Open Access Article
Open Access ArticlepH-controlled DNA- and RNA-templated assembly of short oligomers†
Renaud
Barbeyron
,
Jean-Jacques
Vasseur
* and
Michael
Smietana
*
Institut des Biomolécules Max Mousseron (IBMM) UMR 5247 CNRS-Université Montpellier 1 et Université Montpellier 2, Place Bataillon, 34095 Montpellier, France. E-mail: vasseur@univ-montp2.fr; msmietana@univ-montp2.fr
First published on 28th October 2014
Abstract
In the area of artificial genetics the development of non-enzymatic self-organization of synthetic building blocks is critical for both providing biopolymers with extended functions and understanding prebiotic processes. While reversibility is believed to have played a major role in early functional genetic materials, we previously reported an efficient DNA-templated ligation of suitably designed 5′-end boronic acid and 3′-end ribonucleosidic half-sequences. Here, we report the enzyme-free and activation-free DNA- and RNA-templated assembly of bifunctional hexamers. The reversible assembly was found to be regio- and sequence specific and the stabilities of the resulting duplexes were compared to their nicked counterparts. To go further with our understanding of this unprecedented process we also examined an auto-templated duplex self-assembly representing a key step toward the evolution of sequence-defined synthetic polymers.
Introduction
Reconstructing the dynamic complexity of the living is an emergent field where each biological system is divided into various chemical subsystems. Consequently, controlling the self-assembly of artificial biopolymers has great potential for manipulating their morphology and hence their function. Likewise the interplay of these subsystems might lead to higher organized supersystems enabling pioneering advances with biotechnological applications.1 Apart from their significance for the design of functional materials the suggested systems are also attractive models of processes that may have been relevant in RNA world scenarios.2 To elucidate the complexity of self-replicating systems,3 researchers have designed DNA and RNA template-directed chemical reactions of alternative synthetic mononucleotides or short oligonucleotides in enzyme-free conditions. Phosphodiester formation is however a challenging task which requires the chemical activation of a terminal phosphate group.4–6 In turn, this activation renders the activated monomer prone to hydrolysis and leads to undesired side products. Thus many chemical systems relying on modified linkages have been reported over the past few decades,4,7–22 but only a few are based on reversible connections. Indeed, it has been argued that these dynamic systems are particularly appealing in order to understand the environmental conditions that provided life's original scaffold under enzyme-free conditions and to create new functional stimuli-responsive architectures.10,23–29In this context, we have recently described an enzyme-free and activator-free DNA- and RNA-templated ligation system in which the terminal 5′ phosphate of an oligonucleotide was replaced by a boronic acid.30–34 While boronic acid condensation with diols is a valuable dynamic covalent reaction for the construction of macrocycles and cages after removal of water by Dean–Stark trap,35 in aqueous media there is an equilibrium between the free boronic acid and the corresponding boronate ester that depends on both the substitution of the boronic acid and the cis-diol. Interestingly, that equilibrium can be shifted toward an anionic tetrahedral species in the presence of a nucleophile.36–39 This reversible process has been studied extensively40 and has allowed the remarkable development of boronic acid-based sensors for carbohydrates and anions.36,41–44
Hence in the presence of a 3′-ended ribonucleotide partner, the dynamic and reversible formation of a cyclic five membered boronate internucleosidic linkage was found to provide reversible connectivity able to be activated by external stimuli (T, pH, anions) while maintaining the interstrand electrostatic repulsion (Fig. 1). Herein, we report a pH-controlled DNA- and RNA templated assembly of bifunctional 6mers bearing a 5′-end boronic acid and a 3′-end ribonucleotide. This dynamic and reversible process allows sequence- and chain length-specific reading of the template and displays adaptive behavior.
