 Open Access Article
Open Access ArticleCorrelating antimicrobial activity and model membrane leakage induced by nylon-3 polymers and detergents†
Sara G.
Hovakeemian‡
a,
Runhui
Liu‡
bc,
Samuel H.
Gellman
b and
Heiko
Heerklotz
*ade
aLeslie Dan Faculty of Pharmacy, University of Toronto, Canada
bDepartment of Chemistry, University of Wisconsin-Madison, WI, USA
cState Key Laboratory of Bioreactor Engineering and School of Materials Science and Engineering, East China University of Science and Technology, China
dInstitute of Pharmaceutical Sciences, University of Freiburg, Germany. E-mail: heiko.heerklotz@pharmazie.uni-freiburg.de
eBIOSS Centre for Biological Signalling Studies, Freiburg, Germany
First published on 3rd August 2015
Abstract
Most antimicrobial peptides act upon target microorganisms by permeabilizing their membranes. The mode of action is often assessed by vesicle leakage experiments that use model membranes, with the assumption that biological activity correlates with the permeabilization of the lipid bilayer. The current work aims to extend the interpretation of vesicle leakage results and examine the correlation between vesicle leakage and antimicrobial activity. To this end, we used a lifetime-based leakage assay with calcein-loaded vesicles to study the membrane permeabilizing properties of a novel antifungal polymer poly-NM, two of its analogs, and a series of detergents. In conjunction, the biological activities of these compounds against Candida albicans were assessed and correlated with data from vesicle leakage. Poly-NM induces all-or-none leakage in polar yeast lipid vesicles at the polymer's MIC, 3 μg mL−1. At this and higher concentrations, complete leakage after an initial lag time was observed. Concerted activity tests imply that this polymer acts independently of the detergent octyl glucoside (OG) for both vesicle leakage and activity against C. albicans spheroplasts. In addition, poly-NM was found to have negligible activity against zwitterionic vesicles and red blood cells. Our results provide a consistent, detailed picture of the mode of action of poly-NM: this polymer induces membrane leakage by electrostatic lipid clustering. In contrast, poly-MM:CO, a nylon-3 polymer comprised of both cationic and hydrophobic segments, seems to act by a different mechanism that involves membrane asymmetry stress. Vesicle leakage for this polymer is transient (limited to <100%) and graded, non-specific among zwitterionic and polar yeast lipid vesicles, additive with detergent action, and correlates poorly with biological activity. Based on these results, we conclude that comprehensive leakage experiments can provide a detailed description of the mode of action of membrane permeabilizing compounds. Without this thorough approach, it would have been logical to assume that the two nylon-3 polymers we examined act via similar mechanisms; it is surprising that their mechanisms are so distinct. Some, but not all mechanisms of vesicle permeabilization allow for antimicrobial activity.
Introduction
Physics of membrane models meets chemistry of new antibiotic materials, but will they also meet biology? Specifically, can the fungicidal activity of new nylon-3 polymers be understood and predicted in terms of biophysical measurements and models of membrane leakage? Our answer is yes, to some extent, but not without much caution and more information than that provided by traditional assays.In recent years, there has been a considerable effort to develop new antibiotics in light of the emergence of antibiotic-resistant bacteria. Host-defense peptides (HDPs) have been suggested to represent a good stepping-stone to developing new therapies because of their ability to kill a wide array of microbes.1 The majority of HDPs and their analogs are believed to act at least in part by permeabilizing the cellular membranes of target organisms; some of these peptides may act via other mechanisms as well. A wide variety of membrane-targeted mechanisms has been proposed and reviewed.2–7 Amphiphilic peptides that induce positive spontaneous monolayer curvature (similar to effects of detergents) can cause non-specific defects or leaks by inducing membrane thinning and disordering,8–11 form toroidal pores,12–14 or solubilize the lipid locally.15 Other peptides may induce negative spontaneous curvature16 or form oligomeric channels (barrel-stave model) that do not involve membrane curvature, at least once the channel is formed. Finally, polycationic compounds as the polymers studied here can also damage membranes by electrostatic lipid clustering as described by Epand and co-workers.1 A correlation between anionic lipid clustering and selective cytotoxic action on bacterial species has, for example, been reported for a synthetic α/β-peptide,17 a sequence-random copolymer,18 a synthetic acyl lysine oligomer,19 a 12-residue fragment of the human HDP LL-37,20 and the HDP cateslytin.21 Upon superficial adsorption or binding of these polycationic compounds to a bilayer, anionic lipids are believed to cluster, giving rise to a domain with high charge density and non-average composition. Such lipid clustering may, in turn, induce membrane leakage by inducing a local lipid composition that fails to maintain a stable bilayer, or by creating mismatch and stress along the domain boundaries.22 The fact that certain polycationic species, such as oligo acyl-lysines (OAKs), can not only cluster lipids23 but also induce the formation of cochleates24,25 implies that such polycation–lipid interactions can induce positive or negative membrane curvature.
Mimics of HDPs have been developed to improve activity, selectivity, and stability in the body, and to reduce the costs of production and formulation. Synthetic peptides26,27 and sequence specific oligomers28–31 were some of the first developed HDP mimics. However, cheaper and more readily prepared polymers have recently emerged as promising alternatives.32–42 Here we characterize one of the few selectively fungicidal polymers developed so far,43–45 the nylon-3 polymer poly-NM, and two hydrophobically modified analogs (poly-MM and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 MM
60 MM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CO, Fig. 1), and compare their behaviour with the behaviours of a series of detergents.
CO, Fig. 1), and compare their behaviour with the behaviours of a series of detergents.
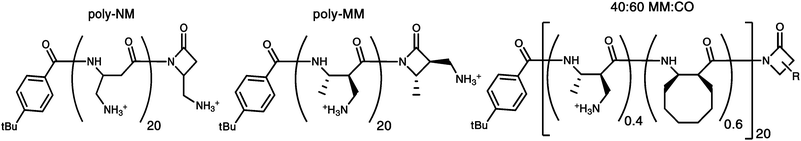 | ||
Fig. 1 Chemical structures of the nylon-3 polymers poly-NM, poly-MM and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 MM 60 MM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CO. All the polymers have an average chain length of approximately 20 subunits, and all are heterochiral. CO. All the polymers have an average chain length of approximately 20 subunits, and all are heterochiral. | ||
The hypothesis that an antimicrobial compound acts by inducing membrane leakage has long been tested by vesicle leakage experiments. These assays are usually based on the de-quenching of an entrapped dye upon release from the vesicle interior.46,47 Unfortunately, the mere observation that certain vesicles are permeabilized at some concentration of a peptide or other antimicrobial agent does not constitute strong evidence that membrane permeabilization is the primary mode of antibiotic action. Instead, membrane permeabilization could be a prerequisite of the ability of the antimicrobial molecule to enter microbial cells and act by inhibiting a specific cytoplasmic protein or interacting with DNA. Vesicle permeabilization could even be simply an artifact, unrelated to biological activity because the permeabilization mechanism does not apply to the target cell, or because antibiotic action is manifested at concentrations well below that needed for vesicle permeabilization.
