Open Access Article
Open Access ArticleEnzymatically crosslinked dendritic polyglycerol nanogels for encapsulation of catalytically active proteins†
Changzhu
Wu
a,
Christoph
Böttcher
b and
Rainer
Haag
*a
aInstitut für Chemie und Biochemie, Freie Universität Berlin, Takustraße 3, 14195 Berlin, Germany. E-mail: haag@chemie.fu-berlin.de; Fax: +49-30-83853357; Tel: +49-30-83852633
bForschungszentrum für Elektronenmikroskopie, Institut für Chemie und Biochemie, Freie Universität Berlin, Fabeckstr. 36a, 14195 Berlin, Germany
First published on 11th December 2014
Abstract
The enormous potential of nanogel scaffolds for protein encapsulation has been widely recognized. However, constructing stable polymeric nanoscale networks in a facile, mild, and controllable fashion still remains a technical challenge. Here, we present a novel nanogel formation strategy using horseradish peroxidase (HRP) catalyzed crosslinking on phenolic derivatized dendritic polyglycerol (dPG) in the presence of H2O2 in an inverse miniemulsion. This “enzymatic nanogelation” approach was efficient to produce stable 200 nm dPG nanogel particles, and was performed under physiological conditions, thus making it particularly beneficial for encapsulating biological proteins. Purification of the nanogels was easy to handle and practical because there was no need for a post-quenching step. Interestingly, the use of dPG resulted in higher HRP laden nanogels than for linear polyethylene glycol (PEG) analogs, which illustrates the benefits of dendritic backbones in nanogels for protein encapsulation. In addition, the mild immobilization contributed to the enhanced thermal stability and reusability of HRP. The nanogel preparation could be easily optimized to achieve the best HRP activity. Furthermore, a second enzyme, Candida antarctica lipase B (CalB), was successfully encapsulated and optimized for activity in dPG nanogels by the same enzymatic methodology, which shows the perspective applications of such techniques for encapsulation of diverse proteins.
Introduction
Nanogels that are composed of swollen hydrophilic or amphiphilic polymer networks on the nanoscale have attracted increased interest as nanocarrier scaffolds for applications in drug/protein delivery, biocatalysis, antibacterial agents, sensors, and biotechnology.1–5 The nanoscale networks are typically generated by either physical interactions or chemical crosslinking of structure-forming synthetic or natural polymers.1,6 Self-assembly is the classical physical approach based on a controlled aggregation of hydrophobic7 or electrostatic interactions8,9 and/or hydrogen bonding.6,10 These physical interactions usually take place under mild conditions, which is appealing for encapsulation of biologically sensitive proteins/drugs. However, the non-covalently formed networks are prone to disassemble in particular if crossing biological barriers in living systems or under extreme conditions such as high temperature and extreme pH, which compromises their functions during biological or industrial use. Alternatively nanogels can also be produced by a template-assisted chemical crosslinking of hydrophilic polymers or monomers.1,6 The advantages of the chemical approach are the covalently connected networks which enable nanogels stably transported during biomedical applications or withstand extreme industrial application conditions. The used templates can be a heterogeneous colloidal environment, such as an inverse miniemulsion where chemical crosslinking is restricted to the volume of aqueous nano-droplets,11–13 or it can be a defined mold to shape the network formation in nanoscale.14 However, the main concern in many chemical approaches is the presence of residual, cytotoxic crosslinking agents or an unavoidable exposure to harsh experimental conditions. Therefore, most physical and chemical approaches are not suitable for the preparation of stable nanogels under physiological conditions.Unlike traditional physical and chemical methods, biological enzymes can mediate polymer crosslinking with high selectivity and specificity under physiologic conditions, which avoids unwanted side products and minimizes the damage of surrounding bioactive agents.15 As a result, enzymatic crosslinking for in situ bulk hydrogel formation is becoming a rapidly growing area of research in the fields of biomaterials,16,17 tissue engineering,15,18 drug delivery,19 and regenerative medicine.20 Despite these obvious advantages, enzymatic crosslinking for nanogel preparation has been relatively under-explored, although it has a high potential for significantly promoting the development of biomacromolecule loaded nanogels. The only example reported to date was from Groll and co-workers who formulated nanogels with redox-sensitive disulfide networks by horseradish peroxidase (HRP) in the absence of hydrogen peroxide.3 This system has proven to be particularly effective for encapsulating active β-galactosidase. But apart from this approach, the combined use of HRP and H2O2 (HRP/H2O2) has been widely used to form bulk hydrogel because of its faster and in particular more controllable gelation process.15,16,18–21 Although the presence of H2O2 has the potential to create a cytotoxic environment for cells or to deactivate proteins, the rate of H2O2 consumption can be explicitly controlled by manipulating its initial concentration for crosslinking.16 Up to now, however, such efficient HRP/H2O2 system has never been exploited for nanogel fabrication.
