Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Encapsulated enzymes with integrated fluorescence-control of enzymatic activity†
Pascal K.
Harimech‡
a,
Raimo
Hartmann‡
a,
Joanna
Rejman
a,
Pablo
del Pino
b,
Pilar
Rivera-Gil
a and
Wolfgang J.
Parak
*ab
aFachbereich Physik, Philipps Universität Marburg, Marburg, Germany. E-mail: wolfgang.parak@physik.uni-marburg.de
bCIC Biomagune, San Sebastian, Spain
First published on 18th February 2015
Abstract
A fluorescence-based particle sensor for oxaloacetic acid is presented. In the presence of nicotinamide adenine dinucleotide as a cofactor, oxaloacetic acid is converted by malate dehydrogenase into L-malic acid. The progress of the reaction is monitored by sensing of proton consumption with an integrated pH sensor. The kinetics of this sensor are investigated on a single particle level. This work demonstrates the feasibility to detect analytes upon their enzymatic conversion into a product, which in turn can be sensed with a fluorophore responding to changes in the concentration of this product. Integration of enzymes and fluorophores into one carrier particle, as demonstrated here for the case of polyelectrolyte polymer capsules, allows the range of analytes that can be detected with fluorescence to be extended, as it enhances selectivity. This coupled system allows enzymatic activity, as well as the kinetics of malate dehydrogenase, to be monitored.
Introduction
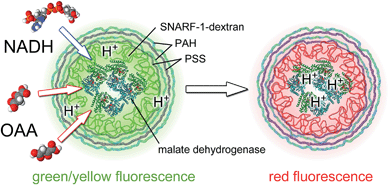
Analyte-sensitive fluorophores are common tools for measuring the concentration of analytes in solution by means of fluorescence measurements.1,2 Fluorophores which selectively respond to the presence of a large number of analytes have been reported in the literature and many of them are also commercially available,3,4 such as Ru(ddp), FluoZin, tetraphenylethene-based diboronic acid, [Ru(phen)3]2+, etc.1,2,5,6 Selectivity to a target analyte however is often limited, as the presence of other (similar) analytes triggers an unwanted crosstalk response. In addition, for many analytes, no fluorophores which show a response upon their presence, have been described. One strategy to bypass these shortcomings is the use of enzymes. Enzymes are very selective to their target substrates. When fluorophores responsive to the enzymatic product are used, analytes which serve as a target for the enzyme can be selectively detected.7–12 Ideally, enzymes and fluorophores (in addition to organic fluorophores and quantum dots and luminescent metal nanoclusters have also been used) should be coupled together into one entity, which is possible for example by linking both of them to colloidal particles. One convenient carrier particle system, which allows embedding of analyte-sensitive fluorophores,13–16 as well as enzymes,17–24 is polyelectrolyte capsules fabricated by layer-by-layer assembly.25–27 The walls of the capsules can be made semi-permeable, which allows small analytes to diffuse inside the capsule to reach the embedded fluorophores and enzymes, while retaining the fluorophores and enzymes inside the cavity of the capsule. If the fluorophores are not retained due to their small size, they can be linked to larger molecules.19 Combination of analyte-sensitive fluorophores and enzymes allows for selective detection of analytes/substrates.28,29 For example, Kazakova et al. co-embedded urease and the pH-sensitive fluorophore seminaphtharhodafluor (SNARF-1) in polyelectrolyte capsules with the intention of designing an urea-selective fluorescence sensor.29 Upon enzymatic processing of urea by urease, the local pH increases due to the production of ammonia, which is detected with the pH-sensitive fluorophore (i.e. consumption of protons). In principle, such enzyme-based fluorescence sensors could also be applied for intracellular sensing, similar to intracellular pH-sensing, which has been demonstrated to monitor cellular reactions.30 In particular, this would be interesting for target molecules which are relevant for cellular function. Besides real-time determination of the intracellular concentrations of such target molecules, these capsules could be seen as theranostic devices. In cases where the concentration of an intracellular target is decreased by the enzymes delivered with the capsules, this decrease could be monitored directly by co-delivered fluorophores selective to one of the enzymatic products.In this work, a proof-of-concept of such an encapsulated enzyme–fluorophore couple for a biologically relevant target analyte is demonstrated. Specifically, we encapsulated malate dehydrogenase, an enzyme involved in many metabolic pathways, including the Krebs cycle, which reversibly converts oxaloacetic acid (OAA) into L-malic acid:
| OAA + NADH + H+ ⇌ L-malic acid + NAD+ |
This reaction requires nicotinamide adenine dinucleotide (NADH) as a cofactor. During the enzymatic reaction, protons are consumed/produced, depending on the direction of the reaction. Based on the response of the co-encapsulated pH-sensitive fluorophore, SNARF-1, the reaction can be monitored by pH-sensing, cf.Fig. 1. The kinetics of this combined system, and in particular that of its fluorescence read-out, are discussed in this work.
