 78, 211 CrossRef CAS PubMed.
- S. C. Davis, V. C. Sheppard, G. Begum, Ye. Cai, Y. Fang, J. D. Berrigan, N. Kröger and K. H. Sandhage, Adv. Funct. Mater., 2013, 23, 4611 CrossRef CAS.
- N. Kröger, M. B. Dickerson, G. Ahmad, Y. Cai, M. S. Haluska, K. H. Sandhage, N. Poulsen and V. C. Sheppard, Angew. Chem., Int. Ed., 2006, 45, 7239 CrossRef PubMed.
- Y. Jiang, D. Yang, L. Zhang, L. Li, Q. Sun, Y. Zhang, J. Li and Z. Jiang, Dalton Trans., 2008, 4165 RSC.
- R. R. Naik, L. L. Brott, S. J. Clarson and M. O. Stone, J. Nanosci. Nanotechnol., 2002, 2, 95 CrossRef CAS.
- M. R. Knecht and D. W. Wright, Chem. Commun., 2003, 3038 RSC.
- J. N. Cha, G. D. Stucky, D. E. Morse and T. J. Deming, Nature, 2000, 403, 289 CrossRef CAS PubMed.
- Y. Zhang, H. Wu, J. Li, L. Li, Y. Jiang, Y. Jiang and Z. Jiang, Chem. Mater., 2008, 20, 1041 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Tables S1 and S2, Fig. S1 and S2. See DOI: 10.1039/c5tb00333d |
| This journal is © The Royal Society of Chemistry 2015 |