Besides the difficulty in finding suitable crosslinking conditions, another big obstacle that prohibits practical applications of nanogels is the requirement for a tedious nanogel separation. For example, nanogel preparation in a classic inverse emulsion requires multiple washing and dialysis steps to transfer particles from the organic phase into water.3,11–13 In particular, post-quenching residual crosslinkable groups on the nanogel surface is essential for avoiding further nanogel crosslinking in the separation steps, which turns out to be expensive and is toxic for the encapsulated bioactive units. Therefore, there has been a quest to establish a reliable and effective methodology to facilitate nanogel purification for biomedical or industrial use.
Herein we describe a new nanogel preparation method for protein encapsulation by HRP-catalyzed oxidative crosslinking on 3-(4-hydroxyphenyl) propionic acid functionalized dendritic polyglycerol (dPG–HPA) in the presence of H2O2 in an inverse miniemulsion (Scheme 1). Compared with traditional inverse emulsion systems for the preparation of linear polymer network nanogels, the current method has four distinct advantages: (1) mild crosslinking conditions, (2) a facile nanogel preparation without post-quenching, (3) efficient protein loading derived from the closely-packed dendritic backbone of networks, and (4) multiple ways to optimize enzyme activity in scaffolds because of controllable gelation process and multi-functionality of dPG scaffolds. To the best of our knowledge, this is the first example of a nanogel fabrication combining HRP and H2O2 in a controllable fashion. Moreover, we demonstrate dPG nanogels provide more efficient scaffolds for active protein encapsulation for biocatalysis compared to linear analogs.
 | ||
| Scheme 1 Schematic representation of dPG nanogel preparation via HRP crosslinking on dPG–HPA in the presence of H2O2 in an inverse miniemulsion. | ||
Results and discussion
dPG nanogel fabrication
The HRP-catalyzed oxidative crosslinking procedure was used to generate nanogels in a water-in-oil inverse miniemulsion template stabilized by the bioinert surfactants Span 80 and Tween 80 (Scheme 1). Hydrogen peroxide (H2O2) was used to react with HRP to form highly reactive intermediates for crosslinking of polymers that contain phenolic moieties.22 In order to minimize the residual H2O2 content in nanogels to facilitate separation and reduce harmful effects, an excessive amount of dPG–HPA and a sufficient reaction time were applied for the crosslinking (Experimental section). After 2 hours, no residual H2O2 could be detected by quantitative peroxide assay (see the ESI†), which indicates that nearly all the H2O2 was consumed in the aqueous phase, making further crosslinking of nanogels impossible. Based on this fact, transferring nanogels from the continuous organic phase into water was directly conducted by stepwise washing out organic solvents and surfactants by cyclohexane, methanol, and water, respectively. In comparison to traditional emulsion polymerization approaches, the current method does not require any post-quenching step to terminate remaining functional groups before nanogel separation, which greatly facilitates the preparation process, avoids further use of toxic reagents, and is definitely beneficial for bio-related nanogel applications.The nanogels were characterized by dynamic light scattering (DLS) and cryogenic transmission electron microscopy (cryo-TEM), respectively. The DLS data in Fig. 1a shows that no crosslinking took place in the absence of either H2O2 or HRP for two hours and only polymeric precursors or enzymes with diameters smaller than 10 nm were present in water after removal of the organic solvents. In contrast, the enzymatic crosslinking was observed in the presence of both HRP and H2O2, resulting in the formation of water-soluble nanogel particles with diameters of approximately 195 ± 5 nm and PDI 0.075 ± 0.04. The formed nanoparticles were of perfect spherical geometry and an average diameter of around 200 nm was further confirmed by cryo-TEM image data (Fig. 1b, ESI†). Stereo cryo-TEM image pairs (Fig. S17, ESI†) confirmed spherical shape and porous network architecture of nanogel particles which are particularly beneficial for a diffusive uptake of substrates in the context of biomedical applications. The obtained nanogels via HRP/H2O2 crosslinking have much narrower PDI and smaller size comparing to those ones formed with HRP in the absence of H2O2 by Groll et al.3 This improvement should be attributed to a more controllable gel formation and facile separation of the current method. Moreover, dPG nanogels were found to be stable in water for over a week without any size change (shown in Fig. 1c), which reflects their strong gel networks and makes them suitable carriers for durable encapsulation of biomacromolecules.