Prior to this work, rather few studies have aimed to employ sophisticated analysis of vesicle leakage for identifying a specific mode of permeabilization. Blumenthal and coworkers were the first to classify leakage from vesicles as all-or-none (in contrast to graded) by monitoring the release of a self-quenching dye during scans that span the lipid phase transition temperature.47 This classification was taken further by Wimley et al.'s re-quenching protocol.48 Then, the additional information provided by time-resolved fluorescence spectroscopy49 was utilized to detect the leakage mechanism by monitoring the amount and local concentration of both free and entrapped dye.50–52 An alternative, statistical evaluation of the influx of a dye into giant vesicles53,54 is much more demanding with respect to time and instrumentation but can provide a histogram of leakage of a vesicle population directly. Several models have been introduced to model leakage kinetics. Matsuzaki described the slowing down of leakage rates due to the translocation of peptide to the inner leaflet.55,56 Almeida and coworkers validated a model based on rates of binding and formation and dissociation of all-or-none pores.57 For surfactin, we discussed a superposition of two major leakage phenomena and described these empirically with exponentials.58 The size of pores or defects has also been assessed by comparing the efflux of dyes of different sizes, including fluorescently labelled dextran molecules.59 Finally, concerted activity tests (CATs) combining peptides and detergents have been proposed to test whether the mode of action of the peptides involved detergent-like effects.60 A detergent-like mode of action was found to be valid for magainin, a helical peptide that forms toroidal pores, as well as for surfactins and fengycins from Bacillus subtilis.
The general aim of our work is to improve the validity and depth of the interpretation of vesicle leakage experiments to understand the specific mode of membrane permeabilization by antimicrobial compounds. To this end, we have carried out a comprehensive set of leakage tests that provides a more detailed picture, rather than simply determining whether or not leakage occurs. Our analysis includes leakage mechanism, kinetics, and concerted activity tests with a membrane-permeabilizing detergent. We have compared these results with antimicrobial activity data. The specific objectives of this paper are to (i) provide, to our knowledge for the first time, a detailed characterisation of the leakage behaviour of an antimicrobial agent believed to act by electrostatic clustering, (ii) elucidate the relationship between structure and mode of action for fungicidal nylon-3 polymers, and (iii) develop hypotheses and strategies that will be broadly useful for assessing the relevance of vesicle leakage data for antimicrobial action.
Materials and methods
Materials
The lipids, 1-palmitoyl-2-oleoyl-sn-3-phosphatidylcholine (POPC) and yeast polar lipid extract (YPLE) were purchased from Avanti Polar Lipids (Alabaster, AL). Calcein, MOPS, and NaCl, of the highest purity available, were purchased from Sigma. The non-ionic detergents n-octyl-β-D-glucopyranoside (OG), octaethylene glycol monodecyl ether (C12EO8), and 6-cyclohexyl-1-hexyl-β-D-maltoside (CYMAL-6) were purchased in Anagrade purity (99% HPLC) from Affymetrix (Santa Clara, CA). The ionic detergents N-dodecyl trimethylammonium chloride (DTAC), cetyl trimethyl ammonium bromide (CTAB), sodium dodecyl sulfate, (SDS), N-lauroyl sarcosine (sodium salt) (NLS) were also purchased from Affymetrix (Santa Clara, CA) at the highest purity available. The ionic detergents 1-dodecylpyridinium chloride hydrate (DPyrC) and benzalkonium chloride (BAC) were purchased from Sigma at the highest purity available. The K1 strain of Candida albicans was a generous gift from Prof. David Andes at the University of Wisconsin-Madison. Escherichia coli JM 109, B. subtilis BR151, and Staphylococcus aureus 1206 (methicillin-resistant) were obtained from Prof. Bernard Weisblum at University of Wisconsin-Madison. RPMI 1640 (31800-089) was obtained from Life Technologies (Grand Island, NY).Vesicle experiments were carried out with standard buffer (130 mM NaCl and 25 mM MOPS, adjusted to a pH of 7.0) and isoosmotic calcein buffer (70 mM calcein, 25 mM MOPS, pH 7.0). Osmotic pressures were checked using a Wescor Vapro 5520 osmometer. All C. albicans cell culture and antifungal studies used RPMI 1640 medium (containing L-glutamine but not sodium bicarbonate) buffered with MOPS to pH 7.0.
Polymer preparation
All nylon-3 polymers were prepared in a moisture-controlled glove box using THF (poly-MM and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 MM
60 MM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CO) or DMAc (poly-NM) as the reaction solvent by following the protocol reported previously.45 Side-chain NH-Boc protected polymers were characterized by GPC using THF (poly-MM and 40
CO) or DMAc (poly-NM) as the reaction solvent by following the protocol reported previously.45 Side-chain NH-Boc protected polymers were characterized by GPC using THF (poly-MM and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 MM
60 MM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CO) or DMAc (poly-NM) as the mobile phase by following the protocol reported previously.45
CO) or DMAc (poly-NM) as the mobile phase by following the protocol reported previously.45
Antifungal assay
The MIC assays toward C. albicans cells were conducted by using a previously described protocol45 suggested by the Clinical and Laboratory Standards Institute (CLSI). C. albicans cells were inoculated in adjusted RPMI medium (buffered with 0.145 M MOPS to pH 7.0) and incubated at 30 °C for 2–3 days. The cultured C. albicans cells were collected by centrifugation and suspended in 0.145 M NaCl at 2.5 × 106 cells per mL to generate the stock suspension. The working suspension for C. albicans cell was prepared as a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of the stock suspension using adjusted RPMI medium (buffered with 0.145 M MOPS to pH 7.0). Two-fold serial dilution of nylon-3 polymers was conducted in a 96-well plate using adjusted RPMI to obtain concentrations from 400 to 0.4 μg mL−1. Each sample well had 100 μL of compound solution after the two-fold serial dilution. Then, 100 μL of the cell working suspension was added to each well (except the cell-free blank control), followed by gentle shaking of the plate for 10 s. The plate was then incubated at 37 °C for 2–3 days, and fungal cell growth was evaluated visually. On the same plate, wells containing only RPMI medium were used as the negative control (cell-free blank); wells containing cells in RPMI without any polymer or antifungal drug were used as the positive control. The antifungal MIC was the lowest concentration of a polymer or antifungal drug that completely inhibited fungal cell growth. Each experiment was performed in duplicate on a given day, and experiments were repeated on two different days.
1000 dilution of the stock suspension using adjusted RPMI medium (buffered with 0.145 M MOPS to pH 7.0). Two-fold serial dilution of nylon-3 polymers was conducted in a 96-well plate using adjusted RPMI to obtain concentrations from 400 to 0.4 μg mL−1. Each sample well had 100 μL of compound solution after the two-fold serial dilution. Then, 100 μL of the cell working suspension was added to each well (except the cell-free blank control), followed by gentle shaking of the plate for 10 s. The plate was then incubated at 37 °C for 2–3 days, and fungal cell growth was evaluated visually. On the same plate, wells containing only RPMI medium were used as the negative control (cell-free blank); wells containing cells in RPMI without any polymer or antifungal drug were used as the positive control. The antifungal MIC was the lowest concentration of a polymer or antifungal drug that completely inhibited fungal cell growth. Each experiment was performed in duplicate on a given day, and experiments were repeated on two different days.
The MIC assays toward C. albicans spheroplasts were conducted in adjusted RPMI using the same protocol as described above for C. albicans cells. To prepare C. albicans spheroplasts, harvested C. albicans cells were centrifuged and redispersed in spheroplasting buffer (10 mM TRIS, pH 7.4, 1 mM EDTA, 1 M sorbitol, and 30 mM 2-mercaptoethanol) at 0.3 g wet cell per mL. Then Zymolyase-20T (200 units per g wet cell) was added to the cell suspension, and the mixture was shaken gently for 1.5 h. After treatment, these cells were washed with RPMI and resuspended in PRPMI at 5 × 106 cells per mL and then were further diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in RPMI to give the final working suspension.
1000 in RPMI to give the final working suspension.
