Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Self-assembling dual component nanoparticles with endosomal escape capability†
Adelene S. M.
Wong
ab,
Sarah K.
Mann
ab,
Ewa
Czuba
b,
Audrey
Sahut
a,
Haiyin
Liu
b,
Tiffany C.
Suekama
abc,
Tayla
Bickerton
a,
Angus P. R.
Johnston
bd and
Georgina K.
Such
*a
aDepartment of Chemistry, The University of Melbourne, Parkville, Victoria 3010, Australia. E-mail: gsuch@unimelb.edu.au
bDrug Delivery, Disposition and Dynamics, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, Victoria 3052, Australia
cChemical & Petroleum Engineering, University of Kansas, Lawrence, Kansas 66045, USA
dARC Centre of Excellence in Convergent Bio-Nano Science and Technology, Monash University, Parkville, Australia
First published on 24th February 2015
Abstract
This study reports a novel nanoparticle system with simple and modular one-step assembly, which can respond intelligently to biologically relevant variations in pH. Importantly, these particles also show the ability to induce escape from the endosomal/lysosomal compartments of the cell, which is integral to the design of efficient polymeric delivery systems. The nanoparticles were formed by the nanoprecipitation of pH-responsive poly(2-(diethylamino)ethyl methacrylate) (PDEAEMA) and poly(2-(diethylamino)ethyl methacrylate)-b-poly(ethylene glycol) (PDEAEMA-b-PEG). Rhodamine B octadecyl ester perchlorate was successfully encapsulated within the hydrophobic core of the nanoparticle upon nanoprecipitation into PBS at pH 8. These particles disassembled when the pH was reduced below 6.8 at 37 °C. Cellular experiments showed the successful uptake of the nanoparticles into the endosomal/lysosomal compartments of 3T3 fibroblast cells. The ability to induce escape from the endosomes was demonstrated by the use of calcein, a membrane-impermeable fluorophore. The modular nature of these particles combined with promising endosomal escape capabilities make these dual component PDEAEMA nanoparticles useful for drug and gene delivery applications.
Introduction
The use of nanoparticles for drug delivery has received widespread attention in recent years.1–4 Soft nanoparticle carriers have the potential to improve therapeutic outcomes compared to therapeutics alone, due to their more targeted drug release.1,5 However, a greater understanding of the relationship between material properties and biological outcomes is still needed to develop next generation materials. Nanocarriers that are responsive toward biological stimuli such as pH, redox, and enzymatic variations are integral for achieving therapeutic release at a specific target site.6 In particular, pH has been a widely studied stimulus for the design of nanocarriers, as it has been well documented that polymer nanoparticles are typically internalized through acidic (endosomal/lysosomal) compartments of the cell.7,8 The pH drop that occurs during this process (from pH 7.4 in the bloodstream to ∼pH 5 in the late endosome/lysosome9) has provided researchers with a convenient but effective variable for the design of responsive nanocarriers.Charge shifting polymers, such as those containing tertiary amines, have been widely studied for the synthesis of pH-responsive nanoparticles.6 These polymers undergo a change from a hydrophobic (deprotonated) state to a hydrophilic (protonated) state upon a decrease in pH. This pH change is closely aligned with the pH variation that occurs during cellular internalization. These charge shifting properties can therefore be used to design nanoparticles that disassemble upon a decrease in pH, causing degradation of the carrier and subsequent release of the therapeutic payload. For example, Lomas et al. have demonstrated the successful encapsulation and delivery of DNA using pH-sensitive poly(2-(methacryloyloxyethyl phosphorylcholine)-co-poly(2-(diisopropylamino)ethyl methacrylate) (PMPC-PDPA) polymersomes. At neutral pH, DNA was encapsulated within the PMPC-PDPA copolymer vesicles but upon acidification these polymersomes dissociated into unimers allowing DNA to escape into the cytosol.10,11 In another example, Zhou et al. have demonstrated the effect of varying the structure of the ionizable tertiary amine groups attached to a poly(ethylene oxide) block copolymer. These polymers formed micelles that were observed to disassemble at varying pHs dependent on the structure of the ionizable tertiary amine groups.12
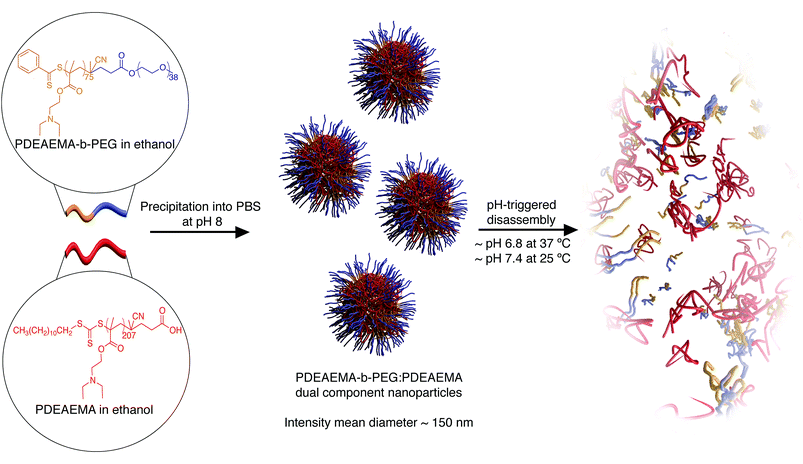
Herein, we report a modular, pH-responsive nanoparticle, which demonstrates disassembly within a biologically relevant pH range. These nanoparticles are synthesised using a modular, one-pot nanoprecipitation method combining two responsive charge shifting polymers, the homopolymer poly(2-(diethylamino)ethyl methacrylate) (PDEAEMA), and the diblock copolymer poly(2-(diethylamino)ethyl methacrylate)-b-poly(ethylene glycol) (PDEAEMA-b-PEG) (Scheme 1). Particle formation occurs due to the charge shifting nature of PDEAEMA, which transitions from hydrophilic to hydrophobic upon an increase in pH above its pKa of 7.0–7.3.13 Dynamic light scattering (DLS) studies demonstrated these particles had an interesting two-stage degradation, based on an early increase in particle size and then subsequent disassembly. At 37 °C the disassembly occurred at a pH of 6.8, making the degradation well suited to cellular delivery.