Release, thermal stability, and reuse of HRP-laden nanogels
Recent studies disclosed the distinct protein leaching profile between nanogels and bulk hydrogels as a result of different mesh size.3,23,24 In order to explore the difference, dPG bulk hydrogels and nanogels were prepared with the same concentration of HRP, H2O2 and dPG–2% HPA and protein release was monitored during the process of gel preparation as well as their further incubation in water. The HRP release profile was evaluated by determining the released HRP activity from an incubation solution of nano-/hydrogels via a chromogenic reaction using ABTS as substrates (Experimental section). Fig. 2a show that up to 85% HRP could be initially encapsulated in dPG bulk hydrogels immediately after gel formation. But these enzymes were quickly released from bulk hydrogel scaffolds when further incubated with water for 72 hours. The complete leaching of HRP was attributed to the larger mesh size of hydrogel networks than HRP, which is in line with our recently revealed rheological data.21 Distinct from bulk gels, only 40% proteins were finally loaded into nanogels after being transferred into water. The reduced encapsulation efficiency in nanogels is thought to be protein loss during nanogel purification, as well as to protein interactions/adsorptions of surfactants that were used for creating miniemulsion.25 Surprisingly, as long as nanogels were transferred into water, there was no further observed protein release for 72 hour incubation (Fig. 2a), eventually resulting in 1.64 μgprotein per mgnanogels protein loading in nanogels. One reason for the negligible protein leaching in dPG nanogels could be that the smaller network mesh size was formed in nanogels than bulk hydrogels as already suggested by Groll et al.3 Another possibility might be due to the globular architecture of dPG backbones which could be connected as closely-packed scaffolds. In order to verify this hypothesis, tyramine conjugated linear polyethylene glycol (PEG–TG, see the synthesis in the ESI†) was enzymatically formulated into nanogels under the same conditions as dPG–2% HPA nanogels. Even though linear PEG had the same molecule weight and number of phenol groups as dPG–2% HPA, the initial HRP encapsulation was four times lower in PEG nanogels than dPG nanogels. Further observation revealed that encapsulated HRP were continuously released from PEG nanocarriers over 72 hours as result of the severe protein leaching profile of linear PEG nanogels. All together, enzymatic crosslinking in dendritic polymers formed more compact nanogel networks, which contributed to a more efficient and stable HRP encapsulation.Enzymes entrapped in a matrix behave significantly different from their native states because of the distinct surrounding microenvironment. In order to examine protein thermal stability, both native and encapsulated HRP were subjected to 50 °C incubation for 42 hours. Fig. 2b shows that encapsulated HRP was quite stable at such high temperature, and nearly 70% HRP activity was eventually retained after incubation. In contrast, native HRP quickly deactivated over 42 hours with an overall 90% activity loss. The calculated data (see the ESI†) further disclosed that the half-life time (t1/2) of encapsulated HRP was 5.6 times enhanced compared to the native HRP. The prolongation of the thermal stability points to the high potential of HRP encapsulated nanogels as practical biocatalysts for asymmetric synthetic transformations, where high operation temperatures are sometimes beneficial.
Industrial applications necessitate not only a high thermal stability of enzymes, but also their multiple usability, which is particularly essential for cost-effective production. The native HRP could hardly be reused because of the difficulty to isolate small proteins with only ∼4 nm diameters from reaction media.3,26 Data from Fig. 1b and 2a indicate that HRP could be easily entrapped into 200 nm dPG nanogels through “self-catalyzed” crosslinking, which eventually equipped them as biocatalytically active nanoparticles with facilitated reuse. The obtained nanogels could be reused up to 5 times and still maintained approximately 50% activity of HRP, as shown in Fig. 2c. Therefore, encapsulating HRP in dPG nanogels makes the biocatalysts reusable, which will reduce synthetic cost and facilitate the separation process for applications.
Optimization of HRP activity in dPG nanogels
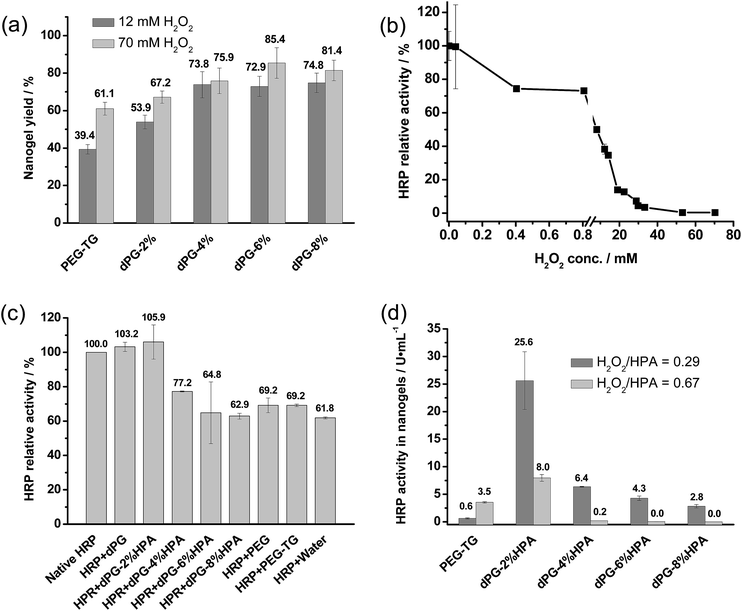
Although there are advantages with respect to thermal stability and reuse of immobilized HRP, the preparation of nanogels has to be optimized in order to achieve an optimal formulation for enzyme encapsulation. For the optimization, a conversion of chromogenic substrate of ABTS by HRP was chosen to determine enzyme activity (U) (Experimental section), and enzyme stability was studied by incubation of HRP in different preparation conditions, and expressed by the change of enzyme relative activity (%). The obtained nanogels were concentrated into one mL, and their total catalytic performance was quantified by the volumetric activity (U mL−1).We first optimized nanogel yield because higher yield is known to benefit protein encapsulation.27 In order to achieve an optimal nanogel yield, crosslinking was conducted with dPG loaded with 2, 4, 6, and 8% phenol groups, respectively. More than 8% phenol loading resulted in the decreased water-solubility of dPG, which made a further increase the degree of substitution (DS) of polymers undesirable for nanogel preparation. 1H NMR spectroscopy was employed to quantify the degree of HPA substitution to polymers, which is calculated by integrating the aromatic peaks at δ 7.09 and 6.80 ppm as well as by the dPG scaffold peaks at δ 3.37–4.10 ppm, as shown in Fig. 3.