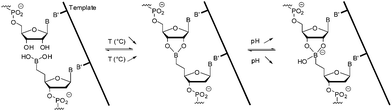 | ||
| Fig. 1 DNA-templated ligation.31 | ||
Results and discussion
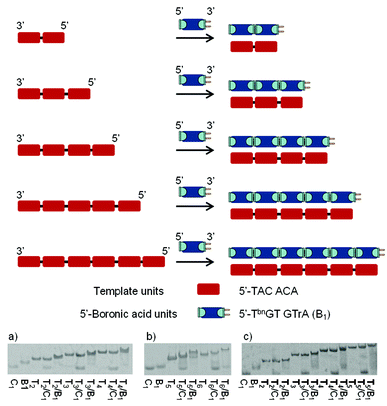
To examine the regio- and sequence specificity of the system, we started by investigating the autoligation of hexanucleotides using a variety of template sequences. Thus, DNA templates of increasing length containing successive repeats complementary to 5′-end boronic 6mer (5′-TbnGTGTrA-3′) were evaluated through thermal-denaturation studies (Fig. 2). All curves featured a single sigmoidal transition that was compared to the one obtained with a non-modified control 6mer (5′-TGTGTrA-3′) forming nicked Watson–Crick duplexes. In the presence of the control hexamer, these transitions increased slightly when increasing the template length probably due to molecular cooperation and stacking strengthening. The observed stabilizations (Table 1) started at Tm = 22.0 °C with one nick and reached Tm = 28.5 °C in the presence of 5 nicks while the resulting nicked duplexes were found to be destabilized in the presence of hydroxide and cyanide ions. On the other hand, compared to the corresponding nicked systems, at pH 7.5 the stabilizations observed with the corresponding boronate-ligated helices ranged from ΔTm = 5.1 °C with one boronate linkage (Table 1, entry 1, Tm = 27.1 vs. 22.0 °C) to ΔTm = 11.1 °C with 5 boronate linkages (Table 1, entry 5, Tm = 39.6 vs. 28.5 °C). Interestingly, the most important stabilization arises when switching from one to two boronate linkages (ΔTm = 6.7 °C). As the number of boronate internucleosidic linkages increases their intrinsic contribution does not add linearly. At pH 9.5 the stabilizations observed with the corresponding boronate-ligated duplexes ranged from ΔTm = 11.0 °C with one boronate linkage (Table 1, entry 1, Tm = 30.2 vs. 19.2 °C) to a remarkable ΔTm = 18.8 °C (Table 1, entry 5, Tm = 42.5 vs. 23.7 °C) with 5 boronate linkages. The same levels of stabilization could also be reached at pH 7.5 in the presence of 3 mM of cyanides ions (Table 1, entries 1–5).| Entry | Template (Tn) | Sequencesa | T m [°C] | |||
|---|---|---|---|---|---|---|
| pH 7.5 | pH 8.5 | pH 9.5 | CN−c | |||
| a Tbn refers to boronothymidine and bold letters represent RNA residues. b Melting temperatures are obtained from the maxima of the first derivatives of the melting curve (A260 vs. temperature) recorded in a buffer containing 1 M NaCl and 10 mM of sodium cacodylate, Template concentration 3 μM; hexamer concentration was adjusted according to the number of repeating units. Curve fits data were averaged from fits of three denaturation curves. c Data were obtained in the presence of 3 mM NaCN. d Not determined. | ||||||
| 1 | 3′-CC(ACACAT)2CC | 5′-TGTGTA | 22.0 | —d | 19.2 | 12.2 |
| 5′-TbnGTGTA | 27.1 | 29.1 | 30.2 | 25.1 | ||
| 2 | 3′-CC(ACACAT)3CC | 5′-TGTGTA | 23.0 | —d | 23.3 | 14.2 |
| 5′-TbnGTGTA | 33.8 | 35 | 37 | 34.1 | ||
| 3 | 3′-CC(ACACAT)4CC | 5′-TGTGTA | 26.0 | —d | 24.3 | 20.2 |
| 5′-TbnGTGTA | 35.9 | 39.1 | 40.8 | 40.2 | ||
| 4 | 3′-CC(ACACAT)5CC | 5′-TGTGTA | 27.0 | —d | 23.3 | 21.3 |