Materials and methods
Chemicals
Poly(sodium 4-styrenesulfonate) (PSS, Mw ≈ 70 kDa, #243051), poly(allylamine hydrochloride) (PAH, Mw ≈ 56 kDa, #283223), calcium chloride dehydrate (CaCl2, #223506), sodium carbonate (Na2CO3, #S7795), ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA disodium salt, #E5134), and mitochondrial malate dehydrogenase from porcine heart (#M2634) were purchased from Sigma-Aldrich (Germany). SNARF-1-dextran (Mw ≈ 70 kDa, #D3304) from Life Technologies (Germany) was used. Sodium chloride (NaCl, #HN00.2), oxaloacetic acid (OAA, #4032.2), and β-nicotinamide adenine dinucleotide disodium salt (NADH, #AE12.1) were obtained from Roth (Karlsruhe, Germany). Phosphate buffered saline (PBS-Dulbecco, #L1825) was purchased from Biochrom (Berlin, Germany). Ultrapure double distilled water (ddH2O) with a resistivity greater than 18.2 MΩ cm was used for all experiments.Synthesis of polyelectrolyte sensor capsules
The synthesis of polyelectrolyte microcapsules was carried out as described previously,14,15,30,31 with the following modifications: CaCO3 microparticles were prepared at room temperature (RT) from solutions of CaCl2 and Na2CO3 under vigorous stirring, in the presence of SNARF-1-dextran and malate dehydrogenase. 615 μL of an aqueous solution of CaCl2 (0.33 M) with NaCl (1.33 M), 600 μL SNARF-1-dextran 70 kDa (1 mg mL−1) and 100 μL malate dehydrogenase (11 mg protein per mL, 7700 U mL−1) were mixed together in a glass vial. During magnetic stirring (1000 rpm), 615 μL of an aqueous solution of Na2CO3 (0.33 M) with NaCl (1.33 M) solution was added quickly. After 30 s the stirrer was turned off and particle growth was stopped after an additional 2 minutes by centrifugation. The particles were washed three times with ddH2O and then directly used for the layer-by-layer assembly of polyelectrolytes. Alternating layers of negatively charged PSS (2 mg mL−1 in 0.5 M NaCl) and positively charged PAH (2 mg mL−1 in 0.5 M NaCl) were deposited onto the charged microparticles until four bilayers of the polyelectrolytes had been established (CaCO3@(PSS/PAH)4). For coating with each layer, the microparticles were suspended in 1 mL of polyelectrolyte solution, shaken gently for 10 minutes and washed three times with ddH2O. The dissolution of the cores was carried out by Ca2+ ion complexation with EDTA (1 mL, 0.2 M, pH 6) for several minutes. The resulting SNARF-1 and malate dehydrogenase containing microspheres were washed three times with ultrapure water to remove excess EDTA and stored at 4 °C. The size of the resulting capsules ranged between 3 and 5 μm.Fluorescence spectroscopy based sensing
To determine the response of SNARF-1 to the enzymatic activity of malate dehydrogenase in free solution, a reaction mixture of 10 mM OAA, 75 μM NADH, 1 μL malate dehydrogenase (11 mg protein per mL, 7700 U mL−1) and 100 μg mL−1 SNARF-1 was prepared. The pH of all stock solutions was adjusted to 6 with HCl and NaOH prior to use. Immediately after preparation, SNARF-1 was excited at 540 nm with a FluoroLog (Horiba, Japan) fluorescence spectrometer and the emission was recorded over time between 560–700 nm.Fluorescence microscopy based sensing
A confocal laser scanning microscope (cLSM) was used (LSM 510 Meta, Zeiss) for local sensing of pH-changes with polyelectrolyte sensor capsules. Drops of capsule solution were examined on a glass slide (approximately 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 capsules per drop of reaction mixture). SNARF-1 was excited simultaneously at 488 nm and 543 nm. The fluorescence signal was recorded at ranges between 550–615 nm (channel 1) and 615–750 nm (channel 2). The ratio of the fluorescence intensity of channel 2 (intensity Ir)/channel 1 (intensity Iy) was calculated for all pixel pairs, where the intensity in both channels was above a threshold. The mean value for all pixel intensity ratios of one image (approximately 300 capsules) was calculated and used to determine the pH changes inside the capsule cavities caused by the enzymatic reaction catalyzed by malate dehydrogenase in the presence of the substrate OAA and the co-enzyme NADH. To determine the reaction kinetics, the fluorescence intensity of SNARF-1 in both channels was measured over time after the addition of OAA and coenzyme consecutively, if not stated otherwise. Image processing was performed with Matlab (Mathworks). All solutions used for the experiments were adjusted to pH 6 with HCl or NaOH immediately before the sensing process was performed. The reaction mixture was buffered with PBS (5%). The sensing experiments were always performed one day after capsule preparation.