 | ||
| Fig. 3 1H NMR spectra (0–12 ppm region) of the dPG–HPA conjugates in D2O with varying phenolic substitutions. | ||
Fig. 4a shows that all the dPG based crosslinking caused a higher nanogel yield compared to the linear PEG analogs, even for dPG–2% HPA, which suggests a preferable enzyme access and hence conversion rate towards the dendritic architecture. The slight increase in the phenolic substitution on dPG from 2% to 4% led to a higher nanogel yield. It was surprising that higher yield was not observable from the nanogels crosslinked with dPG–6% HPA and dPG–8% HPA, which means that some residual phenol groups must have remained unreacted in the polymers. The reason might be that further enzymatic crosslinking was not feasible when protein and substrates (dPG–HPA) were tightly restricted in gel networks to the extent that enzymes could not access substrates anymore.
Apart from the degree of phenolic substitution, H2O2 concentration is another important parameter that can be used to tune the crosslinking for a better nanogel yield. It is found that crosslinking conversion was notably enhanced at elevated H2O2 levels for PEG and dPG–2% HPA, while there was no significant influence upon dPG loaded higher than 2% HPA. This result can be also explained by the full restriction of enzymes in the highly crosslinked gel networks. Nevertheless, the obtained results disclosed that more than 4% phenolic loading and higher H2O2 concentration were beneficial for the nanogel yield.
However, some earlier reports revealed that HRP activity could be irreversibly inactivated in the presence of a high concentration of phenols and H2O2 due to the substrate inhibition.28 The observation was fully supported by our findings for HRP's quick deactivation under the highly concentrated H2O2 conditions (Fig. 4b). HRP lost over 95% activity after 24 hour incubation with >22 mM H2O2 conditions but at low H2O2 level, e.g., less than 40 μM, no HRP deactivation, however, could be observed. This finding demonstrates that the control of initial and residual content of H2O2 for nanogel preparation is essential for obtaining optimal HRP activity. Besides the investigation of H2O2 influence, HRP stability was further studied with the incubation conditions of 100 mg mL−1 dPG loaded with 0, 2, 4, 6, and 8% phenol groups, respectively. Interestingly, it was observed that the addition of dPG alone or only loading 2% phenol groups helped stabilize HRP with 100% activity for at least 24 hours at room temperature, as is shown in Fig. 4c. This stabilization effect illustrates dPG's excellent biocompatibility as well as its high potential as additives or carriers for applications in protein technology. But further increasing of the phenol group loading on dPG impaired the dPG biocompatibility; decreased HRP activity was observed for the incubation with dPG–4%-, 6%- and 8% HPA. This is in accordance with an early observation in the literature that reported HRP deactivation in a highly concentrated phenol solutions.29
Based on the data in Fig. 4a–c, nanogel formulation for HRP activity should consider both the crosslinking yield and HRP's stability. Thus dPG nanogels were prepared with varied concentrations of H2O2 and phenolic loading of polymers while fixing the molar ratio of H2O2/HPA at 0.29 and 0.67, respectively, to assure full consumption of H2O2 in the end. Fig. 4d shows that there was always higher HRP activity in nanogels prepared at low phenolic loading polymers and H2O2 level. This finding revealed that HRP stability is more important than the protein loading efficiency in the pursuit for high HRP activity in dPG nanogels. The nanogels with the highest HRP activity were formulated at dPG–2% HPA with H2O2/HPA at 0.29, where the substrate inhibition was least. This data further demonstrated that HRP activity in nanogels greatly decreased if the H2O2 level was elevated from H2O2/HPA at 0.29 to 0.67 during preparation. Interestingly, dPG–2% HPA nanogels had higher HRP activity than PEG-based nanogels, suggesting that protein loading efficiency took effect when the level of H2O2 and phenol groups was equal. These data helped us to conclude that the best nanogel activity could be accomplished by enzymatic crosslinking of dPG–2% HPA at lower H2O2 level.