| 5′-TbnGTGTA | 37.2 | 40.2 | 42.1 | 42.3 | ||
| 5 | 3′-CC(ACACAT)6CC | 5′-TGTGTA | 28.5 | —d | 23.7 | 18.8 |
| 5′-TbnGTGTA | 39.6 | 41.5 | 42.5 | 39.9 | ||
| 6 | 3′-CC(ACACAU)2CC | 5′-TGTGTA | 14.7 | —d | 17.9 | <5 |
| 5′-TbnGTGTA | 30.6 | 32.5 | 32.5 | 27.6 | ||
| 7 | 3′-CC(ACACAU)3CC | 5′-TGTGTA | 19.6 | —d | 23.7 | 11.7 |
| 5′-TbnGTGTA | 36.8 | 38.7 | 38.7 | 33.8 | ||
| 8 | 3′-CC(ACACAU)4CC | 5′-TGTGTA | 18.6 | —d | 24.7 | 14.7 |
| 5′-TbnGTGTA | 39.7 | 40.8 | 41.7 | 36.6 | ||
| 9 | 3′-CC(ACACAU)5CC | 5′-TGTGTA | 18.8 | —d | 25.4 | 14.7 |
| 5′-TbnGTGTA | 41.8 | 42.7 | 44.6 | 37.8 | ||
In the presence of a RNA template we observed even higher levels of stabilization at pH 7.5 while the system proved to be less sensitive to pH variations (Table 1, entries 6–9). Indeed, with 4 boronate internucleosidic linkages an increase in the Tm value of 23.0 °C at pH 7.5 was observed compared to the control experiment (Table 1, entry 9, Tm = 41.8 vs. 18.8 °C). This observation is in accordance with the one observed for the RNA template boronic ester ligation.31 Native PAGE analysis confirmed these results with the appearance of new bands corresponding to the stabilized duplexes in the presence of the bifunctional 6mer. Under these conditions we observe the disappearance of the 5′-ended boronic units (B1) whereas the bands of the unmodified control (C1) sequences remain mostly unchanged except for the most stable nicked duplexes T5/C1 and T6/C1 with which we observe a small gel shift due to a slower melting of the duplexes on the gel. Nevertheless the presence of the bands corresponding to the unmodified 6mer confirms the absence of any duplex (Fig. 2).
To probe whether the melting at higher temperature represents the dissociation of base-paired strands and/or the boronate internucleosidic linkage a native PAGE analysis was run at 20 °C (see Fig. S2 of the ESI†). Under these conditions neither the control units (C1) nor the boronic units (B1) were able to associate and thus to assemble on the template. Associated with the fact that no hysteresis could be observed between the de- and renaturing profiles displayed in Table 1, this result highlights the importance of base-pairing to convey the boronic acid and cis-diol functionalities in close proximity thus allowing them to react. Moreover, while boronate-based materials are generated and degraded reversibly under dehydrating/rehydrating conditions, the DNA template provide here a unique environment for the emergence of oligomers in water.45
| Entry | Template | Sequencesa | Template/sequence ratio | T m [°C] | |
|---|---|---|---|---|---|
| pH 7.5 | pH 9.5 | ||||
| a Tbn refers to boronothymidine and bold letters represent RNA residues. b Melting temperatures are obtained from the maxima of the first derivatives of the melting curve (A260 vs. temperature) recorded in a buffer containing 1 M NaCl and 10 mM of sodium cacodylate, template concentration 3 μM; complementary units concentrations were adjusted according to stoichiometry. Curve fits data were averaged from fits of three denaturation curves. | |||||
