Open Access Article
Open Access ArticleThe fungal cerato-platanin protein EPL1 forms highly ordered layers at hydrophobic/hydrophilic interfaces
K.
Bonazza
a,
R.
Gaderer
b,
S.
Neudl†
c,
A.
Przylucka
d,
G.
Allmaier
a,
I. S.
Druzhinina
bd,
H.
Grothe
c,
G.
Friedbacher
a and
V.
Seidl-Seiboth
*b
aInstitute of Chemical Technologies and Analytics, Vienna University of Technology, Getreidemarkt 9/164-IAC, 1060 Wien, Austria
bResearch Area Biotechnology and Microbiology, Institute of Chemical Engineering, Vienna University of Technology, Gumpendorfer Strasse 1a, 1060 Wien, Austria. E-mail: verena.seidl@tuwien.ac.at
cInstitute of Materials Chemistry, Vienna University of Technology, Getreidemarkt 9/165, 1060 Wien, Austria
dACIB – Austrian Centre of Industrial Biotechnology, Petersgasse 14, 8010 Graz, Austria
First published on 7th January 2015
Abstract
Cerato-platanin proteins (CPPs) and hydrophobins are two classes of small, secreted proteins that are exclusively found in fungi. CPPs are known as chitin-binding proteins, and were recently also shown to form protein layers at air/water interfaces, but the features of these layers were not investigated on the molecular level yet. In this study, by means of atomic force microscopy (AFM), EPL1, a member of the CPP family was shown to form highly ordered monolayers at a hydrophobic surface/liquid-interface. Furthermore, two new hydrophobins were analysed, and the influence of EPL1 on hydrophobin layers was studied in situ. Hydrophobins are amphiphilic proteins that are able to self-assemble at hydrophobic/hydrophilic interfaces, thereby inverting the polarity of the surface. This renders fungal growth structures such as spores water repellent. The combination of AFM data and wettability experiments led to the conclusion that in presence of both, hydrophobins and EPL1, a previously unknown hybrid layer is formed. This mixed protein layer is on one hand not inverting but enhancing the hydrophobicity of HOPG (highly oriented pyrolytic graphite), typical for EPL1, and on the other hand, it is stable and water insoluble, which is reminiscent of hydrophobin layers.
Introduction
Self-assembly of molecules into ordered structures is one of the most remarkable preconditions for life. One of the most prominent three-dimensional examples for oriented molecular assemblies are lipid monolayers in cell walls. Scientists of multiple disciplines have investigated self-assembly phenomena and even the fundamental studies of aliphatic self-assembled monolayers (SAM), such as octadecylsilane (ODS) or octadecyltrichlorosilane (OTS),1–3 which are meanwhile the most understood systems, can be seen in this general context. While SAMs are often covalently bound to the surface, as shown by reflection infrared spectroscopy,3,4 Langmuir Blodget (LB) films, which grow at air–liquid interfaces prior to be deposited onto a substrate, usually lack anchor groups. However, it has been shown by atomic force microscopy (AFM) that also ODS layers first assemble in liquid and then attach to the surface as pre-formed flakes.4Another type of amphiphilic surface active molecules that has already been studied in considerable detail is the hydrophobin protein family. Hydrophobins are small secreted fungal proteins, which were also found to self-assemble at hydrophobic/hydrophilic interfaces.5,6 Due to hydrophobic and hydrophilic amino-acid patches, creating an amphiphilic protein surface, they are able to invert the polarity of surfaces on which they self-assemble very effectively. Therefore, they are handled as candidates for large scale applications, spanning from non-wetting coatings to biocompatible surfactants. Biologically their surface activity-altering properties are for example relevant for fungal hyphae that emerge from an aqueous growth environment to form aerial hyphae and produce spores, which are then covered with a non-wettable layer of hydrophobins. This facilitates the dispersal of fungal conidia (spores). The common feature of all these layers is that they consist of amphiphilic molecules which are uniformly oriented at hydrophilic/hydrophobic interfaces; hence their growth is self-terminating, generating monolayers.
Here, a member of the cerato-platanin protein family (CPP), namely EPL1 from the fungus Trichoderma atroviride, was studied in view of its ability to form ordered self-assembled layers. CPPs are not related to hydrophobins with respect to their protein sequences or structure, but we recently found that EPL1 is also able to form protein biofilms at air/water interfaces.7 However, EPL1 rather enhances the polarity effects of surfaces and solutions instead of inverting them, which is the opposite of what has been described for hydrophobins.
Similar to hydrophobins, CPPs can only be found in filamentous fungi (moulds), i.e. fungi that produce hyphae as growth structures, or in fungi which have at least a pseudo-hyphal growth stage.8 They are small, secreted proteins that are released into the culture filtrate, but they have also been found within the cell wall of fungal hyphae and spores.9–11 CPPs are important factors in fungal–plant interactions. In plant pathogenic fungi they have been reported to act as phytotoxins and were also shown to induce plant defence responses in plant-beneficial fungi of the genus Trichoderma.12–15 However, the presence and abundant expression of CPPs in fungi with all types of life-styles suggests that the main biological functions are not solely related to fungal–plant interactions but to other, more general aspects of fungal growth.7,16,17
Structural analyses revealed that CPPs have a protein fold that is similar to expansins.18 Expansins are non-enzymatic proteins that aid in plant cell wall extension and plant growth by loosening up the cellulose scaffold of the cell wall.19
In analogy to that, CPPs can bind carbohydrates but are also not enzymatically active. They were found to have an N-acetylglucosamine binding pocket. Binding to chitin, which is a biopolymer consisting of N-acetylglucosamine subunits, was already shown for some CPPs including EPL1.7,20,21 Since chitin is a structural component of the fungal cell wall, it was suggested that CPPs might exhibit similar functions in the fungal cell wall as expansins do in plants.7,20
In addition to these carbohydrate-binding properties, we were recently able to show that EPL1 readily self-assembles at air/water interfaces and is able to form protein layers on the surface of aqueous liquids.7
In this study we investigated protein layers of EPL1 and two new hydrophobins HFB9a and HFB9b with tapping mode atomic force microscopy (TM-AFM). The results from this study significantly increase our understanding of EPL1 protein layers and reveal that upon self-assembly they indeed form a regularly patterned protein biofilm surface. Furthermore, since both, CPPs and hydrophobins, can be found on/in fungal cell walls, we tested whether the formation and patterning of protein layers of hydrophobins might be influenced or perturbed by EPL1 and vice versa.
Results
Features of EPL1 protein layers on HOPG
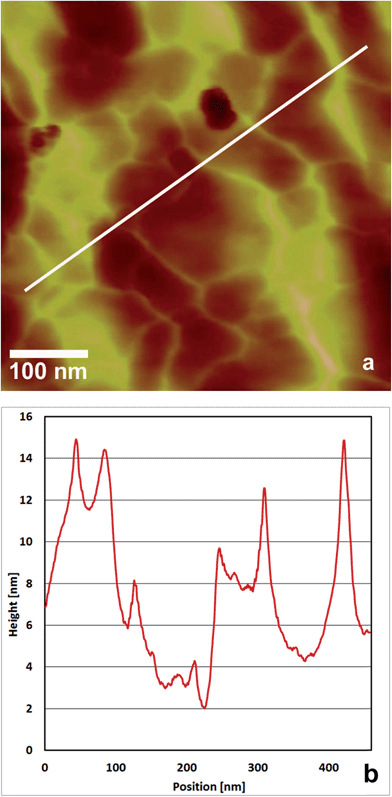
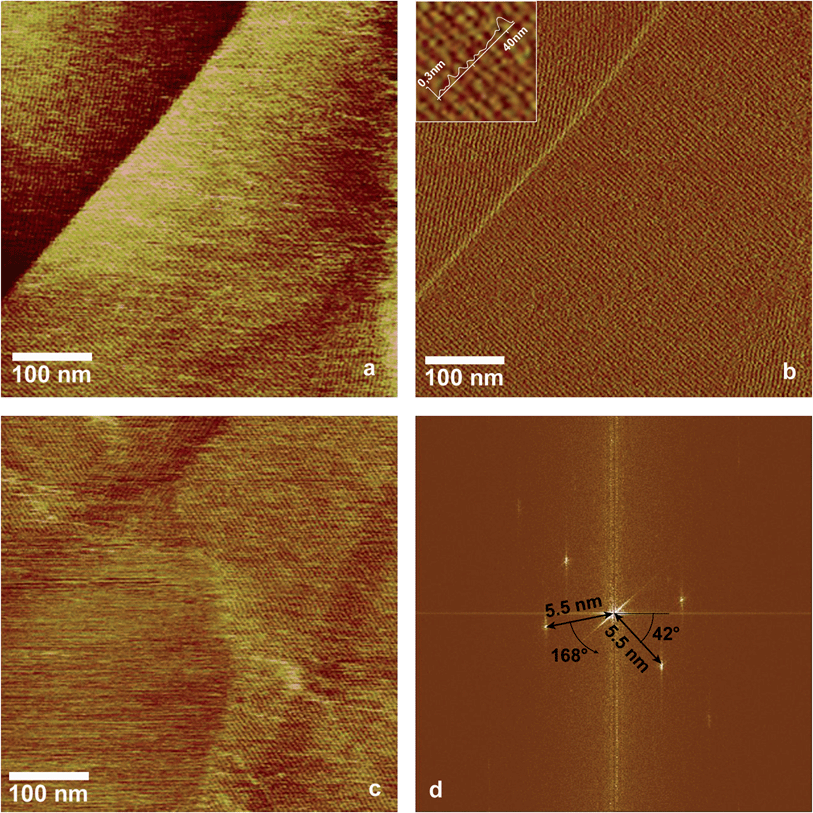
In a previous study we showed that EPL1 forms irregular, meshwork-like or large granular structures upon drying of a droplet of protein solution (0.06 μg mL−1) on a mica surface.7 Following up on these results we investigated the topography of EPL1 protein layers on HOPG (highly orientated pyrolytic graphite) by preparation and imaging in air as well as in situ preparation an imaging directly in liquid (PBS buffer). It turned out that such irregular, meshwork-like protein patches can also be observed on HOPG when imaging samples prepared by drying a droplet in air (Fig. 1a).In order to circumvent limitations in imaging quality on such uneven surfaces (see also Fig. 1b), the following experiment was performed: HOPG was first imaged with AFM under PBS buffer, then a small amount of concentrated EPL1 solution (120 nmol mL−1) was added in situ to achieve a final concentration of approximately 30 nmol mL−1 in the AFM liquid cell. As shown in Fig. 2, the protein immediately formed highly ordered, thin layers (most probably monolayers as indicated by the image of an edge of such an ordered EPL1 domain in Fig. 2c) on the hydrophobic HOPG surface. Fig. 2a shows a topographical image and Fig. 2b an amplitude error image of the same area. It can clearly be seen that the recognisability of the ordered domains is strongly increased in the amplitude error image. This is particularly valid when e.g. steps on the substrate surface are encountered in an image. As shown in the height profile (inset in Fig. 2b) the grooves between protein rows have a depth of only 0.2–0.3 nm.
For that reason only amplitude error images are shown in the following figures. In contrast to the experimental approach shown in Fig. 1, where during drying of the droplet rather large amounts of protein are deposited on the surface, the images shown in Fig. 2 were recorded directly in liquid and here solely a thin layer of EPL1 is deposited on an atomically flat surface. This facilitates high-resolution imaging of the formed protein layer. The Fourier transformation in Fig. 2d shows a periodicity of ∼5.5 nm and the height profile in Fig. 2b displays a groove depth of 0.2–0.3 nm. Previous works reported that hydrophobin layers adhere much stronger to HOPG surfaces than to mica,22 indicating that the hydrophobic interactions are stronger than their hydrophilic counterparts. This is in accordance with our experiments insofar as no formation of EPL1 protein layers could be observed on mica under liquid. Ex situ AFM measurements performed on EPL1 layers on hydrophilic mica and on a hydrophobic gold (111) surface in air yielded no ordered structures.
Influence of EPL1 on hydrophobin protein layers
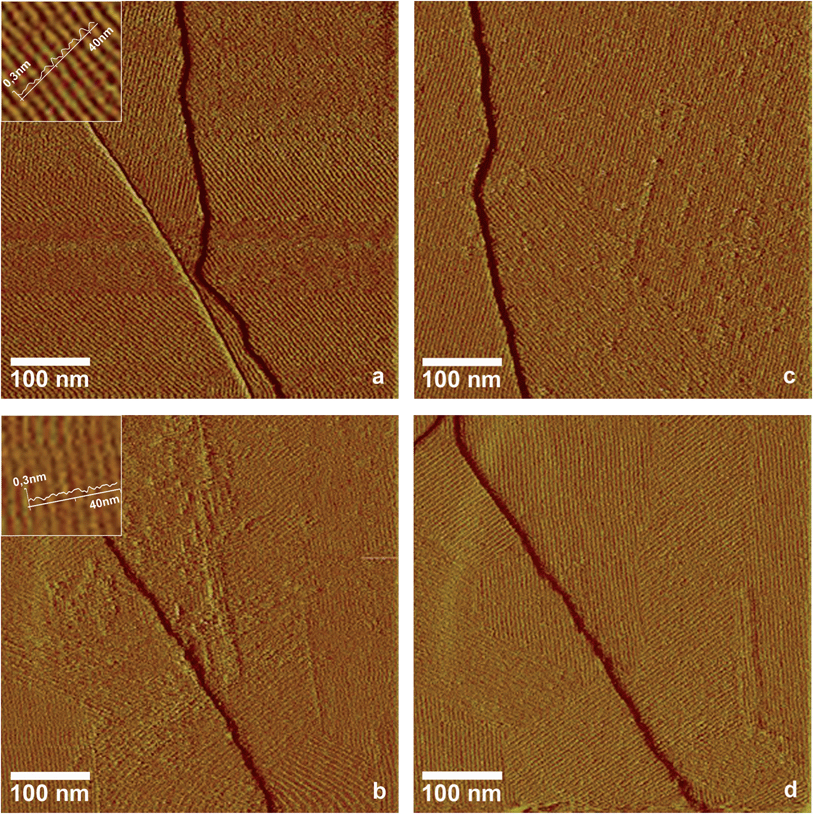
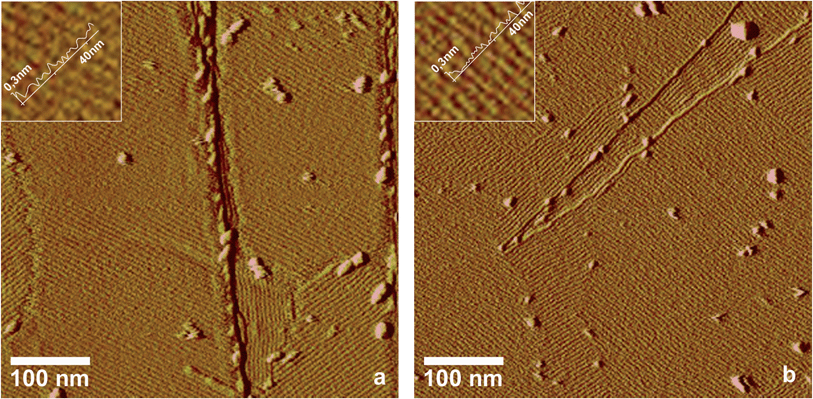
Some members of the classes I and II of the hydrophobin family have been shown to form ordered monolayers,5,6,22,23 but most hydrophobins that were identified in fungal genomes in the last years have not been characterized by high resolution imaging methods. In this study, two hydrophobins, HFB9a and HFB9b from Trichoderma virens, belonging to a phylogenetically new section of class I hydrophobins in ascomycete fungi,24 were analysed in this respect. Self-assembled layers of these two hydrophobins were produced using the droplet method, which is explained in the experimental section.AFM images showed that both hydrophobins are able to form highly ordered lattices on hydrophobic surfaces (Fig. 3a and b). The surface pattern exhibits a riffled morphology, very similar to the EPL1 layers described above, with a separation of 5.3 nm and 5.5 nm (extracted from the Fourier transformed images) between the protein rows, which reflects the similar molecular mass of the mature proteins (EPL1 11 kDa, HFB9a 13 kDa, HFB9b 11 kDa). Since both, EPL1 and HFBs, are present in/on fungal cell walls, we were interested whether the formation and patterning of protein layers of hydrophobins might be influenced or perturbed by EPL1. In order to elucidate this hypothesis, the effect of EPL1 on HFB9a and HFB9b was observed in situ. First, the effect of EPL1 on pre-formed hydrophobin layers was tested. As a representative example of several independent replications of these experiments, Fig. 3c and d show the samples already shown in Fig. 3a and b, respectively, after addition of EPL1 solution to the AFM's liquid cell. Significant structural changes could not be observed even after one hour of scanning. Thus, it can be concluded that EPL1 neither destroys ordered HFB layers nor sticks onto their surface to grow layers on top of them. Evidently, already existing HFB layers are stable under this treatment. Next, the effect of EPL1 on the growth of HFB9a/b layers was studied. The respective hydrophobins were mixed with EPL1 in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the droplet method (see Experimental section) was applied. As shown in Fig. 4, periodic structures were formed, which, at least by AFM, cannot be distinguished from those of either pure HFB9a/b or EPL1 alone. Image analysis by Fourier transformation (analogous to Fig. 2d) revealed a periodicity of 6.2 nm and 5.3 nm and the height profiles showed groove depths <1 nm.
1 and the droplet method (see Experimental section) was applied. As shown in Fig. 4, periodic structures were formed, which, at least by AFM, cannot be distinguished from those of either pure HFB9a/b or EPL1 alone. Image analysis by Fourier transformation (analogous to Fig. 2d) revealed a periodicity of 6.2 nm and 5.3 nm and the height profiles showed groove depths <1 nm.
| Type of layer | Contact angle [°] |
|---|---|
| HOPG surface (without protein) | 59 ± 10 |
| HFB9a | 22 ± 9 |
| HFB9b | 27 ± 5 |
| Hybrid layer EPL1/HFB9a | 53 ± 8 |
| Hybrid layer EPL1/HFB9b | 61 ± 7 |
Discussion
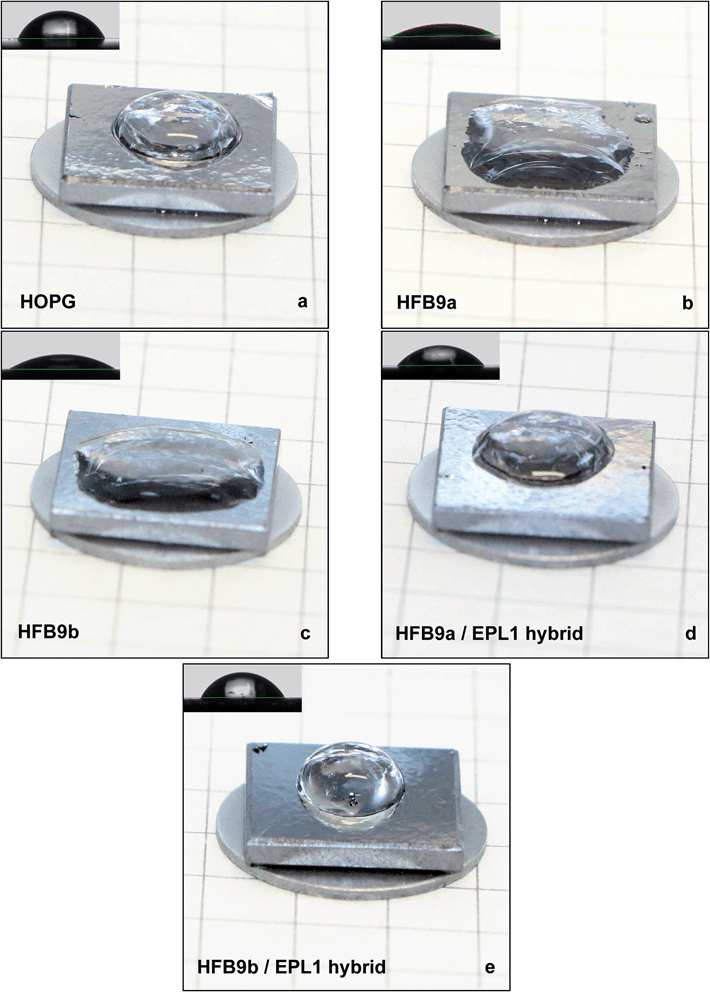
In this study, we analysed the formation of EPL1 protein layers in situ during their formation in buffer, i.e. at solid/liquid interfaces. Up to now it was only known that EPL1 self-assembles at air/water interfaces7 but highly ordered structures on the molecular level were not observed. We were able to show that upon self-assembly of EPL1 indeed highly ordered layers, most likely monolayers (see Fig. 2c) are formed. In the light of the structural analysis of CPPs,18,21 which did not reveal any significant similarities to the amphiphilic surface architecture of hydrophobins,18 it is a curiosity that EPL1 nevertheless shows surface active properties. The coarse meshwork-like structure of the EPL1 protein layer observed in Fig. 1 might result from drying of the EPL1 protein droplet and could be caused by a rather stiff protein film that is already formed on the surface of the droplet (prior to adsorption on the substrate) which is subsequently deformed by the shrinkage of the droplet during drying and adsorption. The stiffness of this surface film is indicative for a stable protein assembly at the air/water interface.In this study also two new hydrophobins were analysed, and the influence of EPL1 on hydrophobin layers was observed. Both, EPL1 and HFBs used in this study, were derived from the fungal genus Trichoderma. Hydrophobins are conventionally grouped into two classes (class I and II) according to their solubility in solvents, hydropathy profiles, and spacing between their conserved cysteines. In general, protein aggregates of class I hydrophobins are more stable and can only be dissolved in strong acids such as TFA, while class II aggregates can be dissolved using aqueous dilutions of organic solvents.25 Previously, in ascomycete fungi only class II hydrophobins were detected, but the wealth of fungal genomes that became available in the past few years led to the discovery of large numbers of new hydrophobins. This included the description of a novel sub-set of class I hydrophobins in ascomycetes, such as Trichoderma24 and Aspergillus species.26 These hydrophobins show also the typical eight cysteines which are characteristic for hydrophobin sequences. Further, structural predictions revealed the typical amphipathic surface that is responsible for the biochemical properties of hydrophobins.24 We found that, in analogy to what has been shown for other hydrophobins before, also HFB9a and HFB9b are able to form highly ordered (mono-)layers. These layers dramatically increase the wettability of HOPG surfaces, as was shown by the strong alteration of droplet shapes (Fig. 5). This can be explained much more straightforward than in the case of EPL1, because these proteins indeed have hydrophilic and hydrophobic surface patches and therefore behaved like typical hydrophobins.
With respect to EPL1 layers, several aspects of our results indicate that the formed protein layers have a monomolecular thickness. At the margins of the highly ordered EPL1 layers (see Fig. 2c) higher step heights which would be indicative for thicker layers could not be observed. Moreover, the flat films with an exceptional long distance order (up to micrometers) suggest a self-terminating growth, most likely leading to monolayers. Furthermore, in case of the amphiphilic hydrophobins the monolayer nature has already been shown.22 Nevertheless, for EPL1 layers a less probable alternative assembly (e.g. double layers) cannot be fully excluded.
The addition of an EPL1 solution had no effect on pre-formed hydrophobin layers, whereas when HFB9a/9b and EPL1 were present in the adsorption solution simultaneously, the resulting layers showed interesting, mixed properties. We suggest that an alternating pattern of single proteins, subunits or protein rows is more likely than a sandwich stack of hydrophobins and EPL1, because sandwiches would probably also be formed if EPL1 was added to an already formed hydrophobin layer, which was not the case.
The distances of periodical rows of HFB9a- and HFB9b-surfaces is 5.3 nm and 5.5 nm, and therefore slightly smaller than those of previously published HFB I and II surfaces, which had lattice parameters ranging from 5.9 nm to 6.1 nm.22,23 EPL1 structures revealed a very similar periodicity of 5.5 nm. The HFB9a/EPL1 hybrid layers had a slightly increased distance between protein rows (6.2 nm) in contrast to the HFB9b/EPL1 hybrids (5.3 nm). However the differences are not significant enough to allow any interpretations on the organization of these hybrid layers. This is also the case for height differences, as profiles show similar sub-nanometer grooves for both, pure-protein and hybrid-layers. The organisation and distribution of the two protein species in those hybrid layers will be an important and interesting aspect for further investigations. The fact that the wettability of hybrid layers was the opposite of what was observed for hydrophobin layers suggests the orientation of hydrophilic/hydrophobic patches of the proteins was at least slightly altered. Protein–protein interactions between EPL1 and the hydrophobins are the likely driving force for that, but detailed investigations on the single-molecule level will probably be necessary to elucidate that further.
The observation of hybrid layers has interesting implications for potential biotechnological applications as well as for the biological roles of EPL1 and CPPs in general. Hybrid layers, which are not as water-soluble as pure EPL1 layers, could be used to enhance the wettability-properties of surfaces in applications where a uniform moistening of a moderately hydrophobic surface is of interest, e.g. in cleaning agents or in spraying applications of plant protection products.
Concerning the biological functions of CPPs it has previously been suggested that they might play a role in fungal growth and development due to their chitin-binding properties and abundant expression during many different growth conditions.7,16,21 Interestingly the epl1 gene in T. atroviride, as well as its orthologues in other fungi, are abundantly expressed during hyphal growth whereas expression of hydrophobins is usually rather related to sporulation, but this has not been tested for the specific case of hfb9a and hfb9b yet. However, since EPL1 is in our experience relatively stable, also in fungal cultivations, and in addition other CPPs such as epl2 are also expressed during sporulation,7 it is presumable that CPPs and hydrophobins also interact in vivo. In this context it is interesting that EPL1 does not affect existing hydrophobin layers, but modifies surface properties in a more subtle way. The biological consequence of mixed hydrophobin/CPP layers would be that fungal growth structures could, under certain conditions, be covered with a protein biofilm which does not invert the polarity of the surface. For polar or hydrophilic molecules, such as those found in the fungal cell wall (carbohydrates and proteins), this would mean an increase of the wettability and hydrophilicity. Most fungi prefer moist growth conditions and therefore an enhancement of the wettability of fungal hyphae might be advantageous for the fungus. Such properties could also aid in the adherence of hyphae to certain surfaces and thus in a better adaptation of the fungus to a growth environment that has zones with varying water content, as might be found in natural environments such as soil.
With respect to the potential roles of CPPs and/or mixed hydrophobin/CPP layers in fungal growth it has to be mentioned that, e.g. for epl1 gene knockout strains so far no growth defects or phenotypes related to the formation of hyphae or spores were detected, also not during stress growth conditions such as osmotic stress.7 This shows that epl1 is a non-essential gene despite the abundant production of EPL1 during fungal growth. In contrast to that, epl1 knockout strains, as well as knockout strains of its orthologue in T. virens, sm1, show a reduced induction of plant defense responses.27,28 Whether this is solely due to the fact that CPPs such as EPL1 or SM1 effectively signal the presence of a fungus for plants or whether CPPs are directly involved in the physical interaction of fungal hyphae with plant roots remains to be investigated. While Trichoderma species are not pathogenic for plants but rather plant-beneficial fungi, in plant pathogens it was already reported that knockout strains of CP-genes showed reduced virulence and necrosis of plant tissue.15,17,29 The attachment and interaction of fungal hyphae with plants, e.g. during the infection process of plant leaves or roots by plant pathogenic fungi could be influenced by either mixed layers of hydrophobins and CPPs, as shown in this paper, which would have a direct effect on the attachment-abilities of hyphae, or possibly also by the interaction of CPPs with plant surface proteins, which could alter the surface properties of the plant leave, thereby aiding in the pathogenic attack of the fungus. The findings of this study provide a possible starting point towards understanding the mechanistic effects of CPPs in fungal–plant interactions in more detail.
Experimental
Production and purification of EPL1
The protein EPL1 was purified from culture supernatants of Trichoderma atroviride P1 as described in ref. 9. Briefly, culture supernatants were concentrated via ultrafiltration using a membrane with 10 kDa cut-off and subsequently purified via cation-exchange chromatography. Purification steps were checked with SDS-PAGE.30 The protein was stored at 4 °C in sodium acetate buffer, pH 4.5. This pH is close to that of T. atroviride cultivations from which EPL1 is purified and the protein was found to be very stable under these conditions.Overexpression and purification of hydrophobins
The hydrophobins that were used in this study are Trichoderma virens HFB9a and HFB9b. The respective NCBI/EMBL/DDBJ accession number, derived from the genome sequencing project of T. virens Gv29-8 v2.0 (http://genome.jgi-psf.org/TriviGv29_8_2/TriviGv29_8_2.home.html) are EHK16816 for HFB9a and EHK25899 for HFB9b. In order to amplify cDNA fragments of T. virens hfb9a and hfb9b the primers hfb9a-fw (CAACAAGGGCAAAGGTGGCAA), hfb9a-rv (TCGTAGATGTTGATGGTGATGGG) and hfb9b-fw (CAACAACAACTGGCAGAGCAAC) and hfb9b-rv (GTAAACGACCTTGGACTGTCCG) were used. The hydrophobin genes were fused between an N-terminal pelB leader directing the proteins to the bacterial periplasm and a C-terminal 6xHis-Tag for rapid purification by affinity chromatography. Overexpression of hydrophobins was carried out in Escherichia coli BL21 DE3 strain (GE Healthcare, Amersham, England). Bacterial strains were cultivated in LB broth containing 40 μg mL−1 kanamycin at 37 °C and 170 RPM, expression was induced by the addition of isothiopropyl-β-D-galactoside at a final concentration of 0.05 mM. The culture was incubated for 5 hours and the cells were harvested by centrifugation (5000g, 4 °C, 10 min). Cell pellets were resuspended in 5 mL buffer and purified using the HisTALON™ Gravity Column Purification Kit (Takara Bio Company, Mountain View, CA, USA). Purification was carried out according to the manufacturer's instructions. Sonification was used to lyse the cells (10 cycles; 30 s pulse with 1 min on ice between pulses) and the supernatant separated by centrifugation at 5000g for 30 min at 4 °C. The pellet was washed twice with the equilibration buffer containing 2 M urea. After the final centrifugation (5000g, 10 min at 4 °C) inclusion bodies were solubilised in the equilibration buffer containing 8 M urea and refolded while bound to the column. The proteins were loaded onto 2 mL of Co2+-charged affinity resin (TALON® Metal Affinity Resin, Takara Bio Company, Mountain View, CA, USA) and subsequently washed with 10 column volumes of equilibration buffer containing 8 M urea. An on-column refolding was performed using a step-wise gradient from 8 M to 0 M urea using a refolding buffer that contained 1 mM reduced glutathione and 0.1 mM oxidized glutathione. For each step 2 column volumes were used until the buffer was free of urea. The beads were then washed with 10 column volumes of equilibration buffer containing 20 mM imidazole to remove histidine-rich impurities. The elution was performed using 300 mM imidazole elution buffer. PD-10 desalting columns (GE Healthcare, Amersham, England) were used to exchange the buffer to 100 mM K2HPO4/KH2PO4 buffer at pH 7. Protein concentration was determined using the Bio-Rad Protein Assay (Biorad, Hercules, CA, USA) and bovine serum albumin as standard. The purified proteins were analysed with SDS-PAGE and staining was performed with Coomassie Brilliant Blue R-250.30AFM sample preparation and imaging
Samples were prepared by 3 different methods: (i) completely drying a drop of protein solution on the HOPG substrate, (ii) droplet method: ordered hydrophobin layers were obtained by the following procedure: a 50 μL drop of 1 nmol mL−1 solution was first pipetted onto a freshly cleaved HOPG surface. After 2 h of incubation in humid environment the hydrophobin layer, which had self-assembled on the drop surface, was transferred and thereby inversed, by gently touching the drop with another freshly cleaved HOPG substrate. Before imaging, the samples were extensively washed with buffer. (iii) In situ preparation: ordered layers of EPL1 were generated in situ adding a small amount of concentrated EPL1 solution (120 nmol mL−1) to the liquid cell to achieve a final concentration of approximately 30 nmol mL−1. For this purpose scanning was interrupted for approximately 30 seconds at a tip-sample separation of 300 nm.AFM images where recorded in tapping mode with a NanoScope V (Bruker, Santa Barbara, CA, USA) either in air or under PBS (phosphate buffered saline) buffer. In both cases the same type of cantilevers was used in order to increase comparability: etched single crystal silicon probes (NCH from Nanoworld, Neuchatel, Switzerland) with a spring constant of 42 N m−1. A free oscillating amplitude of approximately 50–100 mV and a drive frequency of either ∼180 kHz (liquid) or 298 kHz (air) were chosen. Images were taken with set-points corresponding to a damping of approximately 90% of the free amplitude. The pixel size of 500 nm × 500 nm images was set to 512 × 512. In order to enhance the contrast and to increase the visibility of the ordering in the domains, instead of topography images amplitude error images have been chosen for presentation in this paper. The scanner was frequently calibrated and confirmed to have a tolerance of less than 5%. When periodical surface structures were imaged, artefacts due to oscillation were excluded by controlling the correct size scaling and by changing the scanning angle. AFM experiments described in this study were repeated at least two times on independent samples and representative images are shown.
Surface contact angle measurements
Contact angle measurements were performed on a contact angle device DSA 100 (KRÜSS, Hamburg, Germany) using the sessile drop method. For the analysis of the surface properties of protein layers, 2 μL drops of ultrapure water were set down on a HOPG substrate which was either freshly cleaved or coated with hydrophobins or hybrid layers. Drop shapes were modelled with the software program DSA1 (KRÜSS) using a polynomial function. From each surface at least 2 independent samples were prepared, the formation of layers was checked by AFM and 7–15 drops were analysed.Conclusions
In this study we showed with AFM that the CPP EPL1 readily forms highly ordered (mono)layers at liquid/solid interfaces which is remarkable, considering that this molecule has no evidently amphiphilic structural features. This is a clear indication for fast and efficient self-assembly of EPL1. Furthermore, from a phenomenological point of view it was observed that hybrid layers with mixed properties are formed upon simultaneous presence of both, hydrophobins and EPL1, in solution, whereas pre-formed hydrophobin layers are not perturbed by subsequent addition of EPL1. This unprecedented mixture of properties provides a promising starting point for future investigations of their detailed structure and organisation as well as for potential biotechnological applications of CPPs. Furthermore, CPPs might act as antagonists for hydrophobins by subtly modifying their surface activity-altering properties.Acknowledgements
We thank Ernst Pittenauer (Research Group Bio- and Polymer Analysis, Institute of Chemical Technologies and Analytics, Vienna University of Technology) for the MALDI mass spectrometric analyses. This work was funded by the Austrian Science Fund (FWF), grants T390 and V263 to V.S.-S. The work of AP and ISD is supported by ACIB – Austrian Centre of Industrial Biotechnology. The authors wish to thank Oliver Spadiut (Research Area Biochemical Engineering, Institute of Chemical Engineering, Vienna University of Technology) for help with large scale protein ultrafiltration and concentration.Notes and references
- R. Resch, M. Grasserbauer, G. Friedbacher, T. Vallant, H. Brunner, U. Mayer and H. Hoffmann, Appl. Surf. Sci., 1999, 140, 168–175 CrossRef CAS.
- T. Leitner, G. Friedbacher, T. Vallant, H. Brunner, U. Mayer and H. Hoffmann, Microchim. Acta, 2000, 133, 331–336 CrossRef CAS.
- J. Foisner, A. Glaser, T. Leitner, H. Hoffmann and G. Friedbacher, Langmuir, 2004, 20, 2701–2706 CrossRef CAS.
- T. Vallant, H. Brunner, U. Mayer, H. Hoffmann, T. Leitner, R. Resch and G. Friedbacher, J. Phys. Chem. B, 1998, 102, 7190–7197 CrossRef CAS.
- M. B. Linder, G. R. Szilvay, T. Nakari-Setälä and M. E. Penttilä, FEMS Microbiol. Rev., 2005, 29, 877–896 CrossRef CAS PubMed.
- H. A. B. Wösten, Annu. Rev. Microbiol., 2001, 55, 625–646 CrossRef PubMed.
- A. Frischmann, S. Neudl, R. Gaderer, K. Bonazza, S. Zach, S. Gruber, O. Spadiut, G. Friedbacher, H. Grothe and V. Seidl-Seiboth, J. Biol. Chem., 2013, 288, 4278–4287 CrossRef CAS PubMed.
- H. Chen, A. Kovalchuk, S. Keriö and F. O. Asiegbu, Mycologia, 2013, 105, 1479–1488 CrossRef CAS PubMed.
- V. Seidl, M. Marchetti, R. Schandl, G. Allmaier and C. P. Kubicek, FEBS J., 2006, 273, 4346–4359 CrossRef CAS PubMed.
- S. Boddi, C. Comparini, R. Calamassi, L. Pazzagli, G. Cappugi and A. Scala, FEMS Microbiol. Lett., 2004, 233, 341–346 CrossRef CAS PubMed.
- R. González-Fernández, K. Aloria, J. Valero-Galván, I. Redondo, J. M. Arizmendi and J. V. Jorrín-Novo, J. Proteomics, 2014, 97, 195–221 CrossRef PubMed.
- S. Djonovic, M. J. Pozo, L. J. Dangott, C. R. Howell and C. M. Kenerley, Mol. Plant-Microbe Interact., 2006, 19, 838–853 CrossRef CAS PubMed.
- W. A. Vargas, S. Djonovic, S. A. Sukno and C. M. Kenerley, J. Biol. Chem., 2008, 283, 19804–19815 CrossRef CAS PubMed.
- M. Frías, N. Brito and C. González, Mol. Plant Pathol., 2013, 14, 191–196 CrossRef PubMed.
- A. Scala, L. Pazzagli, C. Comparini, A. Santini, S. Tegli and G. Cappugi, J. Plant Pathol., 2004, 86, 27–33 CAS.
- I. Baccelli, C. Comparini, P. P. Bettini, F. Martellini, M. Ruocco, L. Pazzagli, R. Bernardi and A. Scala, FEMS Microbiol. Lett., 2012, 327, 155–163 CrossRef CAS PubMed.
- M. Frías, C. González and N. Brito, New Phytol., 2011, 192, 483–495 CrossRef PubMed.
- A. L. de Oliveira, M. Gallo, L. Pazzagli, C. E. Benedetti, G. Cappugi, A. Scala, B. Pantera, A. Spisni, T. A. Pertinhez and D. O. Cicero, J. Biol. Chem., 2011, 286, 17560–17568 CrossRef CAS PubMed.
- J. Sampedro and D. J. Cosgrove, Genome Biol., 2005, 6, 242 CrossRef PubMed.
- I. Baccelli, S. Luti, R. Bernardi, A. Scala and L. Pazzagli, Appl. Microbiol. Biotechnol., 2014, 98, 175–184 CrossRef CAS PubMed.
- M. R. d. O. d. Barsottini, J. F. de Oliveira, D. Adamoski, P. J. Teixeira, P. F. do Prado, H. O. Tiezzi, M. L. Sforca, A. Cassago, R. V. Portugal, P. S. de Oliveira, M. Z. A. C. de, S. M. Dias, G. A. Pereira and A. L. Ambrosio, Mol. Plant-Microbe Interact., 2013, 26, 1281–1293 CrossRef CAS PubMed.
- G. R. Szilvay, A. Paananen, K. Laurikainen, E. Vuorimaa, H. Lemmetyinen, J. Peltonen and M. B. Linder, Biochemistry, 2007, 46, 2345–2354 CrossRef CAS PubMed.
- A. Paananen, E. Vuorimaa, M. Torkkeli, M. Penttilä, M. Kauranen, O. Ikkala, H. Lemmetyinen, R. Serimaa and M. B. Linder, Biochemistry, 2003, 42, 5253–5258 CrossRef CAS PubMed.
- V. Seidl-Seiboth, S. Gruber, U. Sezerman, T. Schwecke, A. Albayrak, T. Neuhof, H. von Döhren, S. E. Baker and C. P. Kubicek, J. Mol. Evol., 2011, 72, 339–351 CrossRef CAS PubMed.
- M. B. Linder, G. R. Szilvay, T. Nakari-Setälä and M. E. Penttilä, FEMS Microbiol. Rev., 2005, 29, 877–896 CrossRef CAS PubMed.
- B. G. Jensen, M. R. Andersen, M. H. Pedersen, J. C. Frisvad and I. Søndergaard, BMC Res. Notes, 2010, 3, 344 CrossRef CAS PubMed.
- S. Djonovic, W. Vargas, M. Kolomiets, M. Horndeski, A. Wiest and C. Kenerley, Plant Physiol., 2007, 145, 875–889 CrossRef CAS PubMed.
- R. Gaderer, N. L. Lamdan, A. Frischmann, M. Sulyok, R. Krska, B. A. Horwitz and V. Seidl-Seiboth, BMC Microbiology, 2015 DOI:10.1186/s12866-014-0333-0.
- J. S. Jeong, T. K. Mitchell and R. A. Dean, FEMS Microbiol. Lett., 2007, 273, 157–165 CrossRef CAS PubMed.
- J. Sambrook and D. W. Russell, Molecular Cloning: a Laboratory Manual, Cold Spring Harbor Laboratory Press, Plainview, NY, 2nd edn, 2001, pp. 7.1–7.74 and A.78.40–A.78.51.
Footnote |
| † Present address: Department of Anaesthesia, General Intensive Care and Pain Management, Medical University of Vienna, Währinger Gürtel 18-20, 1190 Wien, Austria. |
| This journal is © The Royal Society of Chemistry 2015 |