Encapsulation of the second enzyme CalB in dPG nanogels
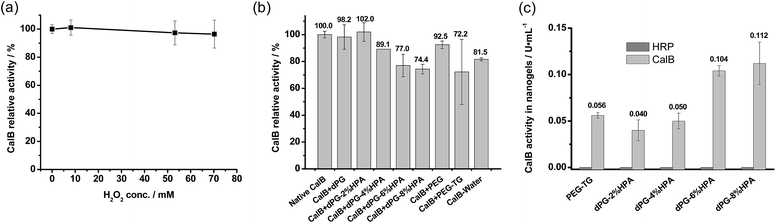
In order to exploit the universality of the currently investigated method for protein encapsulation, a second enzyme, Candida antarctica lipase B (CalB), which is not involved in crosslinking reactions, was also added during nanogel preparation. DLS data (Fig. S19, ESI†) displayed that addition of the second enzyme to nanogels did not change the particle size. Furthermore, pronounced CalB activity could be detected in the nanogels (Experimental section), which confirmed that the “enzymatic nanogelation” approach was versatile for the encapsulation of different enzymes.Similar to HRP encapsulation, CalB activity in nanogels was also needed to be optimized. But unlike HRP, CalB is a robust enzyme against many aggressive operation conditions, thus an associated inhibition by H2O2 and phenol groups might not be expected. To test this hypothesis, native CalB was incubated with H2O2 solutions with a large concentration gradient. The data in Fig. 5a revealed that CalB maintained nearly 100% activity after overnight incubation at 72 mM concentration of H2O2, a level far exceeding the amount needed for nanogel preparation. Based on this finding, it can be concluded that the encapsulation of CalB can be operated even at higher H2O2 level.
Moreover, CalB stability was investigated by incubating the enzymes in solutions of 100 mg mL−1 dPG, phenolic derivatized dPG, PEG, and PEG–TG, respectively. Although deactivation of CalB by the incubation of polymer solutions was also found (Fig. 5b), the stability of CalB was notably enhanced beyond that of HRP with the same incubation conditions. The better CalB stability has profited from its intrinsic stability, while the overloaded phenolic moieties on the dPG levelled off dPG's biocompatibility, thus decreasing CalB activity accordingly. Therefore, CalB activity of dPG nanogels could be optimized by tuning both the crosslinking yield and dPG's phenolic functionality degree.
As what mentioned above, the best HRP activity of nanogels could be obtained under the crosslinking conditions with a low H2O2 concentration and less functionalized dPG. This condition will be particularly favourable for encapsulating a second enzyme, e.g., glucose oxidase, for coupled enzymatic reactions in biochemical assays.30,31 However, in some cases, the active version of HRP in nanogels is unwanted; for example, it may compete with CalB for substrates during formation of peroxycarboxylic acids in the presence of H2O2.32,33 Consequently, in order to demonstrate the possibility to fully suppress HRP activity nanogels were fabricated with 72 mM H2O2 for encapsulation with two enzymes. Surprisingly, the nanogels were obtained at such high level of H2O2 concentrations, as seen in Fig. S20 (see the ESI†); the separation was conducted without using any quenching steps. This indicated that all HRP was inactivated after crosslinking due to the strong H2O2 inhibition. Further activity assays of both CalB and HRP in nanogels confirmed the HRP's deactivation, and disclosed a prominent CalB activity, as shown in Fig. 5c. The highest CalB activity was reached when 8% of phenol groups on dPG were used for the nanogel preparation, which suggested that the nanogel yield was a predominant factor in the increased CalB activity for preparation. This is also in agreement with previous findings of better CalB stability in regard to the influence from dPG–HPA and H2O2 than HRP's stability.
Conclusions
In summary, spherical dPG nanogels have been successfully fabricated by HRP-catalyzed oxidative crosslinking with an inverse miniemulsion template. The preparation method, which can be performed under mild physiological conditions, is particularly beneficial for bio-related delivery systems including drug and/or protein delivery. The success of this method opens up new horizons in nanogel research as it provides controlled crosslinking conditions so that quenching steps can be avoided, thus making it applicable for other polymeric nanogel preparation approaches.Moreover, the encapsulation provides a robust and biocatalytically active nano-architecture that can withstand high temperatures and continuous operation, which suggests that it might be worthy of being tried for applications in other adverse conditions like organic solvents, extreme pH and high ionic strength. In addition, the successful loading of CalB into the carriers indicates the high potential of “enzymatic nanogelation” for encapsulation of diverse enzymes, in particular for applications related to coupled-enzyme reactions.
Last but not least, HRP and CalB activity in nanogels can be optimized, depending on the preparation conditions. Conceivably, the system can be further improved, for example, by employing a wide range of two parameters (H2O2 concentration, and polymer functionality), which should be of use for rational enzyme activity.
Experimental section
Materials
In general, chemicals were purchased from Sigma-Aldrich and used as received. Anhydrous pyridine and dimethylformamide (DMF) were purchased from Acros Organics. Deuterated solvents were bought from Deutero GmbH. dPG (Mw 6 kDa) was synthesized by anionic, ring-opening multi-branching polymerization of glycidol under slow monomer addition.34,35 Polyethylene glycol (PEG, Mw 6 kDa) was obtained from Acros Organics. Horseradish peroxidase (HRP, type VI-A, lyophilized powder) was purchased from Sigma-Aldrich. Candida antarctica lipase B (CalB, Lipozyme CALB L, protein content 0.24% w/w) was a generous gift from Novozymes and was further purified through dialysis in water and lyophilized to powder before use.dPG–HPA loaded with 2, 4, 6, and 8% phenol groups was synthesized in five steps according to our previously reported procedure.21 Typically, dPG was initially converted to dPG–NH2 in three steps: mesylation, nucleophilic substitution, reduction (see the ESI†). Subsequently, HPA was conjugated to dPG–NH2 through amide coupling reactions. The final compound dPG–HPA was purified through dialysis (membrane Mw cut off 1.5 kDa) again methanol for three days (see synthetic details and characterization in the ESI†).
Sonicator™ (Model W-220f, from Heat Systems-Ultrasonics, Inc.) was employed to emulsify aqueous solutions in cyclohexane as nano-sized droplets. Dynamic light scattering (DLS) measurements were carried out using a Zetasizer Nano-ZS from Malvern Instruments equipped with a 633 nm He–Ne laser. Nanogels were visualized through transmission electron microscopy (TEM, a Philips CM12 electron microscope) with cryo-TEM techniques. 1H NMR spectra (500 MHz) were recorded on a Delta JEOL Eclipse at 25 °C using D2O, CD3OD, and CDCl3 as solvents. The spectra were calibrated using the solvent residual peak. FTIR and UV-Vis were carried out on Jasco FT/IR-4100 and Perkin-Elmer Lambda2S, respectively.
Fabrication of dPG nanogels
An inverse miniemulsion was created for making nanogels according to the modified procedure.13 Generally, a 190 μL mixture of dPG–HPA and HRP solutions was pre-emulsified by magnetic stirring for 20 min in 5 mL cyclohexane containing 20 mg mL−1 Span 80 and 67 mg mL−1 Tween 80. The mixture under ice bath was completely emulsified by a tip sonicator for 4 min, and during the course, 60 μL volume of H2O2 solutions was introduced to initiate the enzymatic crosslinking. The amount of dPG–HPA and HRP used was excessive with final concentrations in aqueous phase at 100 mg mL−1 and 0.25 mg mL−1, respectively; the H2O2 level was varied on demand. A sufficient period of time, e.g., more than two hours, was given to allow the enzymatic crosslinking to complete, which, on the other hand, fully consumed H2O2.In order to study the possibility of encapsulating a second enzyme, a mixture was prepared with 100 mg mL−1 polymer and 1 mg mL−1 HRP and 0.6 mg mL−1 CalB. The mixture was then emulsified and crosslinked as nanogels with the above mentioned procedure.
hPG nanogels were purified first by centrifuging the miniemulsion at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 minutes before removing the supernatant. The pellet was further washed 3 times with 15 mL cyclohexane to remove surfactants. Subsequently, residual cyclohexane was removed by washing it 3 times in methanol (totally 15 mL). Finally, pure nanogels were obtained by washing twice with water and then diluted up to 10 mg mL−1 as stock solutions for characterization. The yield was quantified in regard to the nanogel net weight to a totally used up mass after lyophilizing the nanogels.
000 rpm for 30 minutes before removing the supernatant. The pellet was further washed 3 times with 15 mL cyclohexane to remove surfactants. Subsequently, residual cyclohexane was removed by washing it 3 times in methanol (totally 15 mL). Finally, pure nanogels were obtained by washing twice with water and then diluted up to 10 mg mL−1 as stock solutions for characterization. The yield was quantified in regard to the nanogel net weight to a totally used up mass after lyophilizing the nanogels.
Assessment of the catalytic performance of free and encapsulated HRP
The HRP activity was determined by the conversion of chromogenic substrate of ABTS (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)). The obtained nanogels in each preparation batch were concentrated into one mL for activity determination. Typically, a 1.4 mL mixture of substrate solutions was prepared to contain 2.5 mM ABTS and 0.26 mM H2O2 in 100 mM phosphate buffer (pH 7). Subsequently, 50 μL droplets of aqueous solutions of native HRP or nanogels were added into the mixture to initiate the reaction at room temperature. The absorbance increase was recorded at 436 nm by UV-Vis to determine an initial reaction rate period within 2 min. All the reactions were performed in triplicate. In order to simplify the calculation, HRP activity (U) was defined as the absorbance increase per min of free HRP or nanogels. As indicated in the literature,36,37 a relative activity (%) was always used to show the tendency of activity changes during enzyme stability and reuse studies, where 100% was set for the activity measured directly from HRP stock solutions. Volumetric activity (U mL−1) was calculated to reflect the total catalytic performance of nanogels.Assessment of the catalytic performance of free and encapsulated CalB
Prior to the catalytic assessment, the nanogels prepared during optimization were concentrated into one mL as stock solution for use. Determination of the CalB activity was conducted by hydrolysis of 4-nitrophenyl propionate (p-NPB).38 In general, 50 μL enzyme solutions (free CalB or nanogels) were added to 1.45 mL substrate solutions which contained 2 mM p-NPB in 40% (wt/wt) ethanol solutions at room temperature. The absorbance increase was recorded at 410 nm to obtain the initial reaction rate period within 5 min. All reactions were repeated in triplicate. Similar to HRP activity definition, CalB activity (U) was expressed as the absorbance increase per min of the free or encapsulated enzymes. Again, the relative activity (%) was employed for the CalB stability study, and volumetric activity (U mL−1) was used for the expression of total catalytic performance of nanogels.Cryogenic transmission electron microscopy
A droplet (5 μL) of the sample solutions was placed on hydrophilized (plasma treatment using a BALTEC MED 020 device (Leica Microsystems, Wetzlar, Germany)), perforated carbon filmed grids (Quantifoil Micro Tools GmbH, Jena, Germany). The excess fluid was blotted off to create an ultra-thin layer (typical thickness of 200–300 nm) of the solutions, which spans the holes of the support film. The prepared samples were immediately vitrified by propelling the grids into liquid ethane at its freezing point (−184 °C). The vitrified sample grids were transferred under liquid nitrogen by the use of a Gatan (Pleasanton, CA, USA) cryo-holder (Model 626) into a Tecnai F20 TEM (FEI company, Oregon, USA) equipped with FEG and operated at 160 kV acceleration voltage. Microscopy was carried out at −175 °C sample temperature using the microscope's low dose protocol at calibrated primary magnifications of 29 k. The defocus was set to be 9.81 μm. Images were recorded by the use of a 4k-Eagle CCD camera (FEI Company, Oregon, USA) at 2 k resolutions (binning factor 2).Acknowledgements
We thank the BMBF IGCST project, the DFG (Core-Facility BioSupraMol) and the Cluster of Excellence “Unifying Concepts in Catalysis” (UniCat) for financial support. We are grateful to Dr Florian Paulus (FU Berlin) for providing hyperbranched polyglycerol and to Dr Pamela Winchester (FU Berlin) for proofreading this manuscript.Notes and references
- A. V. Kabanov and S. V. Vinogradov, Angew. Chem., Int. Ed., 2009, 48, 5418–5429 CrossRef CAS PubMed.
- W. Chen, F. Meng, J. Feijen and Z. Zhong, J. Controlled Release, 2013, 172, e115 CrossRef CAS PubMed.
- S. Singh, F. Topuz, K. Hahn, K. Albrecht and J. Groll, Angew. Chem., Int. Ed., 2013, 52, 3000–3003 CrossRef CAS PubMed.
- K. Malzahn, W. D. Jamieson, M. Droge, V. Mailander, A. T. A. Jenkins, C. K. Weiss and K. Landfester, J. Mater. Chem. B, 2014, 2, 2175–2183 RSC.
- H.-s. Peng, J. A. Stolwijk, L.-N. Sun, J. Wegener and O. S. Wolfbeis, Angew. Chem., Int. Ed., 2010, 49, 4246–4249 CrossRef CAS PubMed.
- Y. Sasaki and K. Akiyoshi, Chem. Rec., 2010, 10, 366–376 CAS.
- J. Jing, D. Alaimo, E. De Vlieghere, C. Jerome, O. De Wever, B. G. De Geest and R. Auzely-Velty, J. Mater. Chem. B, 2013, 1, 3883–3887 RSC.
- S. Yu, P. Yao, M. Jiang and G. Zhang, Biopolymers, 2006, 83, 148–158 CrossRef CAS PubMed.
- K. Kim, B. Bae, Y. J. Kang, J.-M. Nam, S. Kang and J.-H. Ryu, Biomacromolecules, 2013, 14, 3515–3522 CrossRef CAS PubMed.
- K. Akiyoshi, S. Kobayashi, S. Shichibe, D. Mix, M. Baudys, S. Wan Kim and J. Sunamoto, J. Controlled Release, 1998, 54, 313–320 CrossRef CAS.
- D. Steinhilber, A. L. Sisson, D. Mangoldt, P. Welker, K. Licha and R. Haag, Adv. Funct. Mater., 2010, 20, 4133–4138 CrossRef CAS.
- D. A. Heller, Y. Levi, J. M. Pelet, J. C. Doloff, J. Wallas, G. W. Pratt, S. Jiang, G. Sahay, A. Schroeder, J. E. Schroeder, Y. Chyan, C. Zurenko, W. Querbes, M. Manzano, D. S. Kohane, R. Langer and D. G. Anderson, Adv. Mater., 2013, 25, 1449–1454 CrossRef CAS PubMed.
- D. Steinhilber, S. Seiffert, J. A. Heyman, F. Paulus, D. A. Weitz and R. Haag, Biomaterials, 2011, 32, 1311–1316 CrossRef CAS PubMed.
- S. E. A. Gratton, P. D. Pohlhaus, J. Lee, J. Guo, M. J. Cho and J. M. DeSimone, J. Controlled Release, 2007, 121, 10–18 CrossRef CAS PubMed.
- L. S. Moreira Teixeira, J. Feijen, C. A. van Blitterswijk, P. J. Dijkstra and M. Karperien, Biomaterials, 2012, 33, 1281–1290 CrossRef CAS PubMed.
- D. J. Menzies, A. Cameron, T. Munro, E. Wolvetang, L. Grøndahl and J. J. Cooper-White, Biomacromolecules, 2012, 14, 413–423 CrossRef PubMed.
- L.-S. Wang, C. Du, W. S. Toh, A. C. A. Wan, S. J. Gao and M. Kurisawa, Biomaterials, 2014, 35, 2207–2217 CrossRef CAS PubMed.
- R. Jin, L. S. M. Teixeira, P. J. Dijkstra, Z. Zhong, C. A. v. Blitterswijk, M. Karperien and J. Feijen, Tissue Eng., Part A, 2010, 16, 2429–2440 CrossRef CAS PubMed.
- B. Y. Kim, J. W. Bae and K. D. Park, J. Controlled Release, 2013, 172, 535–540 CrossRef CAS PubMed.
- K. M. Park, Y. Lee, J. Y. Son, D. H. Oh, J. S. Lee and K. D. Park, Biomacromolecules, 2012, 13, 604–611 CrossRef CAS PubMed.
- C. Wu, C. Strehmel, K. Achazi, L. Chiappisi, J. Dernedde, M. C. Lensen, M. Gradzielski, M. B. Ansorge-Schumacher and R. Haag, Biomacromolecules, 2014, 15, 3881–3890 CrossRef CAS PubMed.
- S. Kobayashi, H. Uyama and S. Kimura, Chem. Rev., 2001, 101, 3793–3818 CrossRef CAS PubMed.
- X. Xu, A. K. Jha, D. A. Harrington, M. C. Farach-Carson and X. Jia, Soft Matter, 2012, 8, 3280–3294 RSC.
- W. Wu, S. Chen, Y. Hu and S. Zhou, J. Diabetes Sci. Technol., 2012, 6, 892–901 CrossRef PubMed.
- A. Dan, C. Kotsmar, J. K. Ferri, A. Javadi, M. Karbaschi, J. Kragel, R. Wustneck and R. Miller, Soft Matter, 2012, 8, 6057–6065 RSC.
- M. N.-I. Walters, J. M. Papadimitriou, T. A. Robertson and J. M. Archer, J. Pathol., 1970, 100, 31–37 CrossRef CAS PubMed.
- A. Prokop, E. Kozlov, G. W. Newman and M. J. Newman, Biotechnol. Bioeng., 2002, 78, 459–466 CrossRef CAS PubMed.
- K. J. Baynton, J. K. Bewtra, N. Biswas and K. E. Taylor, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 1994, 1206, 272–278 CrossRef CAS.
- K. J. Baynton, J. K. Bewtra, N. Biswas and K. E. Taylor, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 1994, 1206, 272–278 CrossRef CAS.
- W. M. Aumiller, B. W. Davis, N. Hashemian, C. Maranas, A. Armaou and C. D. Keating, J. Phys. Chem. B, 2014, 118, 2506–2517 CrossRef CAS PubMed.
- F. Costantini, R. Tiggelaar, S. Sennato, F. Mura, S. Schlautmann, F. Bordi, H. Gardeniersb and C. Manetti, Analyst, 2013, 138, 5019–5024 RSC.
- F. Björkling, S. E. Godtfredsen and O. Kirk, J. Chem. Soc., Chem. Commun., 1990, 1972–1995 Search PubMed.
- S. Warwel and M. R. g. Klaas, J. Mol. Catal. B: Enzym., 1995, 1, 29–35 CrossRef CAS.
- A. Sunder, R. Mülhaupt, R. Haag and H. Frey, Adv. Mater., 2000, 12, 235–239 CrossRef CAS.
- A. Sunder, R. Hanselmann, H. Frey and R. Mülhaupt, Macromolecules, 1999, 32, 4240–4246 CrossRef CAS.
- J. Ratzka, L. Lauterbach, O. Lenz and M. B. Ansorge-Schumacher, J. Mol. Catal. B: Enzym., 2012, 74, 219–223 CrossRef CAS PubMed.
- C. Wu, S. Bai, M. B. Ansorge-Schumacher and D. Wang, Adv. Mater., 2011, 23, 5694–5699 CrossRef CAS PubMed.
- D. Pérez, S. Martín, G. Fernández-Lorente, M. Filice, J. M. Guisán, A. Ventosa, M. T. García and E. Mellado, PLoS One, 2011, 6, e23325 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, synthesis of dPG–HPA, synthesis of PEG–TG, nanogel characterization, H2O2 detection, and calculation of the half life time of HRP. See DOI: 10.1039/c4sm01746c |
| This journal is © The Royal Society of Chemistry 2015 |