The ability of nanoparticles to induce trafficking out of acidic endosomal/lysosomal compartments, without causing cytotoxicity, is a major bottleneck in achieving successful cytosolic delivery of therapeutics, and has been the subject of many reviews.14–16 Some research groups have developed materials capable of endosomal escape. The most common endosomal escape mechanisms reported in these studies are the “proton sponge effect”,14 pH-induced swelling,13,17 and disassociation of the carrier into unimers within endosomal/lysosomal compartments leading to a large increase in osmotic pressure and subsequent rupture of the endosome/lysosome.10,18 Interestingly, these PDEAEMA dual component nanoparticles were observed to induce endosomal escape as demonstrated by a calcein green assay. We have previously demonstrated a related self-assembled nanoparticle system based on poly(2-(diisopropylamino)ethyl methacrylate) (PDPA) stabilized with a PEG–doxorubicin amphiphile.19 However, the system described by Liang et al. does not demonstrate the endosomal escape-inducing properties of these PDEAEMA dual component nanoparticles. Several research groups have synthesized PDEAEMA-based nanocarriers that are capable of endosomal escape. However, these nanocarriers are functionally different to the dual component system we have presented here. For example, Hu et al. have reported an elegant pH-responsive core–shell nanoparticle system comprised of a cross-linked PDEAEMA core and an aminoethyl methacrylate (AEMA)-rich shell.13,17 Their system showed a distinct swelling behaviour upon a change in pH from pH 7.4 to pH 5. This swelling behaviour is closely linked to their ability to traffic out of the endosomal/lysosomal compartments. PDEAEMA nanogel systems have also been discussed in a recent review.20 In contrast, the nanoparticle system reported herein is a dual component self-assembled system that rapidly disassociates upon acidification. In another example, Cheng et al. have reported the synthesis of a polyplex composed of dimethylaminoethyl methacrylate (DMAEMA), hydrophilic poly(ethylene glycol) methyl ether methacrylate (PEGMA) and a copolymer of DEAEMA and butyl methacrylate (BMA) for intracellular messenger RNA delivery.21 Polyplexes rely on electrostatic interactions for their formation and can have stability issues with other anionic compounds in vivo.22 In contrast, the PDEAEMA dual component nanoparticles reported here can be easily tuned using their blend composition, as demonstrated by loading a model cellular marker. We believe that the modular, one-step assembly and endosomal escape capability of these PDEAEMA nanoparticles make them attractive for use in therapeutic delivery systems.
Experimental
Materials
2-(Diethylamino) ethyl methacrylate (DEAEMA), (Sigma-Aldrich, 99%) was passed over aluminium oxide (activated, basic) (Sigma-Aldrich) to remove inhibitors prior to use. 4-Cyano-4-[(dodecylsulfanyl thiocarbonyl) sulfanyl] pentanoic acid (Sigma-Aldrich, 97%), poly(ethylene glycol) 4-cyano-4-(phenyl carbonothioylthio) pentanoate (Sigma-Aldrich, 98%, average Mn 2000), Trypsin, no phenol red (Life Technologies, 2.5%), Hoechst 33342, trihydrochloride, trihydrate (Life Technologies, 10 mg mL−1 solution in water) and Rhodamine B octadecyl ester perchlorate (Sigma-Aldrich, ≥98.0%), and all solvents were used as received. The water used in all materials experiments was water obtained from the reverse osmosis (RO) tap while all cell experiments used Milli-Q water (prepared using the Millipore Milli-Q Advantage A10 purification system with a resistivity greater than 18 MΩ cm). Phosphate buffered saline (PBS) was prepared using PBS tablets (Sigma-Aldrich). Calcein (Sigma-Aldrich) was dissolved in Milli-Q water at a concentration of 1 mg mL−1 and stored at 4 °C prior to use. Thiazoyl Blue Tetrazolium Bromide (MTT) (Sigma-Aldrich, 98%) was dissolved in PBS at a concentration of 5 mg mL−1 and stored at 4 °C prior to use. Bafilomycin A1 from Streptomyces griseus (Sigma-Aldrich, ≥90%) was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 100 μM and stored at −20 °C prior to use. CellLight® Lysosomes-GFP, BacMam 2.0 and BacMam enhancer kit were purchased from Life Technologies. Gibco® Dulbecco's Modified Eagle Medium (DMEM) (high glucose, GlutaMAX™ Supplement) was purchased from Life Technologies and treated with 10% fetal bovine serum (FBS), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin prior to use.Cell lines
3T3 mouse embryonic fibroblast wild type cells (3T3 MEFs WT) were purchased from ATCC and cultured in DMEM medium supplemented with 10% fetal bovine serum, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin.Synthesis of poly(2-(diethylamino)ethyl methacrylate) (PDEAEMA)
PDEAEMA was prepared by reversible addition–fragmentation chain transfer (RAFT). DEAEMA (1.0 g, 5.4 mmol), AIBN (0.16 mg, 0.97 μmol) and 4-cyano-4-[(dodecylsulfanylthiocarbonyl)sulfanyl]pentanoic acid (4.5 mg, 11.1 μmol) were dissolved in 1,4-dioxane (2.0 g) and placed into a Schlenk flask equipped with a magnetic stirrer bar. The flask was degassed over three freeze–thaw–pump cycles and back-filled with nitrogen. The degassed reaction mixture was stirred in an oil bath at 60 °C for 16 hours. The reaction was terminated by exposing the reaction mixture to air. The mixture was purified by precipitation into deionized water. The precipitate was dried in vacuo to yield PDEAMA as a tacky solid. 1H-NMR (400 MHz, CDCl3): δ (ppm) 4.1–3.9 (–COO–CH2–CH2–), 2.7–2.6 (–CH2–CH2–N–), 2.6–2.5 (–N–CH2–CH3), 2.0–1.7 (backbone –S–CH2–C–), 1.25–1.2 (CH3–C8H16–CH2–), 1.15–0.95 (–N–CH2–CH3), 0.95–0.8 (backbone –CH2–C–CH3–). The molecular weight of PDEAEMA was determined by NMR analysis to be approximately 38 kDa (see ESI† for details).Synthesis of poly(2-(diethylamino)ethyl methacrylate)-b-poly(ethylene glycol) (PDEAEMA-b-PEG)