000 capsules per drop of reaction mixture). SNARF-1 was excited simultaneously at 488 nm and 543 nm. The fluorescence signal was recorded at ranges between 550–615 nm (channel 1) and 615–750 nm (channel 2). The ratio of the fluorescence intensity of channel 2 (intensity Ir)/channel 1 (intensity Iy) was calculated for all pixel pairs, where the intensity in both channels was above a threshold. The mean value for all pixel intensity ratios of one image (approximately 300 capsules) was calculated and used to determine the pH changes inside the capsule cavities caused by the enzymatic reaction catalyzed by malate dehydrogenase in the presence of the substrate OAA and the co-enzyme NADH. To determine the reaction kinetics, the fluorescence intensity of SNARF-1 in both channels was measured over time after the addition of OAA and coenzyme consecutively, if not stated otherwise. Image processing was performed with Matlab (Mathworks). All solutions used for the experiments were adjusted to pH 6 with HCl or NaOH immediately before the sensing process was performed. The reaction mixture was buffered with PBS (5%). The sensing experiments were always performed one day after capsule preparation.
Results and discussion
Four basic requirements have to be met in order to employ polyelectrolyte microcapsules for fluorescence sensing of products of enzymatic reactions. First, the enzymatic reaction itself should lead to a local change in the concentration of free ions. Second, an ion-sensitive fluorophore-sensor should be loaded together with the catalytically active molecules (i.e. enzymes). Third, the molecular size of the dye and the catalyst should be sufficiently large to retain them within the capsule cavity. Lastly, the substrate molecules should be small enough to penetrate through the capsule wall. Hereby, the precise molecular weight cut-offs depend on the capsule material and architecture.In our experimental set-up we evaluated the sensing capacities of microcapsules carrying malate dehydrogenase (70 kDa, isoelectric point: 10) and the pH-sensitive dye, SNARF-1. SNARF-1 was coupled to dextran (70 kDa) to increase the molecular size of the fluorescent probe and to ensure that it is retained inside the capsule cavity.15 The maximum of the emission spectrum of SNARF-1 shifts from 580 nm in acidic media to 640 nm in alkaline media. As described, the pKa of the fluorophore might change in different environments. While the pKa of the free dye is in accordance with the manufacturer's value of 7.50, it changes when encapsulated.15,29 In the present case, while encapsulated together with malate dehydrogenase, the pKa changes to about 6.80.
Malate dehydrogenase is an enzyme of the citric acid cycle. The molecular weight of the enzyme 70 kDa is sufficient to keep it inside the cavity. The conversion of L-malic acid into oxaloacetic acid is catalysed by the enzyme. This is accompanied by the reduction of the cofactor NAD+ into NADH and the release of one hydrogen ion per reaction. The reaction constant of the enzyme depends on the concentration of the respective substrate, as well as on the pH of the solution. Depending on the initial conditions, the reaction can proceed in both directions. In the case of low pH-values (i.e. when many protons are available) and a low concentration of L-malic acid, the reaction runs to the right, protons are consumed, and thus the local pH increases (cf.Fig. 1): OAA + NADH + H+ → L-malic acid + NAD+. In the case of high pH values, the reaction runs to the left, protons are produced, and thus the local pH decreases: OAA + NADH + H+ ← L-malic acid + NAD+. Note that SNARF-1 is only sensitive to changes in pH around its pKa value, and thus this fluorophore can be used to monitor the reaction in a certain pH range only. The reaction OAA + NADH + H+ → L-malic acid + NAD+ is favoured in the region where SNARF-1 responds most strongly to changes in pH.