For the CAT study to evaluate if nylon-3 polymers and OG can have synergistic or additive effect, polymers were prepared in 2-fold serial dilution with supplementation of OG at designated concentrations. MIC assays of (polymer + OG) toward both C. albicans cells and spheroplasts were performed by following the same protocol described above.
Antibacterial assay
The minimum inhibitory concentration (MIC) assay for bacteria was conducted by following a two-fold broth microdilution protocol previously described.61 Three bacteria were tested: Escherichia coli JM 109, B. subtilis BR151, and Staphylococcus aureus 1206 (methicillin-resistant). Briefly, bacteria were cultured overnight at 37 °C on LB agar plates and then suspended in LB medium at 2 × 106 cells per mL. The cell suspensions (50 μL) were mixed with the same volume of polymer solutions in two-fold serial dilutions (from 400 to 3.13 μg mL−1) in a 96-well plate, which was incubated for 6 h at 37 °C. Optical density (OD) of each well was measured at 650 nm on a Molecular Devices Emax precision microplate reader. Controls included on the same plate: LB medium only (blank) and cells in LB without polymer (uninhibited growth control). The bacterial cell growth in each well was calculated with the equation (% cell growth = (Apolymer − Ablank)/(Acontrol − Ablank) × 100%) and plotted against polymer concentration. The MIC value is the minimum concentration of a given polymer necessary to inhibit bacterial growth completely.Hemolysis assay
Hemolysis assays were conducted as previously described using human red blood cells (RBCs).62,63 Human whole blood (5 mL) was washed three times with TRIS-buffered saline (TBS, pH 7.2) that was composed of 10 mM TRIS and 150 mM NaCl. The collected RBCs were suspended in TBS (250 mL) to obtain a working suspension of 2% RBC relative to total RBC in the whole blood. Two-fold serial dilution of nylon-3 polymers was conducted in a 96-well plate in TBS to obtain concentrations ranging from 800 to 6.25 μg mL−1. Each sample well had 100 μL of compound solution after the two-fold serial dilution. Then 100 μL of the RBC working suspension was added to each well, followed by gentle shaking for 10 s. On the same plate, wells containing TBS without polymer were used as the blank; wells containing Triton X-100 (3.2 μg mL−1 in TBS) were used as the positive control that give 100% RBS lysis. The plate was incubated at 37 °C for 1 h, and then centrifuged at 3700 RPM for 5 min to precipitate the RBCs. Aliquots of 80 μL supernatant from each well were transferred to corresponding wells in a new 96 well plate, and the optical density (OD), at 405 nm, was measured using a Molecular Devices Emax precision microplate reader. Measurements were performed in duplicate, and each measurement was repeated on two different days. The percentage of hemolysis at each polymer concentration was calculated with the equation (% hemolysis = 100% × (Apolymer − Ablank)/(Acontrol − Ablank)) and plotted against polymer concentration to give the dose–response curves for hemolysis for each polymer. The HC10 value for each polymer was defined as the polymer concentration to cause 10% lysis of RBCs.Vesicle preparation
Vesicles were made from POPC or YPLE. The preparation of large unilamellar vesicles (LUVs) is described in detail elsewhere.64 Briefly, lipid was dried from stock solutions in chloroform, hydrated with buffer, freeze–thawed seven times, and extruded ten times through a Lipex Inc. (Burnaby, BC) pressure extruder, equipped with Whatman Nuclepore (GE Healthcare) filters of 100 nm pore size. Then, the buffer outside the calcein-loaded vesicles was exchanged for isotonic standard buffer using a PD-10 desalting column (GE Healthcare). The lipid concentration was quantified by the phosphate colorimetric assay (Biovision). Note that this assay defines lipid concentration on the basis of phosphate content. Thus, one cardiolipin counts as “two lipids” and molecules without phosphate, such as ergosterol, are not considered. Vesicle sizes of 100–130 nm were validated by dynamic light scattering using a Malvern Nano ZS system (Worcestershire, UK).Leakage assay
A detailed protocol of the leakage assay is described elsewhere.50 Vesicles loaded with 70 mM calcein were injected into a solution of the active compound or compounds to be tested. The final lipid concentration was 30 μM, and the sample volume 1.5 mL. Except for kinetics experiments, the samples were then incubated for 1 hour on a gently rocking shaker at room temperature.Time-resolved fluorescence decay curves were acquired at 515 nm using a Fluorolog 3 time-correlated single photon counting (TCSPC) system from HORIBA Jobin Yvon (Edison NJ). A 467 nm laser diode pulsed at 1 MHz was used as the excitation source. Decay curves, F(t), of approximately 104 peak counts were obtained by accumulating photon counts for 180 s. HORIBA's DAS6 software was used to fit decay curves biexponentially by deconvoluting them with the instrument response function as measured with a scattering LUDOX solution.
Entrapped calcein can be distinguished from dilute, free calcein based on their different fluorescence lifetimes, τE and τF, in a free, biexponential fit of the decay:
 | (1) |
 | (2) |
Results
Vesicle leakage as a function of polymer concentration
Fig. 2 shows the results of YPLE and POPC vesicle leakage induced by all three polymers, poly-NM, poly-MM, and poly-MM:CO, for comparison. Fig. 2A shows the dye efflux from YPLE vesicles after 1 h of incubation as a function of polymer concentration. An efflux of, for example, 20% after 1 h is induced by concentrations, cE20, about 2 μg mL−1 poly-MM:CO, 4 μg mL−1 poly-MM, and 4.6 μg mL−1 poly-NM, respectively, in 30 μM YPLE vesicles. All experimental series were repeated several times; repeats are represented by different symbols of the same outline and color but different fillings. Poly-MM plateaus at 6 μg mL−1 at an efflux of around 43%.Leakage experiments performed on POPC vesicles (chosen as a generic, uncharged model membrane because it is composed of purely zwitterionic lipid) were also conducted (Fig. 2C). The homocationic polymers, poly-NM and poly-MM, only caused ≤4% leakage up to 200 μg mL−1, which is, for example, 65 times higher than the MIC of poly-NM against C. albicans (Table 1). However, the co-polymer poly-MM:CO, leaked POPC vesicles more effectively than vesicles made of YPLE, i.e. cE20 decreased to 0.1 μg mL−1 and 2 μg mL−1 in POPC and YPLE vesicles, respectively. The Fig. 2B and D show the efflux of dye as a function of the entrapped fluorescence lifetime on a reciprocal scale. This reveals the leakage mechanism as described in detail elsewhere.50 To briefly summarize; in Fig. 2B, increasing efflux, E, induced by poly-NM and poly-MM leaves the lifetime of entrapped dye (τE) constant, i.e., the local concentration of entrapped dye is unchanged (see blue and green arrows in Fig. 2B). Note that the error of fitting τE at E ≤ 60% is estimated <±0.1 ns. Increased errors at E > 60% (when only ≈4% or less of the counts arise from entrapped dye), some minor variation in τE from one batch to another, and the optical expansion of the low-τ range by the reciprocal scale cause a scatter of the data points; this does not challenge the identification of the leakage mechanism. Increasing E at constant τE is indicative of all-or-none leakage.50,52 That means, some vesicles retain all dye (governing τE) while others lose virtually all entrapped dye through a major pore or defect structure. These leaky vesicles increase E but do not contribute to τE.
| Poly-NM (μg mL−1) | Poly-MM (μg mL−1) | Poly-MM:CO (μg mL−1) | OG (mM) | |
|---|---|---|---|---|
| MIC | ||||
| E. coli | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
>200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
100 | |
| S. aureus | 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
100 | |
| B. subtilis | 6.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
6.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
25 | |
| RBC (HC10) | >400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
>400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
<3 | |
| C. albicans | 3.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 43 |
25 | >75 |
| C. albicans spheroplasts | 3–6 | 12–25 | 19–38 | |
| c E20 observed in vesicle leakage measurements | ||||
| YPLE, 1 h | 4.6 | 4 | 2 | 9 |
| YPLE, 24 h | 2–4 | 2 | 2 | |
| YPLE, mechanism | A-O-N | A-O-N | Graded | Graded |
| POPC, 1 h | >200 | >200 | 0.1 | |
| CAT (polymer + OG) | ||||
| C. albicans spheroplasts | Independent | Independent | ||
| YPLE | Independent | Additive | ||
Poly-MM:CO shows homogeneously graded leakage for both lipids (Fig. 2B and D); all vesicles leak a similar amount of dye, thus increasing free dye and reducing the internal concentration of calcein (which is reciprocally related to τE) in the same proportion.