| 1 | 3′-CC (ACACAT)2 AGTAGT (ACACAT)2 CC | 5′-TGTGTA | 1/4 | 25.0 | 22.9 |
| 5′-TbnGTGTA | 1/4 | 31.1 | 39.3 | ||
| 2 | 3′-CC (ACACAT)2 AGTAGT (ACACAT)2 CC | 5′-TCATCA | 1/1 | <5 | <5 |
| 5′-TbnCATCA | 1/1 | <5 | <5 | ||
| 3 | 3′-CC (ACACAT)2 AGTAGT (ACACAT)2 CC | 5′-TGTGTA + 5′-TCATCA | 1/4/1 | 25.0 | 24.3 |
| 5′-TbnGTGTA + 5′-TbnCATCA | 1/4/1 | 35.2 | 41.1 | ||
| 4 | 3′-CC AGTAGT (ACACAT AGTAGT)2 CC | 5′-TGTGTA | 1/2 | <5 | <5 |
| 5′-TbnGTGTA | 1/2 | <5 | <5 | ||
| 5 | 3′-CC AGTAGT (ACACAT AGTAGT)2 CC | 5′-TCATCA | 1/3 | <5 | <5 |
| 5′-TbnCATCA | 1/3 | <5 | <5 | ||
| 6 | 3′-CC AGTAGT (ACACAT AGTAGT)2 CC | 5′-TGTGTA + 5′-TCATCA | 1/2/3 | 17.5 | 14 |
| 5′-TbnGTGTA + 5′-TbnCATCA | 1/2/3 | 31.2 | 35.4 | ||
| 7 | 3′-CC AGTAGT ACACAT AAAAAAA CC | 5′-TCATCA | 1/1 | <5 | <5 |
| 5′-TbnCATCA | 1/1 | <5 | <5 | ||
| 8 | 3′-CC AGTAGT ACACAT AAAAAAA CC | 5′-TGTGTA | 1/1 | <5 | <5 |
| 5′-TbnGTGTA | 1/1 | <5 | <5 | ||
| 9 | 3′-CC AGTAGT ACACAT AAAAAAA CC | 5′-TTTTTTT | 1/1 | 13.9 | 12.7 |
| 5′-TbnTTTTTT | 1/1 | 12.0 | 9.8 | ||
| 10 | 3′-CC AGTAGT ACACAT AAAAAAA CC | 5′-TGTGTA + 5′-TCATCA | 1/1/1 | 13.9 | 13.8 |
| 5′-TbnGTGTA + 5′-TbnCATCA | 1/1/1 | 22.8 | 25.7 | ||
| 11 | 3′-CC AGTAGT ACACAT AAAAAAA CC | 5′-TCATCA + 5′-TTTTTTT | 1/1/1 | 13.8 | 12.6 |
| 5′-TbnCATCA + 5′-TbnTTTTTT | 1/1/1 | 12.7 | 23.6 | ||
| 12 | 3′-CC AGTAGT ACACAT AAAAAAA CC | 5′-TGTGTA + 5′-TTTTTTT | 1/1/1 | 17.9 | 17.8 |
| 5′-TbnGTGTA + 5′-TbnTTTTTT | 1/1/1 | 22.0 | 27.7 | ||
| 13 | 3′-CC AGTAGT ACACAT AAAAAAA CC | 5′-TGTGTA + 5′-TCATCA + 5′-TTTTTTT | 1/1/1/1 | 17.9 | 16.9 |
| 5′-TbnGTGTA + 5′-TbnCATCA + 5′-TbnTTTTTT | 1/1/1/1 | 26.7 | 30.9 | ||
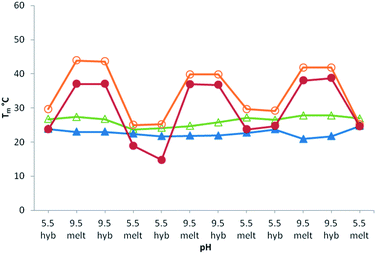
Then, reversibility of the pH-induced assembly was analyzed by thermal denaturation studies on templates T4 and T6. We determined that at pH 5.5 the equilibrium is fully shifted toward the free boronic acid species as can be seen by Tm values of 24 °C and 28 °C respectively, identical to those of the corresponding unmodified controls (Table 1, entries 3 and 5). Fig. 3 shows the melting temperature of systems T4/B1 and T6/B1 as well as their corresponding unmodified analogues T4/C1 and T6/C1 as a function of pH. The pH of each solution was cyclically changed by adding at 0 °C small aliquots of 3 M NaOH or HCl. While no significant change could be observed with the unmodified system, the ΔTm observed with the boronic units is constant even after 3 cycles and demonstrate the ability of the system to be repeatedly assembled/disassembled under enzyme-free conditions in response to pH stimuli. Again, since no hysteresis could be observed between the de- and renaturing curves, pH regulation at 0 °C means that the system is highly reactive and can be reversibly controlled in its duplex state. As such pH variations applied on these systems mimic either ligase or nickase activities. The possibility to efficiently promote the formation of multiple dynamic covalent linkages of 5′-ended boronic acid sequences without any activation prompted us to examine its regio- and sequence specificity.