In order to verify that the reaction ensures a significant impact on the pH, we first tested the reaction using free substrate (OAA), free cofactor (NADH), free enzyme (malate dehydrogenase), and free SNARF-1-dextran as a pH indicator in a solution of initial pH = 6. Upon addition of the substrate (10 mM OAA) and the cofactor (75 μM NADH) to the enzyme (7700 units per mL), a time-dependent change of fluorescence signal of the solution from dominant emission in the yellow region (580 nm) to dominant emission in the red region (640 nm) was observed, which indicates that the solution became more alkaline upon consumption of the protons, cf.Fig. 2a. A calibration curve, which allows the pH to be obtained from the ratio of red-to-yellow fluorescence (Ir/Iy), is shown in Fig. 2b. These data demonstrate that the reaction catalysed by malate dehydrogenase can be followed in situ by monitoring the pH.
In addition to the fluorescent pH indicator, SNARF-1-dextran, the pH indicator thymol blue was also utilized, which exhibits absorption spectrum changes in response to pH. Using UV/vis absorption spectroscopy allowed the pH during the reaction (by monitoring changes in the spectrum of thymol blue) to be measured, in addition to directly measuring the changes in the NADH concentration. Thymol blue changes its colour from blue (pH > 9) to yellow (pH < 7.4) (cf. Fig. S1†). The visual change is attributed to an increase in absorption at 430 nm and a decrease at 595 nm (cf. Fig. S2†). The change in absorption at 595 nm is more sensitive and shows the sigmoidal slope that is typical for a pH indicator. In contrast, the cofactor NADH shows two absorption bands at 260 nm and 340 nm, whilst the oxidized form NAD+ absorbs only at 260 nm. Therefore, we analysed the change in NADH concentration in the solution by UV/vis absorption spectroscopy at 340 nm. This method is common in systems involving NAD+/NADH for the determination of enzyme activities. These measurements cannot be performed in the presence of the dye, because of its absorption of UV light. The full set of data using UV/vis absorption spectroscopy is presented in the ESI.† In agreement with experiments involving SNARF-1-dextran, the data verify that the enzymatic activity of malate dehydrogenase can be followed by in situ pH measurements, in which a pH indicator and an enzyme are free in solution.
For the construction of a sensor system, we attempted to link both the enzyme and the pH-sensitive dye into one carrier particle. As there are multiple molecules in solution (in particular the carrier particles, which absorb and scatter light), we decided to use fluorescence instead of absorption spectroscopy as the most convenient read-out, and employed SNARF-1-dextran as a pH-sensitive reporter. Polyelectrolyte polymer capsules fabricated by layer-by-layer assembly were used as microreactor particles. To prepare the sensing microcapsules, malate dehydrogenase and SNARF-1-dextran were first co-precipitated in calcium carbonate particles. The particles were then coated with PSS and PAH using the well-described layer-by-layer technique.32–34 In a final step, the CaCO3 template was removed by re-dispersing the capsules in EDTA, which forms complexes with the Ca2+ ions. The resulting capsules, filled with the enzyme and SNARF-1-dextran, had a diameter of 3–5 μm. An image of such capsules is presented in Fig. 3. The capsules were spherical and well dispersed. The fluorescent dye, SNARF-1-dextran, was encapsulated and remained within the capsule after removal of the core. As reported in previous studies, the dye is not homogeneously distributed in the capsule cavity, and instead sticks to the inner capsule wall.15 Because SNARF-1 molecules were coupled to 70 kDa dextran, which has the same molecular weight as the enzyme, it is reasonable to assume that the enzyme stays within the cavity as well, in agreement with previous studies,19 though its presence cannot be concluded from the microscopy images.
Finally, we examined the combined system (enzyme and SNARF-1 encapsulated together) by recording the fluorescence intensities of SNARF-1-dextran at suitable wavelengths using a fluorescence microscope. The presence and activity of the enzyme can be proven by recording the changes in pH. Controls (capsules without the enzyme) were first investigated, cf.Fig. 4. The presence of the cofactor NADH has no impact on the Ir/Iy read-out of the dye, cf.Fig. 4A. Although in this case no enzymatic conversion can take place due to the lack of the enzyme, OAA already interacts with SNARF-1, as seen by the increasing Ir/Iy-values over time, independent of whether NADH is present or not, cf.Fig. 4B and C. OAA slowly decomposes in aqueous solution into pyruvic acid and CO2, accompanied by an increase in pH. However, the rate of the alkalinizing effect of this decomposition was determined to be much slower than the observed phenomenon in Fig. 4B and C.