Kinetics of dye release
Fig. 3 shows the dye efflux from YPLE vesicles as a function of time after exposure to detergents and polymers. A control experiment monitoring YPLE vesicles with no polymer added did not exceed 2% efflux after 24 h (not shown). For comparison, we also recorded leakage kinetics for a cationic (DTAC) and a non-ionic (OG) detergent acting on YPLE vesicles. The detergents induce a biphasic leakage behavior that could be fitted by the expression:E(t) = 1 − E1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp{−k1t} − (1 − E1)exp{−k2t}. exp{−k1t} − (1 − E1)exp{−k2t}. | (3) |
 | ||
| Fig. 3 Efflux from 30 μM YPLE induced by different polymers and detergents as indicated (A–C, E and F) as a function of incubation time. The concentrations of the permeabilizers are indicated in the plot. Curves in panels A–C are fit curves according to eqn (3), the resulting parameters E1 and k2 are correlated to each other in panel D. Curves in panels E, F serve to guide the eye. | ||
Given that more specific phenomena can largely be ruled out for detergents, the second, slow but unlimited, leakage process can be assigned to membrane perturbation by monolayer curvature strain. This involves membrane thinning, disordering, and the stabilization of defects or pore-like structures by covering their edges with a detergent-rich rim.67 Leakage kinetics due to this process are governed by the probability of a defect to open, which obviously increases with increasing concentration. However, defects can start from equilibrated membranes so that they re-appear with the same probability.
OG, DTAC, and poly-MM:CO (Fig. 3) as well as surfactin58 all agree with the principal behaviour described by eqn (3), but the balance between the two processes is different. Fig. 3D correlates E1 (a measure for the extent of asymmetry-induced leakage) with the rate of unlimited leakage (k2). It is not surprising that there is a correlation between bilayer curvature strain (governing E1) and monolayer curvature strain (governing k2) induced by an amphiphilic perturbant. In fact, both detergents, OG and DTAC, show a very similar correlation.
The leakage kinetic profiles of poly-NM and poly-MM (Fig. 3E and F) are fundamentally different and cannot be modelled by eqn (3). A well pronounced lag time implies that leakage requires the cooperative formation of a specific structure or feature, by contrast to the apparently homogeneous and potentially instantaneous, detergent-like phenomena. Above a threshold value (2 μg mL−1 for poly-NM), there is an intermediate concentration range where the lag time decreases with increasing polymer concentration. This is what one would expect if the lag time is governed, for example, by the association of the polymer. Finally, at high polymer concentrations (5–20 μg mL−1 for poly-MM, 7–15 μg mL−1 for poly-NM), leakage is not speeding up with increasing concentration any more. One possible reason for such a behavior would be that the molecules superseding a critical concentration form inactive aggregates in solution.52 Note that the limiting leakage curve for 10–20 μg mL−1 in Fig. 3F accounts also for the plateau of E measured after 1 h as seen in Fig. 2A.
Concerted action test (CAT) with the detergent OG
CAT experiments involving poly-NM + OG and poly-MM:CO + OG were conducted in order to shed further light on the leakage mechanism of these compounds as compared to detergent-like mechanisms. OG was selected because it is very well characterized, equilibrates with a membrane quickly, and is unlikely to show any specific interaction with the polymer. To this end, the OG concentration was kept constant within a series of leakage samples and the polymer concentration was varied. The CAT is based on the concept of additivity of fractional activities. The fractional inhibitory concentration index (FICI), also known as the combination index (CI), is used to quantify synergistic or antagonistic deviations from additivity.Eqn (4) below describes the activity of a combination of compound A and B; which in this case would be the detergent OG (A) and poly-NM or MM:CO (B):
 | (4) |
Fig. 4 shows the CAT results of poly-NM combined with the nonionic detergent, OG, against vesicles of 30 μM YPLE. Fig. 4A shows the leakage curve of OG alone, which yields a cE20 of 9 mM and a cE60 of 10 mM. These values obtained at 30 μM lipid are essentially identical with those reported for 300 μM.60 Note that the CMC of OG is 23 mM68,69 and its membrane partitioning is so weak (K ≈ 0.1 mM−1)67 that at these lipid concentrations, >97% of the of the detergent is free in solution. It is only in this case of cL ≪ 1/K that the leakage curve, E(c), does not depend on cL. The crosses in Fig. 4A indicate OG concentrations used in CAT experiments, 5, 6.5, and 8 mM. Alone, 5, 6.5, and 8 mM OG induce only marginal leakage of ∼2, 5, and 10%, respectively, but they represent a substantial fraction of the concentration needed for strong leakage (9–10 mM). The data points in Fig. 4B and C represent CAT experiments, where the 3 sets of fixed OG concentrations were combined with variable polymer concentrations. Again, points of the same outline and color but different fill patterns represent repetitions with new batches of vesicles and polymer stock. The bold blue solid line in Fig. 4B is identical to the blue line of Fig. 2A. It illustrates the effect of poly-NM in the absence of OG. If poly-NM was to act by a similar perturbation as OG, its activity would be increasingly enhanced in the presence of increasing concentrations of OG so that the leakage curves would shift to lower polymer concentrations. We calculated the curves to be expected for exact additivity, as defined by a combination index (CI) of 1 (see eqn (4)). The predictions for CI = 1 are represented by the dashed orange, red, and purple curves for poly-NM + 5, 6.5, and 8 mM OG, respectively.
 | ||
| Fig. 4 CAT of poly-NM combined with OG acting against 30 μM YPLE vesicles. Panel A shows the dye efflux after 1 h, E, as a function of OG concentration in the absence of polymer; crosses denote the concentrations used for the CATs shown in panel B. Panel B: Orange triangles, red diamonds, and purple hexagons denote E measured in CATs with constant OG concentrations of 5, 6.5, and 8 mM, as a function of poly-NM concentration, respectively. The blue line is identical to that in Fig. 2A and illustrates the leakage induced by poly-NM in the absence of OG. Dashed-dot orange, red and, purple lines represent the predicted additive curves (calculated using eqn (4)) when 5, 6.5, and 8 mM OG is added to induce leakage with poly-NM. Thin solid lines are only to guide the eye. Panel C shows efflux as a function of fluorescence lifetime of entrapped dye, τE, on a reciprocal scale. The blue arrow is copied from Fig. 2B, representing poly-NM alone. | ||
Very much in contrast to these predictions, any selected concentration of the poly-NM induces just as much leakage (or even slightly less) in the presence of the detergent (orange triangles, red diamonds, purple hexagons) compared to its absence (blue solid line). This implies that the action of poly-NM is largely independent of the detergent. This is also clearly seen in Fig. 4C, where the effects on E and τE are simply superpositions of those of OG (crosses in Fig. 4C) and the polymer. That means, even in the mixture, the two components retain their individual leakage mechanisms, unaffected by the other component.
Fig. 5 shows the results of the CAT experiment of poly-MM:CO combined with OG against vesicles composed of YPLE. The bold blue solid line (identical to red solid line of Fig. 2A) represents poly-MM:CO leakage in the absence of OG. The 3 OG concentrations used for the CAT experiments are the same as used for poly-NM; see crosses in Fig. 4A. Very much unlike poly-NM, the experimental data (orange triangles, red diamonds, and purple hexagons) fit the curves calculated for CI = 1 (dash-dot lines) within error (Fig. 5A). That means, the membrane-permeabilizing action of poly-MM:CO is additive with that of the detergent. This is supported by the fact that the E(1/τE) data obtained for the mixtures are, roughly, averages of the values for the individual components (Fig. 5B). The additivity between poly-MM:CO and OG suggests that poly-MM:CO works by a similar, detergent-like perturbation. To this end, it needs to insert into the hydrophobic/hydrophilic interface of the membrane bilayer.
 | ||
| Fig. 5 CAT of poly-MM:CO combined with OG acting against 30 μM YPLE vesicles. Panel A: Orange triangles, red diamonds, and purple hexagons denote E measured in 3 concerted action tests with constant OG concentrations of 5, 6.5, and 8 mM, as a function of poly-MM:CO concentration, respectively. The blue solid line is identical to the red solid line of Fig. 2A and illustrates the leakage induced by poly-MM:CO in the absence of OG. Dashed-dot orange, red and, purple lines represent the predicted additive curves (calculated using eqn (3)) when 5, 6.5, and 8 mM OG is added to induce leakage with poly-MM:CO. Panel B shows efflux as a function of fluorescence lifetime of entrapped dye, τE, on a reciprocal scale. | ||
Vesicle leakage induced by nonionic and ionic surfactants
For comparison, we have also tested vesicle leakage as induced by a series of nonionic, zwitterionic, and ionic surfactants against 30 μM YPLE vesicles after 1 h of incubation. The ionic detergents, namely CTAB, BAC, DPyrC, SDS, NLS, and DTAC, were chosen for this study because of their common use as antiseptics. Like CAMPs, they are cationic (except for SDS); so naturally, they are effective at causing leakage of anionic vesicles. The results are summarized in Table 2; see ESI† for the leakage curves and the molecular structures of the detergents. CTAB induces all-or-none leakage that plateaus at about 60% at a concentration up to 1.25 mM. All other detergents exhibit heterogeneously graded leakage (some-or-none) where all vesicles are leaking out dye, some more than others, causing a gradual increase in τE.| Surfactant | Mechanism | c E20 (mM) | c E60 (mM) | MIC C. albicans (mM) | MIC C. albicans spheroplasts (mM) |
|---|---|---|---|---|---|
| a Octaethylene glycol monododecyl ether. b n-Octyl-β-D-glucopyranoside. c 6-Cyclohexyl-1-hexyl-β-D-maltoside. d C316 – CHAPS. e N-Dodecyl trimethylammonium chloride. f Cetyl trimethyl ammonium bromide. g Sodium dodecyl sulfate. h 1-Dodecylpyridinium chloride hydrate. i N-Lauroyl sarcosine (sodium salt). j Benzalkonium chloride. ‘HetG’ denotes ‘heterogeneously graded’ leakage and ‘A-O-N’ denotes ‘all-or-none’ leakage. The asterisks represent leakage from POPC vesicles. | |||||
C12EO8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
Graded | 0.05 | 0.06 | >75 | >75 |
| OGb | Graded | 9 | 10 | >75 | 19 |
| CYMAL-6c | A-O-N | 0.44* | 0.53* | >75 | >75 |
| CHAPSd | A-O-N | 1.7*60 | 2.5*60 | >75 | >75 |
| DTACe | Graded | 0.58 | 1.6 | 0.07 | 0.07 |
| CTABf | A-O-N | 0.03 | 0.08 | 0.04 | 0.04 |
| SDSg | Graded | 0.44 | 0.69 | 0.15 | 0.15 |
| DPyrCh | Graded | 0.25 | 0.61 | 0.07 | 0.07 |
| NLSi | Graded | 1 | 2.2 | 1.17 | 1.17 |
| BACj | Graded | 0.05 | 0.1 | 0.04 | 0.04 |
Antimicrobial activity
Cellular assays presented in a previous study (see ref. 43 in Table 1) had revealed a considerable activity of poly-NM against C. albicans, with a MIC of 3.1 μg mL−1. The low MIC of poly-NM against C. albicans and its inactivity against red blood cells had indicated the potency and selectivity of this polymer as a fungicide. The analog poly-MM was found to be mostly inactive.Here, we tested poly-MM:CO analogously against the same organisms, revealing, by stark contrast, weak antimicrobial but strong hemolytic activity (Table 1). Furthermore, poly-NM and poly-MM:CO were tested against C. albicans spheroplasts, where the cell wall is removed. The agreement of the MICs of cells and spheroplasts indicates that the cell wall does not impose any significant resistance to the polymer. By contrast, the detergent OG was active against spheroplasts at a concentration of the order of its critical micelle concentration (CMC), 23 mM.68,69 However, OG is inactive against C. albicans cells. Finally, we performed concerted activity tests (CAT) for biological activity assays. To this end, several series of MIC experiments for poly-NM and poly-MM:CO against C. albicans spheroplasts were repeated in the presence of 8 mM, 15 mM, and 30 mM of OG. In all series, the MICs were unchanged within error compared to those in the absence of the detergent. The same applied to a test with C. albicans cells and 8 mM OG. We concluded in Table 1 that both poly-NM and poly-MM:CO show an antibiotic activity that is independent of detergent action. The leakage from vesicles induced by the various detergents was compared to MIC values against C. albicans cells and spheroplasts (Table 2). None of the non-ionic and zwitterionic detergents (C12EO8, OG, CHAPS, CYMAL-6) were effective against C. albicans cells (MIC > 75 mM); only OG showed activity (MIC = 19 mM) against spheroplasts. The ionic, or ‘antiseptic’, detergents showed the same activity in C. albicans cells and spheroplasts. Their MIC and cE20 values were similar within an order of magnitude. The existing differences, particularly for DTAC and DPC, imply a somewhat stronger activity against cells than against YPLE vesicles.
Discussion
Leakage behaviour of poly-NM: electrostatic clustering?
In contrast to most antimicrobial peptides and their mimics, poly-NM is expected to be largely hydrophilic rather than amphiphilic. Our experiments have provided the following information:Poly-NM is active against C. albicans (MIC = 3 μg mL−1) and permeabilizes vesicles of YPLE (anionic, leakage threshold between 2 and 4 μg mL−1, see Fig. 2A). This polymer does not permeabilize electrically neutral lipid membranes of red blood cells or POPC vesicles.
Poly-NM acts independently of a detergent, both against vesicles and cells, as seen by the CAT results. Since OG induces leakage by membrane thinning, disordering, and/or defect formation due to positive intrinsic curvature,67 the lack of additivity suggests that the polymer's action does not depend on positive curvature strain in the membrane.
Poly-NM causes all-or-none leakage after a lag time. Moreover, the permeabilization proceeds until all vesicles have lost all of their original contents. Kinetic models for efflux from pores (that open and close/disintegrate) allow for a lag time if pore opening is much slower than efflux through the open pore, but no substantial lag time was observed for the typical, graded and all-or-none, pore formers melittin70 or magainin,57 respectively. It appears that the assembly of a defect-inducing, electrostatic polymer–lipid-cluster requires more time than assembly of a toroidal pore induced by an amphipathic helix. Given the remaining uncertainty regarding the way in which electrostatic clustering leads to leakage, we can conclude from the CAT results that the underlying mechanism apparently does not involve positive curvature effects. Instead, perhaps leakage results from a thickness mismatch between the clusters and the matrix. Alternatively, the slight inhibition of poly-NM-induced leakage by OG might indicate the involvement of negative curvature in this polymer's mode of action. The cationic polymer's ability to bridge between charged lipid head groups might reduce average head group area and, hence, cause a local depression or invagination of the membrane. The good agreement between cE20 and the MIC argues against the alternative possibility that the toxicity of the clustering agent is not based on leakage but rather on interference with putative functional domains in the fungal membrane, which could have deleterious consequences for signaling and other crucial processes.71
Leakage behaviour of poly-MM:CO: asymmetry stress?
The behavior of amphiphilic poly-MM:CO is characterized by the following features.(i) Moderate activity against C. albicans and its spheroplasts (MIC ≥ 12 μg mL−1) but high activity against vesicles (both zwitterionic and anionic) and red blood cells (0.1–3 μg mL−1).
(ii) Transient, graded leakage of vesicles, followed by an almost complete annealing after 1 hour or less.
(iii) Vesicle leakage largely additive with detergent, but no detectable detergent effect on the MIC against C. albicans spheroplasts.
The selectivity and additivity of anti-vesicle action with a detergent are comprehensible, given that this polymer comprises hydrophobic ‘CO’ groups that can intercalate into the interface of the membrane. This intercalation underlies binding to neutral membranes and induction of curvature–strain effects within the membrane. We are aware of only one scenario that has been proposed to explain transient, graded leakage: leakage induced by asymmetric insertion of the permeabilizer into the outer membrane leaflet.65,66,70 If molecules intercalate selectively between the lipids in the outer leaflet, they expand the optimal area of this leaflet while the optimal area of the inner leaflet remains unchanged. As a result, the inner leaflet is stretched (increasing the exposure of hydrophobic surfaces to water), and the membrane tends to bend outward. At a threshold, the stress may relax by a transient “cracking-in” of molecules to the inner leaflet.65,66 This translocation of membrane components may be accompanied by some limited, transient efflux of solutes from the vesicle interior that stops after relaxation of the stress.
The lipopeptide surfactin from B. subtilis shows a leakage pattern similar to that observed for the MM:CO nylon-3 copolymer.58 Surfactin induces graded, transient leakage,58 acts additively with a detergent,60 acts against neutral membranes (POPC vesicles), and causes vesicle leakage in lower the micromolar range,58,60i.e., an order of magnitude below MICs against typical fungal pathogens.72 Unlike surfactin,73 the large poly-MM:CO molecule might not be able to “crack in” to the inner leaflet by itself. Instead, the asymmetry stress could be relaxed by translocating lipid only.
It should be acknowledged that the first asymmetry-based mechanism for limited, graded leakage was proposed by Matsuzaki and coworkers in 1995.55 It is based on the dilution of the local peptide concentration after partial redistribution to the inner leaflet. Given the re-binding from the aqueous solution and the overall concentrations of permeabilizer, transmembrane dilution does not explain our findings. However, in both cases, the annealing after partial, graded leakage results from the dissipation of membrane asymmetry.
Toroidal pore formation
Typical polycationic, antibacterial peptides such as magainin may also involve anionic lipid clustering effects,74 but their action involves the formation of pores by stabilizing their curved edges. This is a positive-curvature phenomenon, as illustrated by the fact that magainin permeabilizes liposomes additively with a detergent.7 Independent action of poly-NM and OG emphasizes a principal difference in the role of clustering between the polymer and the peptide.Magainin induces all-or-none leakage12,57,75 that proceeds, above the threshold peptide concentration, to 100% dye efflux without a significant lag time.57 The concentration range that induces leakage of vesicles mimicking bacterial membranes (e.g., PC–PG and PC–PG–PE mixtures) is 0.3–1 μM,57,60 in very good agreement with MICs against a variety of bacteria.76 This set of leakage parameters for magainin (mechanism, selectivity, kinetics, CAT) is clearly distinguishable from parameters we have obtained for poly-NM (electrostatic clustering mechanism proposed) and poly-MM:CO (asymmetry stress mechanism proposed).
Vesicle leakage versus biological activity
It is traditional to test the hypothesis that a given peptide or other compound exerts antimicrobial action via microbial membrane permeabilization by examining that compound's ability to permeabilize vesicles formed with a lipid mixture that is thought to represent the composition of the biological membrane. Except for poly-MM:CO, all compounds found here to inhibit C. albicans growth also perforated YPLE vesicles at concentrations of the same order of magnitude. In most cases, the agreement of MIC and cE20 was significantly better than 10-fold.From the perspective of illustrating anti-Candida activity, poly-MM and most of the non-ionic detergents tested here could be referred to as “false positives” because they permeabilize YPLE vesicles despite being inactive against the fungus. There are many possible explanations for the inability of these agents to inhibit the growth of C. albicans (see also ref. 4). (i) The specific, highly asymmetric lipid composition of the C. albicans membrane (inner and outer leaflets have distinct compositions) might be more resistant than the symmetric (scrambled) composition that is unavoidable in a model bilayer. (Since fungicides do not act equally against all fungi, it is not possible to identify a generic model membrane for fungi.) (ii) There might be an antagonistic factor present in the biological system that blocks the action of poly-MM and/or nonionic surfactants. Possible active mechanisms of repair or protection include removal (pumps) or degradation of the perturbant or active balancing of the area asymmetry between the lipid leaflets by flippases or permeant membrane constituents. Since the polymers are not subject to enzymatic proteolysis, mechanisms of degradation are not obvious. (iii) The access of poly-MM or nonionic detergents to the cytoplasmic membrane or the specific mode of membrane permeabilization might be inhibited by the presence of the cell wall (see OG, Table 2). Unfortunately, there is no simple, obvious criterion that would predict whether a vesicle-permeabilizing compound exerts activity toward microbes by membrane permeabilization (see Tables 1 and 2). We hypothesize that the distinct mechanisms by which vesicle leakage may be induced vary in the likelihood that they are pertinent to cell membranes. Electrostatic clustering and the formation of large, toroidal pores might be more biologically relevant mechanisms for antibiotic activity than, for example, asymmetry-induced “cracking-in” or budding, because the latter is prevented by the cell wall or by flipases that regulate the lateral pressure difference between the two membrane leaflets. Recalling our above hypothesis (Fig. 3) that the detergent-like mechanism comprises a fast, graded, “cracking-in” contribution and a slow, all-or-none, pore formation, we note that Kuroda and co-workers reported that bactericidal action of amphiphilic polymers require one to several hours.77 So, when discussing vesicle data, one must consider the proper time scale.
Compounds that show some activity against vesicles at very low concentration but become self-inhibited at higher concentration might tend to display only weak activity in vivo. This pattern of behavior has been found for poly-MM (Table 1) in this work, and for fengycin.52,72
In order to test and refine or extend these hypotheses, it will be necessary to conduct additional comprehensive leakage and activity tests, as exemplified here, for a large number of antimicrobial peptides and their mimics. In general, vesicle leakage tests will be much more informative if their correlation with biological action is established more thoroughly than has usually been the case previously.
Conclusions
The fact that an antimicrobial compound permeabilizes vesicles generated with lipids characteristic of a certain microbe is a hint that this compound may inhibit the growth of that microbe; however, this observation does not guarantee that the compound will be active against that microbe, or if the compound is active, that the relevant mode of action (i.e., the mechanism(s) operational at the lowest concentration and in the shortest time) depends upon membrane permeabilization. We hypothesize that the applicability of vesicle leakage studies to suggest the biological mode of action depends on the leakage mechanism. We further propose that it is necessary to perform thorough vesicle and cell tests (including measurements of kinetics and concerted activity tests), as exemplified in this study, in order to validate the relevance of membrane leakage for antimicrobial activity.The cationic nylon-3 polymer poly-NM shows a leakage pattern that is in line with expectations for electrostatic lipid clustering (all-or-none vesicle contents release, reaching 100% after a lag time, selectivity for anionic membranes, independence of detergent). Results for leakage experiments involving poly-NM and YPLE vesicles correlate well with antimicrobial activity against C. albicans. Leakage resulting from electrostatic clustering by poly-NM does not involve positive-curvature effects. In contrast, the cationic-hydrophobic polymer poly-MM:CO appears to permeabilize vesicles by a different mechanism, asymmetry stress (transient and limited release of contents, graded leakage, additive with detergent); this mode of action does not seem to be operational against fungal cells, or only poorly so.
Acknowledgements
Constructive comments on the manuscript by Sebastian Fiedler and Helen Y. Fan are gratefully acknowledged. This work was supported by a grant from the National Science and Engineering Council of Canada to H.H. and by NIH grant R01GM093265 to S.H.G.References
- R. M. Epand and R. F. Epand, J. Pept. Sci., 2011, 17, 298–305 CrossRef CAS PubMed.
- R. E. W. Hancock and H. G. Sahl, Nat. Biotechnol., 2006, 24, 1551–1557 CrossRef CAS PubMed.
- W. C. Wimley, ACS Chem. Biol., 2010, 5, 905–917 CrossRef CAS PubMed.
- K. A. Brogden, Nat. Rev. Microbiol., 2005, 3, 238–250 CrossRef CAS PubMed.
- L. T. Nguyen, E. F. Haney and H. J. Vogel, Trends Biotechnol., 2011, 29, 464–472 CrossRef CAS PubMed.
- A. Tossi, L. Sandri and A. Giangaspero, Pept. Sci., 2000, 55, 4–30 CrossRef CAS.
- M. Zasloff, Nature, 2002, 415, 389–396 CrossRef CAS PubMed.
- B. Bechinger and K. Lohner, Biochim. Biophys. Acta, 2006, 1758, 1529–1539 CrossRef CAS PubMed.
- H. Heerklotz and J. Seelig, Biophys. J., 2001, 81, 1547–1554 CrossRef CAS.
- A. S. Ladokhin and S. H. White, Biochim. Biophys. Acta, 2001, 1514, 253–260 CrossRef CAS.
- A. Wiese, M. Munstermann, T. Gutsmann, B. Lindner, K. Kawahara, U. Zahringer and U. Seydel, J. Membr. Biol., 1998, 162, 127–138 CrossRef CAS.
- S. J. Ludtke, K. He, W. T. Heller, T. A. Harroun, L. Yang and H. W. Huang, Biochemistry, 1996, 2960, 13723–13728 CrossRef PubMed.
- K. Matsuzaki, O. Murase, N. Fujii and K. Miyajima, Biochemistry, 1996, 35, 11361–11368 CrossRef CAS PubMed.
- K. Matsuzaki, K. Sugishita, N. Ishibe, M. Ueha, S. Nakata, K. Miyajima and R. M. Epand, Biochemistry, 1998, 37, 11856–11863 CrossRef CAS PubMed.
- Z. Oren and Y. Shai, Biopolymers, 1998, 47, 451–463 CrossRef CAS.
- K. Lohner, Gen. Physiol. Biophys., 2009, 28, 105–116 CrossRef CAS.
- R. F. Epand, M. a. Schmitt, S. H. Gellman and R. M. Epand, Biochim. Biophys. Acta, 2006, 1758, 1343–1350 CrossRef CAS PubMed.
- R. F. Epand, B. P. Mowery, S. E. Lee, S. S. Stahl, R. I. Lehrer, S. H. Gellman and R. M. Epand, J. Mol. Biol., 2008, 379, 38–50 CrossRef CAS PubMed.
- R. M. Epand, S. Rotem, A. Mor, B. Berno and R. F. Epand, J. Am. Chem. Soc., 2008, 130, 14346–14352 CrossRef CAS PubMed.
- R. F. Epand, G. Wang, B. Berno and R. M. Epand, Antimicrob. Agents Chemother., 2009, 53, 3705–3714 CrossRef CAS PubMed.
- F. Jean-François, S. Castano, B. Desbat, B. Odaert, M. Roux, M.-H. Metz-Boutigue and E. J. Dufourc, Biochemistry, 2008, 47, 6394–6402 CrossRef PubMed.
- R. M. Epand and R. F. Epand, Mol. BioSyst., 2009, 5, 580–587 RSC.
- R. F. Epand, A. Mor and R. M. Epand, Cell. Mol. Life Sci., 2011, 68, 2177–2188 CrossRef CAS PubMed.
- R. F. Epand, H. Sarig, D. Ohana, B. Papahadjopoulos-Sternberg, A. Mor and R. M. Epand, J. Phys. Chem. B, 2011, 115, 2287–2293 CrossRef CAS PubMed.
- L. Livne, R. F. Epand, B. Papahadjopoulos-Sternberg, R. M. Epand and A. Mor, FASEB J., 2010, 24, 5092–5101 CrossRef CAS PubMed.
- Y. Hamuro, J. P. Schneider and W. F. Degrado, J. Am. Chem. Soc., 1999, 121, 12200–12201 CrossRef CAS.
- D. Liu and W. F. DeGrado, J. Am. Chem. Soc., 2001, 123, 7553–7559 CrossRef CAS PubMed.
- L. Arnt, J. R. Rennie, S. Linser, R. Willumeit and G. N. Tew, J. Phys. Chem. B, 2006, 110, 3527–3532 CrossRef CAS PubMed.
- J. Rennie, L. Arnt, H. Tang, K. Nüsslein and G. N. Tew, J. Ind. Microbiol. Biotechnol., 2005, 32, 296–300 CrossRef CAS PubMed.
- G. N. Tew, D. Clements, H. Tang, L. Arnt and R. W. Scott, Biochim. Biophys. Acta, 2006, 1758, 1387–1392 CrossRef CAS PubMed.
- G. N. Tew, D. Liu, B. Chen, R. J. Doerksen, J. Kaplan, P. J. Carroll, M. L. Klein and W. F. DeGrado, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5110–5114 CrossRef CAS PubMed.
- M. A. Gelman, B. Weisblum, D. M. Lynn and S. H. Gellman, Org. Lett., 2004, 6, 557–560 CrossRef CAS PubMed.
- K. Kuroda and W. F. DeGrado, J. Am. Chem. Soc., 2005, 127, 4128–4129 CrossRef CAS PubMed.
- B. P. Mowery, S. E. Lee, D. A. Kissounko, R. F. Epand, R. M. Epand, B. Weisblum, S. S. Stahl and S. H. Gellman, J. Am. Chem. Soc., 2007, 129, 15474–15476 CrossRef CAS PubMed.
- K. Lienkamp, A. E. Madkour, A. Musante, C. F. Nelson, K. Nusslein and G. N. Tew, J. Am. Chem. Soc., 2008, 130, 9836–9843 CrossRef CAS PubMed.
- E. F. Palermo, I. Sovadinova and K. Kuroda, Biomacromolecules, 2009, 10, 3098–3107 CrossRef CAS PubMed.
- F. Nederberg, Y. Zhang, J. P. K. Tan, K. J. Xu, H. Y. Wang, C. Yang, S. J. Gao, X. D. Guo, K. Fukushima, L. J. Li, J. L. Hedrick and Y. Y. Yang, Nat. Chem., 2011, 3, 409–414 CrossRef CAS PubMed.
- F. Costanza, S. Padhee, H. F. Wu, Y. Wang, J. Revenis, C. H. Cao, Q. Li and J. F. Cai, RSC Adv., 2014, 4, 2089–2095 RSC.
- R. Liu, X. Chen, S. Chakraborty, J. J. Lemke, Z. Hayouka, C. Chow, R. A. Welch, B. Weisblum, K. S. Masters and S. H. Gellman, J. Am. Chem. Soc., 2014, 136, 4410–4418 CrossRef CAS PubMed.
- R. Liu, J. M. Suarez, B. Weisblum, S. H. Gellman and S. M. McBride, J. Am. Chem. Soc., 2014, 136, 14498–14504 CrossRef CAS PubMed.
- S. Chakraborty, R. Liu, Z. Hayouka, X. Chen, J. Ehrhardt, Q. Lu, E. Burke, Y. Yang, B. Weisblum, G. C. Wong, K. S. Masters and S. H. Gellman, J. Am. Chem. Soc., 2014, 136, 14530–14535 CrossRef CAS PubMed.
- K. Kuroda and W. F. Degrado, J. Am. Chem. Soc., 2005, 127, 4128–4129 CrossRef CAS PubMed.
- R. Liu, X. Chen, Z. Hayouka, S. Chakraborty, S. P. Falk, B. Weisblum, K. S. Masters and S. H. Gellman, J. Am. Chem. Soc., 2013, 135, 5270–5273 CrossRef CAS PubMed.
- R. Liu, X. Chen, S. P. Falk, K. S. Masters, B. Weisblum and S. H. Gellman, J. Am. Chem. Soc., 2015, 137, 2183–2186 CrossRef CAS PubMed.
- R. Liu, X. Chen, S. P. Falk, B. P. Mowery, A. J. Karlsson, B. Weisblum, S. P. Palecek, K. S. Masters and S. H. Gellman, J. Am. Chem. Soc., 2014, 136, 4333–4342 CrossRef CAS PubMed.
- T. M. Allen and L. G. Cleland, Biochim. Biophys. Acta, 1980, 597, 418–426 CrossRef CAS.
- J. N. Weinstein, R. D. Klausner, Y. Innerarity, E. Ralston and R. Blumenthal, Biochim. Biophys. Acta, 1981, 647, 270–284 CrossRef CAS.
- W. C. Wimley, M. E. Selsted and S. H. White, Protein Sci., 1994, 3, 1362–1373 CrossRef CAS PubMed.
- M. Amaro, R. Šachl, P. Jurkiewicz, A. Coutinho, M. Prieto and M. Hof, Biophys. J., 2014, 107, 2751–2760 CrossRef CAS PubMed.
- H. Patel, C. Tscheka and H. Heerklotz, Soft Matter, 2009, 5, 2849 RSC.
- M. G. Lete, J. Sot, D. Gil, M. Valle, M. Medina, F. M. Goñi and A. Alonso, Biophys. J., 2015, 108, 863–871 CrossRef CAS PubMed.
- H. Patel, C. Tscheka, K. Edwards, G. Karlsson and H. Heerklotz, Biochim. Biophys. Acta, 2011, 1808, 2000–2008 CrossRef CAS PubMed.
- S. A. Wheaten, A. Lakshmanan and P. F. Almeida, Biophys. J., 2013, 105, 432–443 CrossRef CAS PubMed.
- G. Fuertes, A. J. García-Sáez, S. Esteban-Martín, D. Giménez, O. L. Sánchez-Muñoz, P. Schwille and J. Salgado, Biophys. J., 2010, 99, 2917–2925 CrossRef CAS PubMed.
- K. Matsuzaki, O. Murase, N. Fujii and K. Miyajima, Biochemistry, 1995, 34, 6521–6526 CrossRef CAS.
- K. Matsuzaki, O. Murase and K. Miyajima, Biochemistry, 1995, 34, 12553–12559 CrossRef CAS.
- S. M. Gregory, A. Pokorny and P. F. Almeida, Biophys. J., 2009, 96, 116–131 CrossRef CAS PubMed.
- H. Heerklotz and J. Seelig, Eur. Biophys. J., 2007, 36, 305–314 CrossRef CAS PubMed.
- A. S. Ladokhin, M. E. Selsted and S. H. White, Biophys. J., 1997, 72, 1762–1766 CrossRef CAS.
- H. Patel, Q. Huynh, D. Barlehner and H. Heerklotz, Biophys. J., 2014, 106, 2115–2125 CrossRef CAS PubMed.
- B. P. Mowery, A. H. Lindner, B. Weisblum, S. S. Stahl and S. H. Gellman, J. Am. Chem. Soc., 2009, 131, 9735–9745 CrossRef CAS PubMed.
- A. J. Karlsson, W. C. Pomerantz, B. Weisblum, S. H. Gellman and S. P. Palecek, J. Am. Chem. Soc., 2006, 128, 12630–12631 CrossRef CAS PubMed.
- T. L. Raguse, E. a. Porter, B. Weisblum and S. H. Gellman, J. Am. Chem. Soc., 2002, 124, 12774–12785 CrossRef CAS PubMed.
- H. Heerklotz, A. D. Tsamaloukas and S. Keller, Nat. Protoc., 2009, 4, 686–697 CrossRef CAS PubMed.
- S. Esteban-Martin, H. Jelger Risselada, J. Salgado and S. J. Marrink, J. Am. Chem. Soc., 2009, 131, 15194–15202 CrossRef CAS PubMed.
- H. Heerklotz, Biophys. J., 2001, 81, 184–195 CrossRef CAS.
- H. Heerklotz, Q. Rev. Biophys., 2008, 41, 205–264 CrossRef CAS PubMed.
- P. R. Majhi and A. Blume, Langmuir, 2001, 17, 3844–3851 CrossRef CAS.
- S. Paula, W. Siis, J. Tuchtenhagen and A. Blume, J. Phys. Chem., 1995, 99, 11742–11751 CrossRef CAS.
- A. Andersson, J. Danielsson, A. Graslund and L. Maler, Eur. Biophys. J., 2007, 36, 621–635 CrossRef CAS PubMed.
- K. Matsumoto, J. Kusaka, A. Nishibori and H. Hara, Mol. Microbiol., 2006, 61, 1110–1117 CrossRef CAS PubMed.
- J. Liu, I. Hagberg, L. Novitsky, H. Hadj-Moussa and T. J. Avis, Fungal Biol., 2014, 118, 855–861 CrossRef CAS PubMed.
- H. Y. Fan, M. Nazari, G. Raval, Z. Khan, H. Patel and H. Heerklotz, Biochim. Biophys. Acta, 2014, 1838, 2306–2312 CrossRef CAS PubMed.
- P. Wadhwani, R. F. Epand, N. Heidenreich, J. Burck, A. S. Ulrich and R. M. Epand, Biophys. J., 2012, 103, 265–274 CrossRef CAS PubMed.
- E. Grant, T. J. Beeler, K. M. Taylor, K. Gable and M. A. Roseman, Biochemistry, 1992, 31, 9912–9918 CrossRef CAS.
- M. Zasloff, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 5449–5453 CrossRef CAS.
- L. M. Thoma, B. R. Boles and K. Kuroda, Biomacromolecules, 2014, 15, 2933–2943 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sm01521a |
| ‡ S.G.H. and R.L. contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |