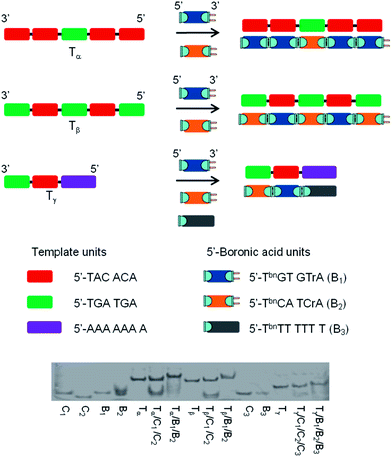
The specificity was probed by performing experiments with templates exhibiting sections complementary to 2 or 3 different 5′-boronic acid building blocks. Again the results were compared to unmodified oligonucleotides. As represented in Fig. 4, the first template, Tα, contained 5 sections complementary to hexameric 5′-TbnGTGTrA (positions 1, 2, 4 and 5) and 5′-TbnCATCrA (position 3) building blocks. Control experiments with stoichiometric amounts of each hexamer (modified and unmodified) against the template, confirmed the Tm values obtained with one nick or one boronate linkage (compare Table 1, entry 1 and Table 2, entry 1) and revealed that neither 5′-TbnCATCrA nor 5′-TCATCrA was able to induce an observable transition in the absence of flanking units (Table 2, entries 2). Remarkably, adding the second building block to the system lead to stabilizations similar to the one observed with either 4 nicks (in the case of the non-modified hexamers) or 4 boronate internucleosidic linkages (compare Table 2, entry 3 and Table 1, entry 4). These results were confirmed, when the template contained alternative complementary sections to 5′-TbnGTGTrA (positions 2 and 4) and 5′-TbnCATCrA (positions 1, 3 and 5) building blocks (Fig. 4, Tβ). Again control experiments in the presence of only one partner confirmed the importance of flanking units to induce an observable transition (Table 2, entries 4 and 5). In the presence of both 5′-boronic acid hexamers, transitions in the range of 4 boronate internucleosidic linkages were displayed while a lower cooperativity was observed for the control sequences (Table 2, entry 6). Finally, a template designed to contain 3 different sections was evaluated (Fig. 4, Tγ). In that particular case, a TbnT6 building block was used to probe both the termination of the process and the possible use of size-different units (Table 2, entry 3). While a low transition could be observed with the 7mers alone (Table 2, entries 9 and 11), the presence of flanking units acting as helpers induced an observable transition with the unmodified sequences (Table 2, entries 10 and 12). Again, at pH 7.5 the melting temperature of the boronate ligated duplex was 8.8 °C higher than that of its nicked analogue and 14.0 °C higher at pH 9.5. In all these experiments, high levels of stabilization could also be reached at pH 7.5 with cyanides ions (see Table S2 of the ESI†). These results were also confirmed by native PAGE analysis (Fig. 4). Taken together these results demonstrate that functional group proximity is required for efficient templated assembly and suggest high degrees of efficiency even in the presence of a mixture of different building blocks.
When mismatches were introduced into position 3 of template Tα thermal-denaturation analysis revealed that the number and the relative placement of these mismatches affect the boronate internucleosidic linkage formation. Indeed, when 4 mismatches were located inside position 3 of Tα, the Tm value at pH 7.5 was almost identical to T2/B (Tm = 28.3 vs. 27.1 °C) while displaying higher levels of stabilization at pH 9.5 (Tm = 34.3 vs. 30.2 °C), thus suggesting the formation of a stabilized bulge between the 5′-end complementary units of position 2 and the 3′-end complementary units of position 4. The same observations were made when 2 mismatches were centrally located inside position 3 of Tα (Tm = 29.7 °C at pH 7.5 and Tm = 33.8 °C at pH 9.5). However, with only one mismatch located either centrally or involved in the formation of a boronate junction, higher degrees of stabilization could be observed though not in the level of perfectly matched duplexes. Indeed, compared to T5/B, the systems were destabilized by 6.6 °C at pH 7.5 (Tm = 30.6 vs. 37.2 °C) and 6.3 °C at pH 9.5 (Tm = 35.8 vs. 42.1 °C) when the mismatch was located centrally, whereas it was destabilized by only 3.5 and 5.4 °C respectively (Tm = 33.7 at pH 7.5 and Tm = 36.7 at pH 9.5) when the mismatch was involved in a boronate junction. These levels of destabilization with one or more mismatches support a dynamic selection of optimal building blocks within a certain pH window. These results were also confirmed by PAGE with the appearance of new retarded bands when one mismatch was located inside position 3 of Tα (see Table S3 and Fig. S1 and S3 of the ESI†).
Finally, we evaluated the ability of this nonenzymatic process to support polymer evolution efforts in the absence of a template. To that purpose two hexamers (5-TbnGACGrC and 5′-TbnCAGCrG) were designed to allow a 3 bases overlap (Fig. 5). While no transition could be observed with unmodified analogues, mixing both 5′-boronic acid hexamers led to a pH-dependent melting temperature demonstrating an unprecedented auto-templated duplex assembly in enzyme free and activator-free conditions.
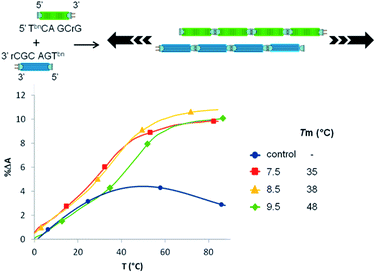 | ||
| Fig. 5 Schematic representation of the autotemplated duplex assembly and UV thermal denaturation curves at different pHs (control experiment represented at pH 7.5). | ||
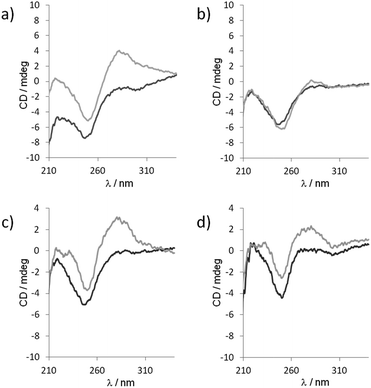
Though the exact distribution of the self-assembled polymer could not be resolved by native PAGE analysis due to only 3 bases overlap of each fragments, the self-assembly behavior was further investigated by circular dichroism (CD). CD spectroscopy is a prevailing method to study the organization of conformational polymorphism of nucleic acids.46 The CD spectra of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of unmodified 5′-TGACGrC and 5′-TCAGCrG sequence recorded at two different temperatures (283 and 293 K) and two different pHs (7.5 and 9.5) exhibited only a strong negative band at 246 nm characteristic of a lack of secondary structure (Fig. 6).46,47 However, when 5′-TbnGACGrC and 5′-TbnCAGCrG were mixed at 283 K in a 1
1 mixture of unmodified 5′-TGACGrC and 5′-TCAGCrG sequence recorded at two different temperatures (283 and 293 K) and two different pHs (7.5 and 9.5) exhibited only a strong negative band at 246 nm characteristic of a lack of secondary structure (Fig. 6).46,47 However, when 5′-TbnGACGrC and 5′-TbnCAGCrG were mixed at 283 K in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio at pH 7.5, the CD spectrum changed dramatically and showed a negative band at 246 nm and a strong positive band at 280 nm characteristic of a B-form46 that disappeared upon heating the sample to 293 K (Fig. 6a and b). In contrast, this positive band could still be observed at 293 K when the pH value was increased to 9.5 (Fig. 6c and d), thus indicating the pH and temperature windows where the self-assembly could be switched on and demonstrate how this organization could be tuned and manipulated.
1 ratio at pH 7.5, the CD spectrum changed dramatically and showed a negative band at 246 nm and a strong positive band at 280 nm characteristic of a B-form46 that disappeared upon heating the sample to 293 K (Fig. 6a and b). In contrast, this positive band could still be observed at 293 K when the pH value was increased to 9.5 (Fig. 6c and d), thus indicating the pH and temperature windows where the self-assembly could be switched on and demonstrate how this organization could be tuned and manipulated.
Conclusion
Based on the formation of reversible and pH-responsive boronate connections, we have developed an efficient non-enzymatic and sequence-specific DNA- and RNA-templated assembly of short oligomers. Their off–on assembly in response to pH triggers and the demonstration that such a system supports an auto-templated duplex self-assembly paves the way toward the possibility of exploring systems with greater complexity as well as the formation of highly defined architectures with self-healing properties. We are currently investigating how to control the size distribution and the thermodynamic factors that governs this process.48 Moreover, these results contribute actively to recent hypothesis highlighting the potential prebiotic relevance of boron.49–52Acknowledgements
We thank the Agence Nationale de la Recherche (ANR “Prody- gy”–11-JS07-005-01) and the Région Languedoc-Roussillon (Programme “Chercheur d'Avenir”).Notes and references
- S. S. Mansy, J. P. Schrum, M. Krishnamurthy, S. Tobe, D. A. Treco and J. W. Szostak, Nature, 2008, 454, 122–125 CrossRef CAS PubMed.
- K. Gorska and N. Winssinger, Angew. Chem., Int. Ed., 2013, 52, 6820–6843 CrossRef CAS PubMed.
- V. Patzke and G. von Kiedrowski, ARKIVOC, 2007, 293–310 CrossRef CAS.
- A. Kaiser and C. Richert, J. Org. Chem., 2013, 78, 793–799 CrossRef CAS PubMed.
- J. Szostak, J. Syst. Chem., 2012, 3, 2 CrossRef CAS.
- L. E. Orgel, Crit. Rev. Biochem. Mol. Biol., 2004, 39, 99–123 CrossRef CAS PubMed.
- M. Hey, C. Hartel and M. W. Gobel, Helv. Chim. Acta, 2003, 86, 844–854 CrossRef CAS.
- J. P. Schrum, A. Ricardo, M. Krishnamurthy, J. C. Blain and J. W. Szostak, J. Am. Chem. Soc., 2009, 131, 14560–14570 CrossRef CAS PubMed.
- C. Deck, M. Jauker and C. Richert, Nat. Chem., 2011, 3, 603–608 CrossRef CAS PubMed.
- Z. Y. Li, Z. Y. J. Zhang, R. Knipe and D. G. Lynn, J. Am. Chem. Soc., 2002, 124, 746–747 CrossRef PubMed.
- D. M. Rosenbaum and D. R. Liu, J. Am. Chem. Soc., 2003, 125, 13924–13925 CrossRef CAS PubMed.
- R. E. Kleiner, Y. Brudno, M. E. Birnbaum and D. R. Liu, J. Am. Chem. Soc., 2008, 130, 4646–4659 CrossRef CAS PubMed.
- B. D. Heuberger and C. Switzer, Org. Lett., 2006, 8, 5809–5811 CrossRef CAS PubMed.
- J. C. Chaput and C. Switzer, J. Mol. Evol., 2000, 51, 464–470 CAS.
- S. L. Zhang, J. C. Blain, D. Zielinska, S. M. Gryaznov and J. W. Szostak, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17732–17737 CrossRef CAS PubMed.
- S. L. Zhang, N. Zhang, J. C. Blain and J. W. Szostak, J. Am. Chem. Soc., 2013, 135, 924–932 CrossRef CAS PubMed.
- P. Hagenbuch, E. Kervio, A. Hochgesand, U. Plutowski and C. Richert, Angew. Chem., Int. Ed., 2005, 44, 6588–6592 CrossRef CAS PubMed.
- M. Rothlingshofer, E. Kervio, T. Lommel, U. Plutowski, A. Hochgesand and C. Richert, Angew. Chem., Int. Ed., 2008, 47, 6065–6068 CrossRef PubMed.
- K. Leu, E. Kervio, B. Obermayer, R. M. Turk-MacLeod, C. Yuan, J. M. Luevano, E. Chen, U. Gerland, C. Richert and I. A. Chen, J. Am. Chem. Soc., 2013, 135, 354–366 CrossRef CAS PubMed.
- M. Dorr, P. M. G. Loffler and P. A. Monnard, Curr. Org. Synth., 2012, 9, 735–763 CrossRef CAS.
- E. Kuruvilla, G. B. Schuster and N. V. Hud, ChemBioChem, 2013, 14, 45–48 CrossRef CAS PubMed.
- E. Kervio, B. Claasen, U. E. Steiner and C. Richert, Nucleic Acids Res., 2014, 7409–7420 CrossRef CAS PubMed.
- Y. Ura, J. M. Beierle, L. J. Leman, L. E. Orgel and M. R. Ghadiri, Science, 2009, 325, 73–77 CrossRef CAS PubMed.
- H. D. Bean, F. A. L. Anet, I. R. Gould and N. V. Hud, Origins Life Evol. Biospheres, 2006, 36, 39–63 CrossRef CAS PubMed.
- A. E. Engelhart and N. V. Hud, Cold Spring Harbor Perspect. Biol., 2010, 2, a002196 CAS.
- V. Patzke, J. S. McCaskill and G. von Kiedrowski, Angew. Chem., Int. Ed., 2014, 53, 4222–4226 CrossRef CAS PubMed.
- A. E. Engelhart, B. J. Cafferty, C. D. Okafor, M. C. Chen, L. D. Williams, D. G. Lynn and N. V. Hud, ChemBioChem, 2012, 13, 1121–1124 CrossRef CAS PubMed.
- X. Y. Li, A. F. Hernandez, M. A. Grover, N. V. Hud and D. G. Lynn, Heterocycles, 2011, 82, 1477–1488 CAS.
- S. I. Walker, M. A. Grover and N. V. Hud, PLoS One, 2012, 7, e34166 CAS.
- D. Luvino, C. Baraguey, M. Smietana and J. J. Vasseur, Chem. Commun., 2008, 2352–2354 RSC.
- A. R. Martin, I. Barvik, D. Luvino, M. Smietana and J. J. Vasseur, Angew. Chem., Int. Ed., 2011, 50, 4193–4196 CrossRef CAS PubMed.
- A. R. Martin, K. Mohanan, D. Luvino, N. Floquet, C. Baraguey, M. Smietana and J. J. Vasseur, Org. Biomol. Chem., 2009, 7, 4369–4377 CAS.
- M. Smietana, A. R. Martin and J. J. Vasseur, Pure Appl. Chem., 2012, 84, 1659–1667 CrossRef CAS.
- R. Barbeyron, J. Wengel, J. J. Vasseur and M. Smietana, Monatsh. Chem., 2013, 144, 495–500 CrossRef CAS PubMed.
- Y. Jin, C. Yu, R. J. Denman and W. Zhang, Chem. Soc. Rev., 2013, 42, 6634–6654 RSC.
- S. D. Bull, M. G. Davidson, J. M. H. Van den Elsen, J. S. Fossey, A. T. A. Jenkins, Y. B. Jiang, Y. Kubo, F. Marken, K. Sakurai, J. Z. Zhao and T. D. James, Acc. Chem. Res., 2012, 46, 312–326 CrossRef PubMed.
- S. Jin, Y. Cheng, S. Reid, M. Li and B. Wang, Med. Res. Rev., 2010, 30, 171–257 CAS.
- R. Nishiyabu, Y. Kubo, T. D. James and J. S. Fossey, Chem. Commun., 2011, 47, 1124–1150 RSC.
- R. Nishiyabu, Y. Kubo, T. D. James and J. S. Fossey, Chem. Commun., 2011, 47, 1106–1123 RSC.
- G. Springsteen and B. H. Wang, Tetrahedron, 2002, 58, 5291–5300 CrossRef CAS.
- R. Badugu, J. R. Lakowicz and C. D. Geddes, Dyes Pigm., 2005, 64, 49–55 CrossRef CAS PubMed.
- D. Luvino, D. Gasparutto, S. Reynaud, M. Smietana and J.-J. Vasseur, Tetrahedron Lett., 2008, 49, 6075–6078 CrossRef CAS PubMed.
- D. Luvino, M. Smietana and J. J. Vasseur, Tetrahedron Lett., 2006, 47, 9253–9256 CrossRef CAS PubMed.
- A. R. Martin, J. J. Vasseur and M. Smietana, Chem. Soc. Rev., 2013, 42, 5684–5713 RSC.
- D. G. Hall, Boronic acids, Wiley-VCH, 2011 Search PubMed.
- J. Kypr, I. Kejnovska, D. Renciuk and M. Vorlickova, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- W. C. Johnson and I. Tinoco, Biopolymers, 1969, 7, 727–749 CrossRef CAS.
- D. Zhao and J. S. Moore, Org. Biomol. Chem., 2003, 1, 3471–3491 CAS.
- R. Scorei, Origins Life Evol. Biospheres, 2012, 42, 3–17 CrossRef CAS PubMed.
- R. Scorei and V. M. Cimpoiasu, Origins Life Evol. Biospheres, 2006, 36, 1–11 CrossRef CAS PubMed.
- A. Ricardo, M. A. Carrigan, A. N. Olcott and S. A. Benner, Science, 2004, 303, 196 CrossRef CAS PubMed.
- B. E. Prieur, C. R. Acad. Sci., 2001, 4, 667–670 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Full experimental details, melting curves and control experiments. See DOI: 10.1039/c4sc03028a |
| This journal is © The Royal Society of Chemistry 2015 |