 | ||
| Fig. 4 Control system. Polyelectrolyte multilayer capsules without malate dehydrogenase were prepared. The capsules carrying SNARF-1-dextran were only exposed to OAA (500 μM) in the absence (B) or in the presence of NADH (50 μM, (C), dashed line). Lane A represents results obtained for the cofactor alone (500 μM). The substrate (OAA) was added at the time point indicated with an asterisk. The cofactor (NADH) was added at the time point marked with two asterisks. The reaction was followed for 25 minutes. The initial pH of all the solutions was adjusted to 6. The corresponding fluorescence images are shown on the right side, illustrating the response of a single capsule in the red (R) and yellow (Y) channel, in brightfield transmission mode (TM) and in a pseudo-coloured image (R/Y) corresponding to the Ir/Iy ratio per pixel (the same colour map as used in the plot window). The correlation of Ir/Iy to pH is shown in Fig. 2b, and is indicated by the colour code presented in the top panel. | ||
In the next set of experiments, capsules carrying malate dehydrogenase were exposed to their substrate OAA, in the presence of different concentrations of the cofactor NADH. The changes in SNARF-1-dextran fluorescence intensities were then recorded. As shown in Fig. 5, the initiation of the enzymatic reaction led to a decrease in the amount of free H+ ions. This was visualised by the change in the Ir/Iy ratio of the fluorescence intensities of SNARF-1-dextran, as recorded in the range from 560–615 nm (Iy, yellow channel) to 615–750 nm (Ir, red channel), cf.Fig. 2a. The reaction only took place in the presence of the substrate (OAA). Without the substrate, the pH of the solution remained constant over time (cf.Fig. 5A), indicating that no reaction occurred. The presence of the cofactor increased the reaction rate. Saturation at high Ir/Iy values is caused by a decrease of the reaction rate and not by limited sensitivity of SNARF-1, as at Ir/Iy > 3.5, SNARF-1 is still responsive to changes in pH, cf.Fig. 2b.
 | ||
| Fig. 5 Concentration-dependent changes in the emission spectra I(λ) of encapsulated SNARF-1-dextran, reported as Ir/Iy (t), in response to malate dehydrogenase activity. The sensor capsules carrying malate dehydrogenase and SNARF-1-dextran were exposed to OAA (500 μM) in the absence (B) or presence of NADH (50 μM (C) or 100 μM (D)). The substrate and cofactor were added at the time point indicated with an asterisk. Lane A represents results obtained for the cofactor alone (500 μM). The reaction was followed for 12 min. The initial pH of all the solutions was adjusted to 6. The corresponding fluorescent images are shown on the right side, illustrating the response of a single capsule in the red (R) and yellow (Y) channel, in brightfield transmission mode (TM) and in a pseudo-coloured image (R/Y) corresponding to the Ir/Iy ratio per pixel (the same colour map as used in the plot window). The Ir/Iy ratio could be directly related to the corresponding pH values according to Fig. 2b, as displayed by the colour code in the panel. | ||
To demonstrate the effect of the cofactor on the kinetics of the enzymatic reaction, the capsules carrying the enzyme were incubated with the substrate first, followed by the addition of NADH. As shown in Fig. 6, the more coenzyme was added, the higher the increase in fluorescence intensities of SNARF-1 in the red channel. Moreover, the sensor capsules showed no significant response to the cofactor alone (cf.Fig. 6A). Addition of the substrate caused a steady increase of the Ir/Iy ratio (cf.Fig. 6B), as already seen for the control particles without encapsulated enzymes, cf.Fig. 4B. The reaction could be enhanced by the addition of NADH (cf.Fig. 6C–EversusFig. 6B). At an initial NADH concentration of 100 μM, the local pH strongly increased temporarily (cf.Fig. 6C and D). After a few minutes, the cofactor was used up and the pH inside the capsules became equilibrated with the outside value. For higher concentrations of the cofactor, the bulk pH was also increased by the sensing reaction (cf.Fig. 6E). Note that the Ir/Iy curves with OAA and with NADH lie on top of the curve with OAA, but without NADH. When small amounts of NADH were added (cf.Fig. 6C), after NADH has been consumed by the enzymatic reaction, the Ir/Iy curve approaches the curve in which NADH was not added (cf.Fig. 6B). The substrate can be enzymatically converted when the cofactor is present.
The data in Fig. 6C and D suggest that the continuance of the enzymatic reaction is limited by the amount of NADH present. Therefore, we tested whether it could be continued by supplying the system with additional NADH. To that end, the sensing capsules carrying malate dehydrogenase were first incubated with the substrate, followed by the addition of the cofactor. When the Ir/Iy ratio has reached a plateau value, additional NADH was added. Indeed, as seen in Fig. 7, this allowed the reaction to continue.
Conclusions
The synthesis and characterization of a particle bound sensor for the detection of OAA, based on enzymatic digestion followed by pH measurements has been demonstrated. As the enzymatic conversion of OAA is a continuous process, as long as the substrate and cofactor are available, this reaction needs to be monitored over time. This has been demonstrated with time lapse recordings of individual polyelectrolyte capsules, which serve as carrier particles for both enzymes and pH sensitive fluorophores. As the present work is a proof-of-concept study, no detailed analysis about selectivity and sensitivity is given. Instead, it is interesting to discuss the potential applications of such sensors for intracellular detection. It is well accepted that polyelectrolyte polymer capsules, as used here, are internalized by adherent cell lines via endocytosis, and that their final intracellular location is the lysosome.31,35–37 This however involves general problems associated with particle based intracellular sensing.16 Though some methods exist to release molecules from capsules (in the size range from ca. 150 nm38 to 3–5 μm39–41) residing inside lysosomes into the cytosol, in a typical scenario, sensors will remain trapped inside lysosomes, which suggests sensing in the lysosome as the most facile application. Besides imposing a hostile environment to particles (lysosomal proteases, highly acidic pH), localization inside the lysosome imposes other limits to particle based intra-lysosomal sensing. The sensing principle, as sketched in Fig. 1, relies on the detection of protons, which are consumed upon enzymatic digestion of the target molecule OAA. However, all pH-sensitive fluorophores have a limited range of operation, centred around their pKa value. While the lysosomal pH can be highly acidic down to pH values of 3 (precise values depend on the cell line), the pKa value of encapsulated SNARF-1-dextran, as used in this study, is 6.8. Thus, inside lysosomes, the pH-indicator SNARF-1 would be barely sensitive to small changes in pH upon digestion of OAA by malate dehydrogenase. In the data shown in this work, the pH of the starting solution was buffered on purpose to a value around 6, which sets an optimal range for observing the increase in pH upon the enzymatic reaction. In order to apply the sensing strategy for the intra-lysosomal detection of OAA, an alternative pH-indicator with a highly acidic pKa value would be required. In the ESI†, thymol blue is presented as an example in this direction, though its pKa value (ca. 1.7) would be too low for sensing inside lysosomes. It also needs to be pointed out, that the particle architecture could be improved. In our work, enzymes and analyte-sensitive fluorophores were co-localized in the cavity of polyelectrolyte polymer capsules. However, in principle, these capsules also allow for a multicompartment structure, in which enzymes and fluorophores could be placed at different locations.42–47 Based on these considerations we suggest that further improvements could help to facilitate intracellular enzyme-based fluorescence sensing using polymer capsules as carrier systems. In addition to intracellular sensing, other potential applications which involve enzymatic or enzymatic-coupled reactions could be envisioned, such as enzymatic microreactors for chemical analysis (e.g. sensing of glucose, food contaminants, etc.), kinetic studies and catalysis. At any rate, it should be highlighted that inhomogeneous and/or variable loading of molecules in the capsules, as well as different diffusion limits of the molecules involved, hampers the application of the current coupled system for absolute detection of concentrations (be it for the proof-of-concept system described here or for other enzyme-based sensing systems). Nevertheless, the proof-of-concept system reported here demonstrates the feasibility of real time monitoring of enzymatic reactions (as well as kinetics) in situ. Further improvements of the material (e.g. homogeneous loading, development of new fluorescence probes, multi-compartmental geometries, etc.) will strengthen the potential applications of enzyme-loaded capsules.Acknowledgements
This work was supported by LOEWE (grant Synchembio to WJP). PRG acknowledges support from the MINECO (RYC-2012-10059 and CTQ2013-45433-P).References
- D. W. Domaille, E. L. Que and C. J. Chang, Nat. Chem. Biol., 2008, 4, 168–175 CrossRef CAS PubMed.
- L. Kazakova, L. Shabarchina, S. Anastasova, A. Pavlov, P. Vadgama, A. Skirtach and G. Sukhorukov, Anal. Bioanal. Chem., 2013, 405, 1559–1568 CrossRef CAS PubMed.
- A. S. Susha, A. Munoz_Javier, W. J. Parak and A. L. Rogach, Colloids Surf., A, 2006, 281, 40–43 CrossRef CAS PubMed.
- M. J. Ruedas-Rama, A. Orte, E. A. H. Hall, J. M. Alvarez-Pez and E. M. Talavera, Analyst, 2012, 137, 1500–1508 RSC.
- W. Qi, Z. Liu, J. Lai, W. Gao, X. Liu, M. Xu and G. Xu, Chem. Commun., 2014, 50, 8164–8166 RSC.
- E. A. Lemke and C. Schultz, Nat. Chem. Biol., 2011, 7, 480–483 CrossRef CAS PubMed.
- M. J. Ruedas-Rama and E. A. H. Hall, Anal. Chem., 2010, 82, 9043–9049 CrossRef CAS PubMed.
- V. Pavlov, B. Shlyahovsky and I. Willner, J. Am. Chem. Soc., 2005, 127, 6522–6523 CrossRef CAS PubMed.
- R. Freeman and I. Willner, Nano Lett., 2009, 9, 322–326 CrossRef CAS PubMed.
- R. Freeman, L. Bahshi, T. Finder, R. Gill and I. Willner, Chem. Commun., 2009, 764–766 RSC.
- F. Khan, T. E. Saxl and J. C. Pickup, Anal. Biochem., 2010, 399, 39–43 CrossRef CAS PubMed.
- F. Wen, Y. Dong, L. Feng, S. Wang, S. Zhang and X. Zhang, Anal. Chem., 2011, 83, 1193–1196 CrossRef CAS PubMed.
- S. Hiller, A. Schnackel and E. Donath, Cytometry, Part A, 2005, 64, 119 Search PubMed.
- M. Semmling, O. Kreft, A. Muñoz Javier, G. B. Sukhorukov, J. Käs and W. J. Parak, Small, 2008, 4, 1763–1768 CrossRef CAS PubMed.
- L. L. del Mercato, A. Z. Abbasi and W. J. Parak, Small, 2011, 7, 351–363 CrossRef CAS PubMed.
- K. Kantner, S. Ashraf, S. Carregal-Romero, C. Carrillo-Carrion, M. Collot, P. del Pino, W. Heimbrodt, D. J. De Aberasturi, U. Kaiser, L. I. Kazakova, M. Lelle, N. M. de Baroja, J. M. Montenegro, M. Nazarenus, B. Pelaz, K. Peneva, P. R. Gil, N. Sabir, L. M. Schneider, L. I. Shabarchina, G. B. Sukhorukov, M. Vazquez, F. Yang and W. J. Parak, Small, 2015, 11, 896–904 CrossRef CAS PubMed.
- O. P. Tiourina, A. A. Antipov, G. B. Sukhorukov, N. L. Larionova, Y. Lvov and H. Mohwald, Macromol. Biosci., 2001, 1, 209–214 CrossRef CAS.
- N. G. Balabushevich, G. B. Sukhorukov and N. I. Larionova, Macromol. Rapid Commun., 2005, 26, 1168–1172 CrossRef CAS.
- M. Ochs, S. Carregal-Romero, J. Rejman, K. Braeckmans, S. C. De Smedt and W. J. Parak, Angew. Chem., Int. Ed., 2013, 52, 695–699 CrossRef CAS PubMed.
- C. S. Karamitros, A. M. Yashchenok, H. Moehwald, A. G. Skirtach and M. Konrad, Biomacromolecules, 2013, 14, 4398–4406 CrossRef CAS PubMed.
- F. Caruso, D. Trau, H. Möhwald and R. Renneberg, Langmuir, 2000, 16, 1485–1488 CrossRef CAS.
- M. Fischlechner, Y. Schaerli, M. F. Mohamed, S. Patil, C. Abell and F. Hollfelder, Nat. Chem., 2014, 6, 791–796 CrossRef CAS PubMed.
- A. M. Pavlov, G. B. Sukhorukov and D. J. Gould, J. Controlled Release, 2013, 172, 22–29 CrossRef CAS PubMed.
- O. S. Sakr and G. Borchard, Biomacromolecules, 2013, 14, 2117–2135 CrossRef CAS PubMed.
- E. Donath, G. B. Sukhorukov, F. Caruso, S. A. Davis and H. Möhwald, Angew. Chem., Int. Ed., 1998, 37, 2202–2205 CrossRef CAS.
- G. B. Sukhorukov, A. L. Rogach, B. Zebli, T. Liedl, A. G. Skirtach, K. Köhler, A. A. Antipov, N. Gaponik, A. S. Susha, M. Winterhalter and W. J. Parak, Small, 2005, 1, 194–200 CrossRef CAS PubMed.