PDEAEMA-b-PEG was prepared by reversible addition–fragmentation chain transfer (RAFT). DEAEMA (0.5 g, 2.7 mmol), AIBN (0.5 mg, 3.0 μmol) and poly(ethylene glycol)4-cyano-4-(phenyl carbonothioylthio) pentanoate (53 mg, 26.5 μmol) were dissolved in 1,4-dioxane (1.0 g) and placed into a Schlenk flask equipped with a magnetic stirrer bar. The flask was degassed over three freeze–thaw–pump cycles. The degassed reaction mixture was stirred in an oil bath at 60 °C for 15 hours. The reaction was terminated by exposing the reaction mixture to air. The mixture was purified by dialysis against sodium acetate buffer at pH 5, then lyophilised. 1H-NMR (400 MHz, D2O): δ (ppm) 7.9–7.3 (C5H5–C–), 4.4–4.0 (–COO–CH2–CH2–), 3.75–3.45 (–COO–CH2–CH2–O–), 3.3–3.15 (–CH2–CH2–N–), 3.2 (–CH2–O–CH3– (under polymer peak)), 3.15–2.8 (–N–CH2–CH3), 2.5–2.4 (–C–CH2–CH2–COO–), 2.0–1.6 (backbone –S–CH2–C–), 1.9 (–CH2–C–CH3), 1.25–1.1 (–N–CH2–CH3), 1.1–0.7 (backbone –CH2–C–CH3). The molecular weight of PDEAEMA-b-PEG was determined by NMR analysis to be approximately 16 kDa (see ESI† for details).Synthesis of PDEAEMA dual component nanoparticles
PDEAEMA (∼38 kDa) and PDEAEMA-b-PEG (∼16 kDa) were co-dissolved into 300 μL of ethanol to a total mass of 3 mg. The PDEAEMA-b-PEG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PDEAEMA ratio was 1
PDEAEMA ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, thus 0.5 mg PDEAEMA-b-PEG was used relative to 2.5 mg PDEAEMA. The polymer mixtures were added dropwise (7.5 μL per drop) into 3 mL of PBS adjusted to pH 8 stirring at 750 rpm. The solutions were left stirring at 300–400 rpm for 1 hour. The particles were purified by dialysis against PBS adjusted to pH 8. The dialysis procedure involved six changes of dialysis solution over 24 hours. The nanoparticle solution was removed from dialysis and left to sit for 48 hours, then filtered with a 0.45 μm filter immediately prior to any characterization procedures. To synthesize Rhodamine B loaded nanoparticles, Rhodamine B octadecyl ester perchlorate (4.86 μg, 6.1 nmol) was co-dissolved with PDEAEMA and PDEAEMA-b-PEG in ethanol to a total polymer mass of 3 mg and a polymer concentration of 10 mg mL−1, nanoprecipitated, and purified as described above.
5, thus 0.5 mg PDEAEMA-b-PEG was used relative to 2.5 mg PDEAEMA. The polymer mixtures were added dropwise (7.5 μL per drop) into 3 mL of PBS adjusted to pH 8 stirring at 750 rpm. The solutions were left stirring at 300–400 rpm for 1 hour. The particles were purified by dialysis against PBS adjusted to pH 8. The dialysis procedure involved six changes of dialysis solution over 24 hours. The nanoparticle solution was removed from dialysis and left to sit for 48 hours, then filtered with a 0.45 μm filter immediately prior to any characterization procedures. To synthesize Rhodamine B loaded nanoparticles, Rhodamine B octadecyl ester perchlorate (4.86 μg, 6.1 nmol) was co-dissolved with PDEAEMA and PDEAEMA-b-PEG in ethanol to a total polymer mass of 3 mg and a polymer concentration of 10 mg mL−1, nanoprecipitated, and purified as described above.
pH-dependent disassembly of PDEAEMA dual component nanoparticles analysis by DLS
PDEAEMA dual component nanoparticles were prepared as described. 300 μL of PDEAEMA particles were dispersed into 2 mL of filtered PBS buffer adjusted to various pHs (ranging from pH 6.0 to pH 8.0 in 0.2 pH increments) for analysis by dynamic light scattering (DLS).Assessing particle association of PDEAEMA dual component nanoparticles with 3T3 fibroblast cells
3T3 mouse embryonic fibroblast wild type cells (3T3 MEFs WT) were seeded on a 24-well plate at a density of 6 × 104 cells per well and incubated overnight in standard cell culture conditions (37 °C and 5% CO2). Rhodamine B loaded nanoparticles were prepared as described and added to the cells at concentrations of 0.27, 0.56, 1.11, 2.23, 4.44 and 8.90 μg mL−1 in triplicates and incubated for 2 hours. After 2 hours, the cells were washed to remove free-floating nanoparticles. The cells were lifted by incubation with 100 μL of 0.25% trypsin at 37 °C for 3 minutes, followed by the addition of 100 μL of PBS. The cells were centrifuged, washed and re-suspended in PBS for analysis by flow cytometry.Assessing the cytotoxicity of PDEAEMA dual component nanoparticles
3T3 MEFs WT cells were seeded on a 96-well plate at a density of 5 × 103 cells per well and incubated overnight in cell culture conditions (37 °C and 5% CO2). The media was replaced with media containing filtered PDEAEMA dual component particles at concentrations of 17.3, 5.8, 1.7, 0.6 and 0.2 μg mL−1 and incubated for 2 hours. The cells were washed to remove free-floating nanoparticles and incubated overnight for 22 further hours. Thiazolyl Blue Tetrazolium Bromide (MTT) was added to the cells at a concentration of 0.5 mg mL−1 and incubated for 3 hours at cell culture conditions. The supernatant was carefully removed, and the MTT product dissolved in 100 μL of dimethyl sulfoxide (DMSO). The cell viability was determined from the absorption at 570 nm relative to non-treated cells (control). All measurements were performed on a FLUOstar OPTIMA microplate reader (BMG Labtech, Germany).Assessing the localization of PDEAEMA dual component nanoparticles in vitro
3T3 MEFs WT cells were seeded on an 8-well plate at a density of 2 × 104 cells per well and incubated for 4 hours in cell culture conditions (37 °C and 5% CO2). CellLight® Lysosomes-GFP were added at a concentration of 50 particles per cell and incubated for an hour. The media was replaced with media containing BacMam enhancer (added according to manufacturer's instructions) and incubated for 1 hour. The enhancer media was then removed and replaced with fresh media then allowed to incubate overnight. Rhodamine B loaded nanoparticles were prepared as described and added to the cells at a concentration of 4.4 μg mL−1 (8.9 × 10−11 g per cell) (an addition of 7.7 μL of particles at 0.23 mg mL−1 to a total media volume of 400 μL) and allowed to incubate for 4 hours. After incubation, the cells were washed 4 times with phenol red-free media to remove free-floating nanoparticles. Hoechst stain (1.3 μg mL−1) was added to the cells and incubated for 10 minutes. The cells were imaged by live cell fluorescence microscopy.Assessing the endosomal escape capability of PDEAEMA dual component nanoparticles using calcein
3T3 MEFs WT cells were seeded on a 96-well plate at a density of 7.5 × 103 cells per well and incubated overnight in cell culture conditions (37 °C and 5% CO2). Rhodamine B loaded PDEAEMA dual component particles were added at a concentration of 2.2 μg mL−1 (4.4 × 10−11 g per cell) (an addition of 1.4 μL of particles at 0.23 mg mL−1 to a total media volume of 150 μL) and allowed to incubate for 4 hours. Calcein was added to the cells at a concentration of 100 μg mL−1 two hours after the addition of the nanoparticles and allowed to incubate for two hours. A control experiment was carried out where only calcein was added to the cells and incubated for two hours. The cells were washed 4 times with phenol red-free media to remove free-floating nanoparticles, and imaged by live cell fluorescence microscopy. For the Bafilomycin A1 experiments, the 3T3 fibroblast cells were seeded as described. 30 minutes prior to addition of the nanoparticles, the cells were treated with 1 μM of Bafilomycin A1. Calcein was added to the cells, washed and imaged as described. The washing and imaging media were also prepared with 1 μM of Bafilomycin A1 to maintain a constant Bafilomycin A1 concentration throughout the experiment.Nuclear magnetic resonance (NMR) spectroscopy
1H NMR spectroscopy was conducted on a Varian 400 MR spectrometer operating at 400 MHz using deuterated chloroform (CDCl3) and deuterium oxide (D2O) as the solvents. Samples were prepared at a concentration of approximately 15 mg mL−1. NMR analyses were performed on MestReNova, with baselines adjusted according to the Bernstein polynomial fit with a polynomial order of 3.Dynamic light scattering (DLS)
Nanoparticle size, stability, and pH-dependent disassociation measurements were performed on a Horiba nanopartica SZ-100 (Horiba Scientific, Japan) operating at a fixed scattering angle of 90 °C.Cryo-electron microscopy (Cryo-EM)
Images were obtained on a Tecnai F30 (FEI, Hillsborough, USA) at the Melbourne Advanced Microscopy Facility (Bio21 Molecular science and Biotechnology Institute). Samples were snap frozen in liquid ethane and imaged at 200 kV under Low Dose mode (∼1000 electron per nm2, Camera Gatan US1000).Flow cytometry
PDEAEMA nanoparticle association with 3T3 fibroblast cells was studied using a BD FACS Canto II flow cytometer (BD Biosciences, USA) using an excitation wavelength of 488 nm and an emission bandpass of 585/42 nm. Data analysis was performed using the FlowJo analysis software to obtain fluorescence intensities of the samples.Live cell fluorescence microscopy
Live cell imaging was performed using an Olympus IX83 microscope with a 40× 0.9 NA air objective (Fig. 5 and S7†) or a 60× 1.3 NA silicone objective (Fig. 4, S4† and S6†) with a standard “Pinkel” DAPI/FITC/CY3/CY5 filter set from Semrock. The cells were imaged in a humidified incubation chamber with 10% CO2 and regulated at a temperature of 37 °C. The 60× images were taken as a series of z-sections with 0.33 μm spacing and deconvolved using the Richardson–Lucy algorithm (100 iterations).23,24 The 40× images were captured as a single slice with a brightfield overlay. All images were processed using the Slidebook 6.0 software.Results and discussion
Design, synthesis and characterization of PDEAEMA dual component nanoparticles
PDEAEMA and PDEAEMA-b-PEG were both synthesized by reversible addition–fragmentation chain-transfer polymerization (RAFT). PDEAEMA-b-PEG was synthesized by polymerizing DEA monomer using a PEG-containing macro-chain transfer (macro-CTA) agent (average Mn = 2000). The molecular weights of PDEAEMA and PDEAEMA-b-PEG were determined by 1H NMR analysis to be approximately 38 kDa and 16 kDa respectively (see ESI†). The nanoparticles were formed by a nanoprecipitation technique, which involved co-dissolving the two polymeric components into 300 μL of ethanol at a PDEAEMA-b-PEG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PDEAEMA mass ratio of 1
PDEAEMA mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
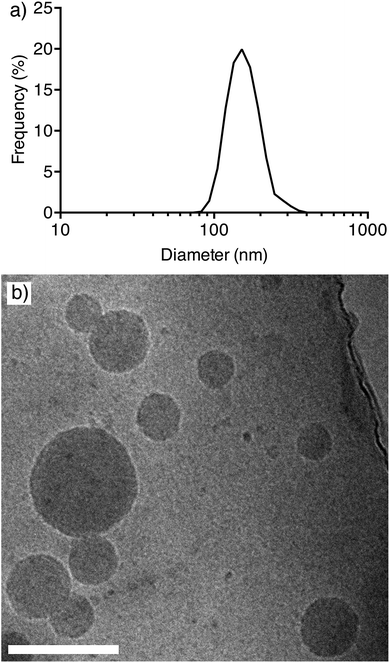
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. The total polymer mass was kept constant at 3 mg. The solution was then added dropwise to phosphate buffered saline (PBS) (pH 8) while stirring. The solution was left to stir for 1 hour, then dialyzed against PBS at pH 8 over 24 hours to remove solvent and uncomplexed polymer.19,25 We hypothesize that when the two components are co-dispersed into a solution with a pH greater than their pKa, hydrophobic interactions drive the assembly of the PDEAEMA components into a hydrophobic core, while the hydrophilic PEG extends into the solution to form a stabilizing hydrophilic corona. The solution was passed through 0.45 μm filters to remove large aggregates prior to any analysis and left to sit for 48 h. The intensity mean diameter of the self-assembled nanoparticles was determined by dynamic light scattering (DLS) to be approximately 150 nm. The intensity mean distribution of the nanoparticles demonstrates reasonable control over particle size as shown in Fig. 1a. To assess the colloidal stability of the nanoparticles, their sizes were assessed by DLS over a period of 10 days. The particles were shown to be stable in size at pH 8 over a period of six days (Fig. S1†). Between the 7th to the 9th day, a significantly lower particle count rate was observed, resulting in unreliable size data. Therefore, subsequent experiments and analysis of these nanoparticles was conducted within 6 days. Cryo-electron microscopy (cryo-EM) of the nanoparticles was performed to confirm the size and polydispersity results obtained by DLS. Cryo-EM images of the nanoparticles show a similar size range to the DLS analysis (Fig. 1b).
5. The total polymer mass was kept constant at 3 mg. The solution was then added dropwise to phosphate buffered saline (PBS) (pH 8) while stirring. The solution was left to stir for 1 hour, then dialyzed against PBS at pH 8 over 24 hours to remove solvent and uncomplexed polymer.19,25 We hypothesize that when the two components are co-dispersed into a solution with a pH greater than their pKa, hydrophobic interactions drive the assembly of the PDEAEMA components into a hydrophobic core, while the hydrophilic PEG extends into the solution to form a stabilizing hydrophilic corona. The solution was passed through 0.45 μm filters to remove large aggregates prior to any analysis and left to sit for 48 h. The intensity mean diameter of the self-assembled nanoparticles was determined by dynamic light scattering (DLS) to be approximately 150 nm. The intensity mean distribution of the nanoparticles demonstrates reasonable control over particle size as shown in Fig. 1a. To assess the colloidal stability of the nanoparticles, their sizes were assessed by DLS over a period of 10 days. The particles were shown to be stable in size at pH 8 over a period of six days (Fig. S1†). Between the 7th to the 9th day, a significantly lower particle count rate was observed, resulting in unreliable size data. Therefore, subsequent experiments and analysis of these nanoparticles was conducted within 6 days. Cryo-electron microscopy (cryo-EM) of the nanoparticles was performed to confirm the size and polydispersity results obtained by DLS. Cryo-EM images of the nanoparticles show a similar size range to the DLS analysis (Fig. 1b).
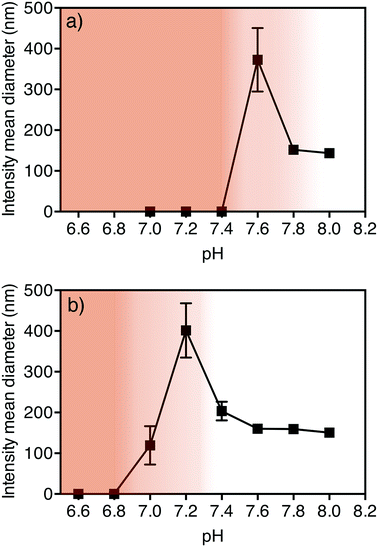
In order for these nanoparticles to be effective drug delivery carriers, it is important that they are able to respond within a biologically relevant pH range. To investigate this, we observed the disassembly of the PDEAEMA dual component nanoparticles in simulated biological conditions. The size and stability of the nanoparticles were assessed over a range of physiological pH values using dynamic light scattering (DLS). To achieve this, PBS buffers ranging from pH 6 to pH 8 were prepared in 0.2 pH increments. 300 μL of filtered particles was added to 2 mL of the pH-adjusted buffers, and analyzed by DLS. The pH-triggered disassembly study was conducted both at room (25 °C) and physiological (37 °C) temperatures, to observe the dependence of nanoparticle stability on temperature. Fig. 2 shows the intensity mean diameters of the particles when dispersed into PBS buffers adjusted to various pHs at 25 °C (Fig. 2a) and 37 °C (Fig. 2b). It can be seen from Fig. 2 that at 25 °C, the particles were stable at pH 8.0 (white unshaded region) but were completely disassembled by pH 7.4 (dark shaded region). We determined that there is a region of instability prior to disassembly, which consistently showed partial aggregation of particles. This region of instability is indicated in Fig. 2 by a shaded gradient from light (start of instability) to dark (point just before complete disassembly). The analysis of DLS results is discussed in detail in the ESI.† Interestingly, the points of instability and disassembly at physiological temperature (37 °C) were at lower pH values compared to those at 25 °C. At 37 °C, the particles were stable from pH 8 to a pH around 7.4, unstable at pH 7.2, and completely disassembled at pH 6.8. Importantly, this data shows that the nanoparticles are stable at physiological conditions (pH 7.4 PBS at 37 °C). These results demonstrate that the disassociation of the PDEAEMA dual component nanoparticles show a mild temperature sensitivity. Hu et al. have previously demonstrated a similar temperature-dependent behaviour on the swelling response of their PDEAEMA-core/poly(aminoethyl methacrylate) (PAEMA)-shell crosslinked nanoparticle system.13 They observed that at 25 °C, the swelling transition of their PDEAEMA core–shell nanoparticles was detected at pH ∼7.4 but at 37 °C was lowered to pH 7.0.
PDEAEMA nanoparticle loading with Rhodamine B octadecyl ester perchlorate
It is important that a delivery system is able to load cargo such as hydrophobic drugs.26 The suitability of these PDEAEMA dual component nanoparticles for loading of hydrophobic cargo was investigated using Rhodamine B octadecyl ester perchlorate as a model dye. In this study the cargo was used as a fluorophore to assess localization of the particles within the cell. To load the nanoparticles, 6.1 nmol of Rhodamine B octadecyl ester perchlorate was co-dissolved with the two polymeric components, PDEAEMA-b-PEG and PDEAEMA in 300 μL of ethanol and added dropwise to PBS at pH 8. We hypothesize that the hydrophobic dye is encapsulated within the core of the dual component nanoparticles due to hydrophobic interactions with the PDEAEMA component upon nanoprecipitation. After dialysis, the efficiency of loading was determined by ultraviolet-visible (UV-vis) spectrophotometry to be ∼30% (Fig. S2†). The successful loading of Rhodamine B octadecyl ester perchlorate within the core of the nanoparticles demonstrates the potential these nanoparticles have for carrying hydrophobic therapeutic cargo in the future.Cellular behaviour and endosomal escape capability of PDEAEMA nanoparticles
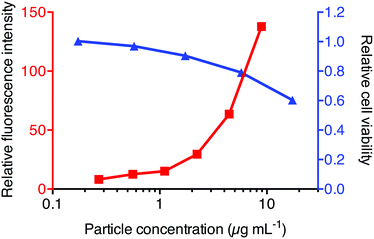
Trafficking of nanoparticles within the cell is a key area of interest for improving polymeric carriers, as efficient delivery to the cytosolic compartment of the cell is known to be key to optimising therapeutic efficacy.14 One critical step in this process is escaping the endosomal/lysosomal compartments where the particles are initially localized. To assess the intracellular behaviour of these PDEAEMA dual component nanoparticles, we investigated their ability to be internalized into 3T3 fibroblast cells as well as their ability to induce escape from the endosomes into the cytosol of the cell. The concentration of nanoparticles after dialysis and filtration was determined using UV-vis spectrophotometry based on their absorbance at 305 nm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 27 (Fig. S3†). The cytotoxic effect of the nanoparticles against 3T3 cells was examined using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell viability assay. The cells showed >80% viability when exposed to nanoparticle concentrations up to 5 μg mL−1 (Fig. 3). We then assessed the association of PDEAEMA dual component nanoparticles with 3T3 fibroblast cells by flow cytometry. Rhodamine B loaded PDEAEMA nanoparticles were incubated with 3T3 fibroblast cells at a range of different concentrations for 2 hours and the fluorescence intensity of the cells were analyzed by flow cytometry. As expected, the relative fluorescence intensity of the cells increased with an increase in particle concentration, indicating greater particle association when particle concentration is increased (Fig. 3). These results show that between particle concentrations of 1 and 5 μg mL−1 there is association of particles with the cells, with limited cytotoxic effects. For all subsequent cellular experiments, we incubated the cells with concentrations of particles less than 5 μg mL−1 to ensure that the effects seen in the cells are not due to the cytotoxicity of the nanoparticles.
27 (Fig. S3†). The cytotoxic effect of the nanoparticles against 3T3 cells was examined using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell viability assay. The cells showed >80% viability when exposed to nanoparticle concentrations up to 5 μg mL−1 (Fig. 3). We then assessed the association of PDEAEMA dual component nanoparticles with 3T3 fibroblast cells by flow cytometry. Rhodamine B loaded PDEAEMA nanoparticles were incubated with 3T3 fibroblast cells at a range of different concentrations for 2 hours and the fluorescence intensity of the cells were analyzed by flow cytometry. As expected, the relative fluorescence intensity of the cells increased with an increase in particle concentration, indicating greater particle association when particle concentration is increased (Fig. 3). These results show that between particle concentrations of 1 and 5 μg mL−1 there is association of particles with the cells, with limited cytotoxic effects. For all subsequent cellular experiments, we incubated the cells with concentrations of particles less than 5 μg mL−1 to ensure that the effects seen in the cells are not due to the cytotoxicity of the nanoparticles.
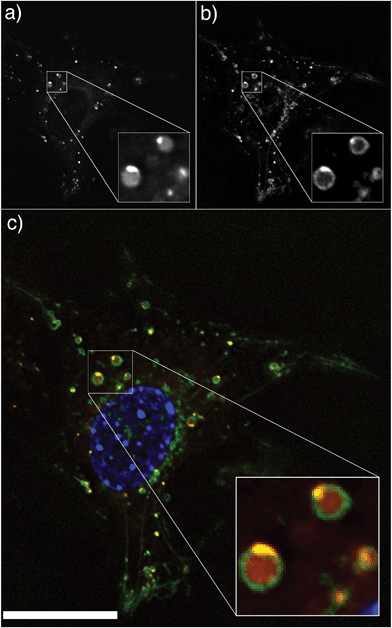
The cellular localization of the nanoparticles was determined by incubating the Rhodamine B loaded PDEAEMA nanoparticles with 3T3 fibroblast cells expressing GFP-tagged lysosomal associated membrane protein 1 (LAMP1). Deconvolved fluorescence microscopy images revealed that the nanoparticles were co-incident with the LAMP1 staining, suggesting endocytosis of these nanoparticles into late endosomal/lysosomal compartments (Fig. 4).
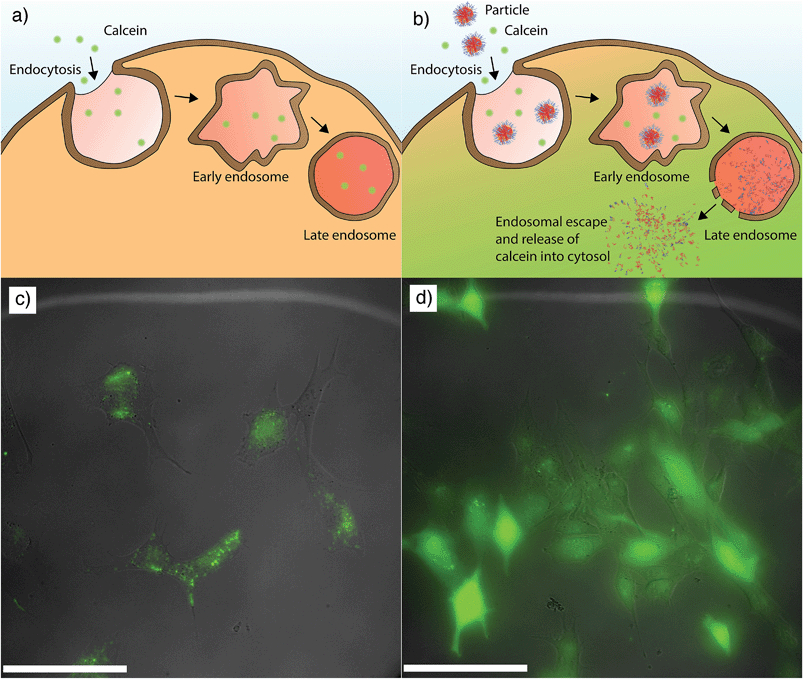
Several research groups have demonstrated that pH-responsive materials are capable of escaping the endosomal/lysosomal compartments, an important characteristic for effective therapeutic delivery. However, the mechanism of escape is still unclear. One well-known mechanism is the “proton-sponge effect” which is a build-up of osmotic pressure associated with chloride accumulation in the presence of a buffering material in these acidic compartments.14 An alternative mechanism proposes that the disassociation of pH-responsive, self-assembled nanoparticles into unimers upon uptake into acidic compartments creates a large increase in osmotic pressure, eventually leading to lysis and release of contents into the cytosol.10,18 Therefore, to investigate if the pH-responsive behaviour of these nanoparticles might facilitate endosomal escape into the cytosol of the cells, we used the calcein green assay. Calcein is a membrane-impermeable fluorophore that shows a punctate fluorescence if entrapped within the endosomal/lysosomal compartments but fluoresces brightly throughout the cytosol if it escapes.13,25,28 Filtered nanoparticles were added to the cells at a concentration of 2.2 μg mL−1 (4.4 × 10−11 g per cell) and incubated for four hours.
Calcein (100 μg mL−1) was added two hours after the addition of the nanoparticles. Four hours after the addition of the nanoparticles (and two hours after addition of calcein), the excess particles and calcein were removed and endosomal escape assessed using fluorescence microscopy. As an illustration of the calcein control experiment, Fig. 5a depicts the entrapment of calcein in the acidic compartments when calcein is delivered without nanoparticles. The corresponding cellular microscopy image (Fig. 5c) shows a punctate distribution of calcein (green) fluorescence indicating entrapment within the endosomal/lysosomal compartments of the cell. In contrast, Fig. 5b illustrates the escape and release of calcein into the cytosol when delivered with nanoparticles. The corresponding cellular image (Fig. 5d) shows the presence of a percentage of cells displaying bright, diffused calcein (green) fluorescence throughout the cell. The cell images demonstrated that 30% of the cells incubated with the PDEAEAMA dual component nanoparticles exhibited endosomal escape, while the cells that were not treated with nanoparticles exhibited escape in less than 1% of the cells (Table 1). Importantly, we demonstrate that these nanoparticles showed endosomal escape at a concentration with limited cytotoxicity. Higher resolution images of the control and escape cells are shown in Fig. S4.† Flow cytometry histograms show a uniform shift in fluorescence intensity of cells incubated with nanoparticles, indicating that more than 30% of cells have associated particles (Fig. S5†). This indicates that there are a significant number of cells with associated nanoparticles that do not demonstrate calcein release into the cytosol. The mechanism by which the particles induce escape of the calcein is not clear, and is the subject of on-going research in our group. It has previously been observed that cell cycle plays a role in transfection efficiency,29 and it is possible that cell cycle could also play a role in endosomal escape. A representative image demonstrating the presence of cells with internalized nanoparticles that have either successfully or unsuccessfully aided the endosomal escape of calcein is shown in Fig. S6.†
| Total number of cellsa | Number of escaped cells (with diffused calcein fluorescence) | |
|---|---|---|
| a The percentage of cells exhibiting diffused fluorescence was assessed by the counting of more than 500 cells from images captured at various locations in the well. | ||
| Cells only (control) | 574 | 0 |
| Cells incubated with nanoparticles | 584 | 182 |
| Cells treated with Baf A1 and incubated with nanoparticles | 896 | 82 |
The results of the calcein assay show that PDEAEMA dual component pH-responsive nanoparticles are able to induce escape form the endosomal/lysosomal compartments of the cell, allowing the transport of calcein into the cytosol. However, the high degree of co-localization of the Rhodamine B octadecyl ester perchlorate loaded nanoparticles with LAMP1 (Fig. 4) suggests that while the PDEAEMA dual component nanoparticles have aided the escape of calcein into the cytosol, the nanoparticles itself have remained entrapped within the endosomes/lysosomes. This suggests that complete endosomal rupture is not occurring; instead, we postulate that membrane destabilization occurs,14 allowing calcein to escape while the polymers remain predominantly trapped. To investigate if the nanoparticle behaviour in response to the acidification of endosomes is important for their endosomal escape-inducing capabilities, we performed the same calcein assay in the presence of Bafilomycin A1 (Baf A1). Bafilomycin A1 is a proton pump inhibitor that inhibits endosomal acidification.30–32 For these experiments, 3T3 fibroblast cells were treated with Bafilomycin A1 at a concentration of 1 μM for 30 minutes, followed by the addition of PDEAEMA nanoparticles and calcein as previously described. Results of these experiments show that the acidification of the endosomes plays a role in inducing the escape of calcein into the cytosol. The cells treated with Bafilomycin A1 show a decrease in cells displaying diffused calcein fluorescence throughout the cytoplasm (∼9%) compared to those not treated with Bafilomycin A1 (∼30%) (Table 1). Representative cellular images from these experiments are shown in Fig. S7.†
The results of the cellular experiments collectively show that the protonation of tertiary amine groups of PDEAEMA in acidified compartments plays a crucial role in aiding the leakage of calcein into the cytoplasm of the cell. However, the co-localization of the Rhodamine B loaded nanoparticles with LAMP1 markers suggests that while the PDEAEMA nanoparticles have provided an avenue of calcein release into the cytosol, the particles have largely remained entrapped within the late endosomes/lysosomes. This work demonstrates the importance of understanding the mechanisms and intricacies involved in endosomal escape for designing effective therapeutic nanocarriers. Simply tracing the localization of nanoparticles in a cell, or using a fluid phase marker such as calcein as an indirect way to measure endosomal escape does not in isolation tell the whole story. The calcein escape demonstrates that the nanoparticles are efficiently disrupting the endosomal/lysosomal compartments, however the localization studies show that while the membrane disruption is sufficient to allow the majority of calcein to escape into the cytoplasm, the polymer remains predominantly trapped. We are currently developing new methods to quantify the efficiency of endosomal escape and hope that this will help better understand the key mechanisms involved.
Conclusions
In summary, we have demonstrated the self-assembly of nanoparticles based on the pH-responsive polymer, PDEAEMA by nanoprecipitation into PBS at pH 8. The nanoparticles were observed to disassemble completely below pH 6.8 at physiological temperature (37 °C). The nanoparticles have a two stage degradation mechanism; firstly, partial instability characterized by some aggregation or swelling of the particles, and then subsequent disassembly. We demonstrated the successful encapsulation of Rhodamine B octadecyl ester perchlorate within the core of the nanoparticles, showing the potential of these nanoparticles for loading therapeutic cargo. Cellular studies showed that the nanoparticles were successfully internalized into the endosomal/lysosomal compartments of the cell. Importantly, the nanoparticles were observed to successfully induce the delivery of calcein into the cytosol of the cell in 30% of cells. Work is currently underway to further investigate the endosomal escape mechanism of these nanoparticles. We believe the nanoparticle system reported herein is a promising drug carrier system as it has a modular one-step assembly, is responsive to changes in physiologically relevant pH, and importantly, has shown the ability to induce endosomal escape into the cytosol.Acknowledgements
This work was supported by the Australian Research Council through the Future Fellowship Scheme (FT120100564 – GKS and FT110100265 – APRJ) and Centre of Excellence in Convergent Bio-Nano Science and Technology (APRJ). APRJ is also supported through the Monash University Larkin's Fellowship Scheme. TCS was supported through the Whitaker International Summer Program.Notes and references
- M. E. Davis, Z. Chen and D. M. Shin, Nat. Rev. Drug Discovery, 2008, 7, 771–782 CrossRef CAS PubMed.
- J. Du and R. K. O'Reilly, Soft Matter, 2009, 5, 3544–3561 RSC.
- M. A. C. Stuart, W. T. S. Huck, J. Genzer, M. Müller, C. Ober, M. Stamm, G. B. Sukhorukov, I. Szleifer, V. V. Tsukruk, M. Urban, F. Winnik, S. Zauscher, I. Luzinov and S. Minko, Nat. Mater., 2010, 9, 101–113 CrossRef PubMed.
- S. De Koker, R. Hoogenboom and B. G. De Geest, Chem. Soc. Rev., 2012, 41, 2867–2884 RSC.
- R. J. Passarella, D. E. Spratt, A. E. van der Ende, J. G. Phillips, H. Wu, V. Sathiyakumar, L. Zhou, D. E. Hallahan, E. Harth and R. Diaz, Cancer Res., 2010, 70, 4550–4559 CrossRef CAS PubMed.
- Y. Li, G. H. Gao and D. S. Lee, Adv. Healthcare Mater., 2013, 2, 388–417 CrossRef CAS PubMed.
- L. M. Bareford and P. W. Swaan, Adv. Drug Delivery Rev., 2007, 748 CrossRef CAS PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- T.-G. Iversen, T. Skotland and K. Sandvig, Nano Today, 2011, 6, 176–185 CrossRef CAS PubMed.
- H. Lomas, M. Massignani, K. A. Abdullah, I. Canton, C. Lo Presti, S. MacNeil, J. Du, A. Blanazs, J. Madsen, S. P. Armes, A. L. Lewis and G. Battaglia, Faraday Discuss., 2008, 139, 143 RSC.
- H. Lomas, I. Canton, S. MacNeil, J. Du, S. P. Armes, A. J. Ryan, A. L. Lewis and G. Battaglia, Adv. Mater., 2007, 19, 4238–4243 CrossRef CAS.
- K. Zhou, Y. Wang, X. Huang, K. Luby-Phelps, B. D. Sumer and J. Gao, Angew. Chem., Int. Ed., 2011, 50, 6109–6114 CrossRef CAS PubMed.
- Y. Hu, T. Litwin, A. R. Nagaraja, B. Kwong, J. Katz, N. Watson and D. J. Irvine, Nano Lett., 2007, 7, 3056–3064 CrossRef CAS PubMed.
- T. F. Martens, K. Remaut, J. Demeester, S. C. De Smedt and K. Braeckmans, Nano Today, 2014, 9, 344–364 CrossRef CAS PubMed.
- A. K. Varkouhi, M. Scholte, G. Storm and H. J. Haisma, J. Controlled Release, 2011, 151, 220–228 CrossRef CAS PubMed.
- D. Ma, Nanoscale, 2014, 6, 6415–6425 RSC.
- Y. Hu, P. U. Atukorale, J. J. Lu, J. J. Moon, S. H. Um, E. C. Cho, Y. Wang, J. Chen and D. J. Irvine, Biomacromolecules, 2009, 10, 756–765 CrossRef CAS PubMed.
- M. Massignani, C. LoPresti, A. Blanazs, J. Madsen, S. P. Armes, A. L. Lewis and G. Battaglia, Small, 2009, 5, 2424–2432 CrossRef CAS PubMed.
- K. Liang, J. J. Richardson, H. Ejima, G. K. Such, J. Cui and F. Caruso, Adv. Mater., 2014, 26, 2398–2402 CrossRef CAS PubMed.
- M. Oishi and Y. Nagasaki, React. Funct. Polym., 2007, 67, 1311–1329 CrossRef CAS PubMed.
- C. Cheng, A. J. Convertine, P. S. Stayton and J. D. Bryers, Biomaterials, 2012, 33, 6868–6876 CrossRef CAS PubMed.
- R. S. Burke and S. H. Pun, Bioconjugate Chem., 2008, 19, 693–704 CrossRef CAS PubMed.
- L. B. Lucy, Astron. J., 1974, 79, 745 CrossRef PubMed.
- W. H. Richardson, J. Opt. Soc. Am., 1972, 62, 55 CrossRef.
- X. Su, J. Fricke, D. G. Kavanagh and D. J. Irvine, Mol. Pharm., 2011, 8, 774–787 CrossRef CAS PubMed.
- A. P. R. Johnston, G. K. Such, S. L. Ng and F. Caruso, Curr. Opin. Colloid Interface Sci., 2011, 16, 171–181 CrossRef CAS PubMed.
- H. Willcock and R. K. O'Reilly, Polym. Chem., 2010, 1, 149 RSC.
- X. Su, N. Yang, K. D. Wittrup and D. J. Irvine, Biomacromolecules, 2013, 14, 1093–1102 CrossRef CAS PubMed.
- S. Brunner, T. Sauer, S. Carotta, M. Cotten, M. Saltik and E. Wagner, Gene Ther., 2000, 7, 401–407 CrossRef CAS PubMed.
- L. S. Johnson, K. W. Dunn, B. Pytowski and T. E. McGraw, Mol. Biol. Cell, 1993, 4, 1251–1266 CrossRef CAS.
- Z. ur Rehman, D. Hoekstra and I. S. Zuhorn, ACS Nano, 2013, 7, 3767–3777 CrossRef CAS PubMed.
- A. Erazo-Oliveras, K. Najjar, L. Dayani, T.-Y. Wang, G. A. Johnson and J.-P. Pellois, Nat. Methods, 2014, 11, 861–867 CrossRef CAS PubMed.
Footnote |
† Electronic supplementary information (ESI) available: Nanoparticle stability DLS analysis, UV-vis absorbance standard curves of Rhodamine B octadecyl ester perchlorate and the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 nanoparticle polymeric formulation measured at 560 nm and 305 nm respectively, higher resolution calcein assay cellular images, flow cytometry histograms of the Rhodamine B loaded nanoparticles in 3T3 cells, representative cellular image showing the presence of endosomal and non-endosomal escaped cells, Bafilomycin A1 experiment cellular images, polymer molecular weight calculations, nanoparticle disassembly analysis by DLS, loading efficiency of octadecyl Rhodamine B and determination of nanoparticle concentration, and calcein assay quantification analyses. See DOI: 10.1039/c5sm00082c 5 nanoparticle polymeric formulation measured at 560 nm and 305 nm respectively, higher resolution calcein assay cellular images, flow cytometry histograms of the Rhodamine B loaded nanoparticles in 3T3 cells, representative cellular image showing the presence of endosomal and non-endosomal escaped cells, Bafilomycin A1 experiment cellular images, polymer molecular weight calculations, nanoparticle disassembly analysis by DLS, loading efficiency of octadecyl Rhodamine B and determination of nanoparticle concentration, and calcein assay quantification analyses. See DOI: 10.1039/c5sm00082c |
| This journal is © The Royal Society of Chemistry 2015 |