- M. F. Bedard, A. Munoz-Javier, R. Mueller, P. del Pino, A. Fery, W. J. Parak, A. G. Skirtach and G. B. Sukhorukov, Soft Matter, 2009, 5, 148–155 RSC.
- O. Kreft, M. Prevot, H. Mohwald and G. B. Sukhorukov, Angew. Chem., Int. Ed. Engl., 2007, 46, 5605–5608 CrossRef CAS PubMed.
- L. I. Kazakova, L. I. Shabarchina and G. B. Sukhorukov, Phys. Chem. Chem. Phys., 2011, 13, 11110 RSC.
- P. Rivera Gil, M. Nazarenus, S. Ashraf and W. J. Parak, Small, 2012, 8, 943–948 CrossRef CAS PubMed.
- R. Hartmann, M. Weidenbach, M. Neubauer, A. Fery and W. J. Parak, Angew. Chem., Int. Ed., 2015, 54, 1365–1368 CrossRef CAS PubMed.
- P. Rivera Gil, L. L. del Mercato, P. del Pino, A. Muñoz-Javier and W. J. Parak, Nano Today, 2008, 3, 12–21 Search PubMed.
- J. W. Cui, M. P. van Koeverden, M. Mullner, K. Kempe and F. Caruso, Adv. Colloid Interface Sci., 2014, 207, 14–31 CrossRef CAS PubMed.
- S. De Koker, R. Hoogenboom and B. G. De Geest, Chem. Soc. Rev., 2012, 41, 2867–2884 RSC.
- A. Muñoz_Javier, O. Kreft, M. Semmling, S. Kempter, A. G. Skirtach, O. Bruns, P. d. Pino, M. F. Bedard, J. Rädler, J. Käs, C. Plank, G. Sukhorukov and W. J. Parak, Adv. Mater., 2008, 20, 4281–4287 CrossRef.
- L. Kastl, D. Sasse, V. Wulf, R. Hartmann, J. Mircheski, C. Ranke, S. Carregal-Romero, J. A. Martínez-López, R. Fernández-Chacón, W. J. Parak, H.-P. Elsaesser and P. Rivera Gil, ACS Nano, 2013, 7, 6605–6618 CrossRef CAS PubMed.
- Y. Yan, A. P. R. Johnston, S. J. Dodds, M. M. J. Kamphuis, C. Ferguson, R. G. Parton, E. C. Nice, J. K. Heath and F. Caruso, ACS Nano, 2010, 4, 2928–2936 CrossRef CAS PubMed.
- J. Ruesing, O. Rotan, C. Gross-Heitfeld, C. Mayer and M. Epple, J. Mater. Chem. B, 2014, 2, 4625–4630 RSC.
- A. Muñoz Javier, P. del Pino, M. F. Bedard, A. G. Skirtach, D. Ho, G. B. Sukhorukov, C. Plank and W. J. Parak, Langmuir, 2008, 24, 12517–12520 CrossRef PubMed.
- S. Carregal-Romero, M. Ochs, P. Rivera Gil, C. Ganas, A. M. Pavlov, G. B. Sukhorukov and W. J. Parak, J. Controlled Release, 2012, 159, 120–127 CrossRef CAS PubMed.
- C. Ganas, A. Weiß, M. Nazarenus, S. Rösler, T. Kissel, P. Rivera_Gil and W. J. Parak, J. Controlled Release, 2014, 196, 132–138 CrossRef CAS PubMed.
- O. Kreft, A. G. Skirtach, G. B. Sukhorukov and H. Mohwald, Adv. Mater., 2007, 19, 3142–3145 CrossRef CAS.
- L. L. del Mercato, A. Z. Abbasi, M. Ochs and W. J. Parak, ACS Nano, 2011, 5, 9668–9674 CrossRef CAS PubMed.
- B. V. Parakhonskiy, A. M. Yashchenok, M. Konrad and A. G. Skirtach, Adv. Colloid Interface Sci., 2014, 207, 253–264 CrossRef CAS PubMed.
- R. Xiong, S. J. Soenen, K. Braeckmans and A. G. Skirtach, Theranostics, 2013, 3, 141–151 CrossRef CAS PubMed.
- M. Delcea, H. Moehwald and A. G. Skirtach, Adv. Drug Delivery Rev., 2011, 63, 730–747 CrossRef CAS PubMed.
- M. Delcea, A. Yashchenok, K. Videnova, O. Kreft, H. Möhwald and A. G. Skirtach, Macromol. Biosci., 2010, 10, 465 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4tb02077d |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |