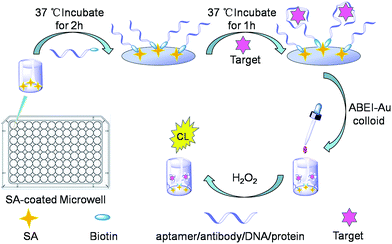
Determination of the binding constant of specific interactions and binding target concentration simultaneously using a general chemiluminescence method†
Lingfeng Gao,
Li Ju and
Hua Cui*
CAS Key Laboratory of Soft Matter Chemistry, Collaborative Innovation Center of Chemistry for Energy Materials, Department of Chemistry, University of Science & Technology of China, Hefei, Anhui 230026, P. R. China. E-mail: hcui@ustc.edu.cn; Fax: +86-551-63600730; Tel: +86-551-63600730
First published on 22nd December 2015
Abstract
The measurement of the binding constant of specific interactions and concentration of a target is of considerable importance in clinical diagnosis, therapy, bioassays and drug design. The development of methods that combine high sensitivity with generalization and simplicity for the measurement of both binding constant and target concentration is highly desirable. Previously, we developed a label-free chemiluminescence (CL) strategy for the measurement of the target concentration and binding constants between DNA aptamers and target simultaneously based on the fact that the target could enhance the CL produced by the reaction of the N-(4-aminobutyl)-N-ethylisoluminol (ABEI) functionalized gold colloid with H2O2. In this study, the enhancement and inhibition effect of various targets on the CL reaction is studied. The generalization of the proposed CL strategy for various targets is also explored. The results demonstrate that the proposed CL strategy is suitable for targets that can cause a change in CL intensity, which includes enhancement and inhibition. It could be applied for the measurement of the dissociation constants of aptamer-binding targets, antibody–antigen complexes, protein-binding small molecules and double-strand DNA hybrids from the millimole to picomole level. It could also be used for the sensitive determination of target concentration, including 2,4,6-trinitrotoluene (TNT), dopamine, tetracycline, human IgG (hIgG), tuberculosis (TB) DNA and mannose, with the detection limit of 0.93 nM–4.1 fM. This strategy is of great potential in fundamental research as well as in applications in life sciences.
Introduction
Studies on specific interactions, such as aptamers with a target, antibody with an antigen, protein with small molecules and DNA hybridization, is of considerable interest for clinical diagnosis and therapy, bioassays, drug design, environmental monitoring and food safety assessment. The determination of the binding constant of specific interactions and binding target concentration is very important in both fundamental research and applications in life sciences. Nowadays, various methods have been developed for the measurement of the binding constant of specific interactions.1 Although these methods have been demonstrated to be efficient in the measurement of binding constants, they mostly either require complicated operations2–4 and expensive instruments5,6 or are time-consuming.7–9 Some methods are only suitable for the interactions either between small-molecules and macromolecules or between macromolecules and macromolecules.10,11 On the other hand, quantitative methods for target concentration based on specific interactions have been studied, including labeling and label-free methods. Labeling methods with high sensitivity, which involve multi-step separations and purifications, are laborious and time-consuming.12,13 Label-free methods are simple, fast and low-cost, but their sensitivity is not ideal.14,15 Usually, the methods for the measurement of binding constants are not suitable for the determination of target concentration due to their low sensitivity, whereas the methods for the determination of target concentration are not suitable for the measurement of binding constants because they are difficult to generate a significant signal change at high target concentrations. Thus, methodologies for the measurement of both of binding constants and target concentration have rarely been reported. Thus, the development of methods that combine high sensitivity with generalization and simplicity for the measurement of both binding constant and target concentration is highly desirable.Recently, we developed a novel chemiluminescence (CL) label-free method for the simultaneous measurement of the DNA aptamer–target dissociation constant (the reciprocal of the binding constant) and target concentration.16 The method was based on the fact that DNA aptamers could capture the corresponding target, which could enhance the CL produced by the reaction of an ABEI functionalized gold (ABEI-Au) colloid with H2O2. It is highly sensitive, selective, simple, fast and low-cost compared with previous methods. We considered that the proposed label-free CL method might be also suitable for targets that could cause a change in CL intensity from the reaction of the ABEI-Au colloid with H2O2. In this study, the effect of various substances on the CL reaction between the ABEI-Au colloid and H2O2 was studied, including bases, sugars, amino acids, proteins, anilines, phenols and nitrotoluenes. It was found that some substances could enhance or inhibit the CL reaction. On this basis, we further explored the applicability of this strategy to measure the binding constant between a DNA aptamer and target that could inhibit the CL signal as well as those between peptide aptamer and target, antibody and antigen, protein and small molecules and DNA hybridization and to determine target concentration. The application scope of this CL strategy was greatly expanded.
Experimental section
Materials and chemicals
Streptavidin (SA), bovine serum albumin (BSA), hIgG, goat-anti-human IgG, tetracycline HCl and amino acids were obtained from Solarbio (Beijing, China). Lysozyme, glucose oxidase (GOD), cytosine, adenosine triphosphate (ATP) and glutathione (GSH) were obtained from Sangon, Inc. (Shanghai, China). 2,4-Dinitrotoluene, 4-nitrotoluene, 2,4,6-trinitrotoluene (TNT) and D-ribose were purchased from Aladdin Reagent (Shanghai, China). Biotinylated concanavalin A was obtained from Sigma-Aldrich (USA). Dopamine was purchased from J&K Scientific Ltd. Guanine and thymine were obtained from Bio Basic Inc. Mannose, adenine, phenol, p-dihydroxybenzene, o-dihydroxybenzene, m-dihydroxybenzene, 1,2,3-benzenetriol, tannic acid, gallic acid, aniline, o-phenylenediamine, m-phenylenediamine, p-phenylenediamine, 2,4-dihydroxybenzoic acid and 3,4-dihydroxybenzoic acid were purchased from Shanghai Reagent Company (Shanghai, China). The sequence of the TNT peptide aptamer is as follows: (N terminus) Trp-His-Trp-Gln-Arg-Pro-Leu-Met-Pro-Val-Ser-Ile-Lys-biotin (C terminus)17 and this TNT peptide aptamer was purchased from GL Biochem Co. Ltd. (Shanghai, China) and purified using high performance liquid chromatography. The dopamine aptamer (5′-GTC TCT GTG TGC GCC AGA GAA CAC TGG GGC AGA TAT GGG CCA GCA CAG AAT GAG GCC C-3′-spacer 93-biotin),18 tetracycline aptamer (biotin-5′-TTT TTC GTA CGG AAT TCG CTA GCC CCC CGG CAG GCC ACG GCT TGG GTT GGT CCC ACT GCG CGT GGA TCC GAG CTC CAC GTG-3′),19 TB capture DNA (biotin-5′-GGT GAC AAA GGC CAC GTA GGC GAA CCC TGC CCA GGT CGA CAC ATA GGT GAG GTC TGC TAC CCA CAG CCG ACC AGG TGC TGG-3′) and TB target DNA (5′-CCA GCA CCT AAC CGG CTG TGG GTA GCA GAC CTC ACC TAT GTG TCG ACC TGG GCA GGG TTC GCC TAC GTG GCC TTT GTC ACC-3′)20 were synthesized by Sangon, Inc. (Shanghai, China) and purified using high performance liquid chromatography. A HAuCl4 stock solution (2‰ w/w) was prepared by dissolving 1.0 g of HAuCl4·4H2O (Shanghai Reagent, China) in 412 mL of purified water, and stored at 4 °C. The ABEI-Au colloid used in this study was synthesized by a seed growth method as described in a previous study.21 The obtained ABEI-Au colloid was stored at 4 °C for further use. All the other reagents were of analytical grade. Ultrapure water was prepared with a Milli-Q system (Millipore, France) and used throughout.Measurement procedure
As shown in Fig. 1, a 100 μL portion of biotinylated aptamers, antibodies, DNA or proteins in a 0.02 M PBS (pH 7.4, containing 0.05 M NaCl) solution was injected into each SA-coated microwell.After incubation at 37 °C for 2 h, the unbounded aptamers, antibodies, DNA or proteins were rinsed with 0.01 M Tris–HCl (pH 7.0, containing 0.05 M NaCl). To control the temperature of the reactions, an incubator (PST-60 HL plus Thermo Shaker, Biosan, Latvia) was used. Then, a 100 μL portion of target solution in 0.02 M PBS (pH 7.4, containing NaCl 0.05 M) was added to each well and incubated at 37 °C for 1 h. The rinsing operation was repeated (0.01 M Tris–HCl, pH 7.0, containing 0.05 M NaCl) to remove the unbound target molecules. Subsequently, a 100 μL portion of the ABEI-Au colloid was added to each well and incubated at 37 °C for 10 min. Then, an aliquot of H2O2 (100 μL, 0.15 M H2O2 in 0.1 M NaOH) was injected into a well, and the CL signal was recorded using a microplate luminometer (Centro LB 960, Berthold, Germany).
To explore the effect of various substances, such as bases, sugars, amino acids, phenols, anilines, nitrotoluenes, peptides and proteins, on the CL reaction between the ABEI-Au colloid and H2O2, these substances were mixed separately with the ABEI-Au colloid and the mixtures were incubated at 37 °C for 10 min before testing.
Result and discussion
Effect of various substances on CL reaction between ABEI-Au colloid and H2O2
In our previous study, it was found that the proposed label-free CL method for the measurement of the dissociation constant of a DNA aptamer–target and target concentration is suitable for targets that can enhance the CL produced by the reaction of the ABEI-Au colloid with H2O2.16 It was considered that the CL method might be generalized for the targets that could lead to a change in CL intensity from the reaction of the ABEI-Au colloid with H2O2. Accordingly, the effect of various substances on the CL reaction between the ABEI-Au colloid and H2O2 was studied by mixing test substance solutions with the ABEI-Au colloid and then injecting H2O2 into the mixture. The tested substances include bases, sugars, amino acids, proteins, anilines, phenols and nitrotoluenes. Fig. 2 shows the effect of four bases, two sugars and ATP on the ABEI-Au colloid CL reaction. The results demonstrate that all of these substances can obviously enhance CL intensity. It was reported that the enhancement of ATP containing adenine and ribose on the ABEI-Au colloids CL reaction is due to the fact that ATP promotes hydroxyl radical production, which accelerates the ABEI-Au colloid CL reaction.16,22 Both bases and sugars are similar to some parts of the ATP structure, thus they might follow a similar enhancement mechanism. Fig. 3 shows the effect of 22 amino acids on CL intensity. All the amino acids exhibited CL enhancement. For most of the amino acids, the CL enhancement was weak. Glutamic acid (Glu), methionine (Met), cysteine (Cys), GSH and homocysteine (Hcy) show a very strong enhancement effect. In earlier studies, the enhancement of amino acids on the luminol CL reaction has been proposed to be due to the reaction of –COO− in their structure with O2˙− to form –CO4˙2−, which accelerates the CL reaction.23 Thus, glutamic acid and GSH with two –COO− groups have strong enhancement. In addition, it was reported that sulfide and some thiol-containing compounds readily reduce the dissolved oxygen to form O2˙−, which results in CL enhancement.24,25 Thus, methionine, cysteine, GSH and homocysteine with thiol or methylthiol groups could strongly enhance the CL reaction. However, cystine, which is formed by the oxidation of two cysteine molecules with a disulfide bond, has a weak enhancement effect. This might be due to the fact that the disulfide bond does not reacts easily with dissolved oxygen to form O2˙−.24 Fig. 4 shows the effect of various proteins and peptides on the CL reaction. Both the proteins and peptides show obvious CL enhancement because they are composed of various amino acids. Fig. 5 shows the effect of anilines, phenols and nitrotoluenes on the CL reaction. Most of these substances show CL inhibition to some extent. Among these, TNT and its derivatives could inhibit the CL reaction obviously. The inhibition effect of aniline, phenol and their derivatives may be due to the competition of the –NH2 or –OH groups in their structures with luminol for O2˙−.23 The different inhibition effects of aniline, phenol and their derivatives might due to their different reducing capabilities. The TNT inhibition effect might be due to the fact that TNT could interact with the luminophor excited-state N-(aminobutyl)-N-(ethylphthalate) formed by the CL reaction, which leads to energy loss and quenching of the CL emission.26 In addition, TNT could consume the ˙OH radical generated in the CL reaction, which leads to a decrease in CL intensity. The derivatives of TNT might follow a similar inhibition mechanism.27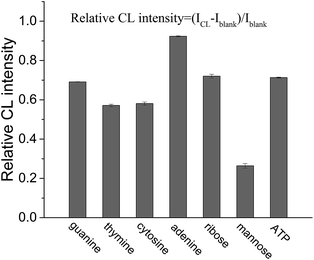 | ||
| Fig. 2 Effect of bases, sugars and ATP on CL reaction between ABEI-Au colloid and H2O2. Reaction conditions: 100 μL 0.15 M H2O2 (pH = 13) and 1 μM substance concentration. | ||
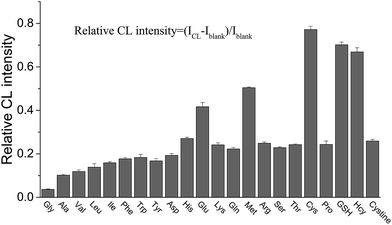 | ||
| Fig. 3 Effect of amino acids on CL reaction between ABEI-Au colloid and H2O2. Reaction conditions: 100 μL 0.15 M H2O2 (pH = 13) and 1 μM substance concentration. | ||
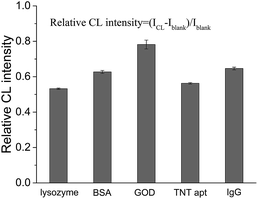 | ||
| Fig. 4 Effect of proteins and peptide aptamers on CL reaction between ABEI-Au colloid and H2O2. Reaction conditions: 100 μL 0.15 M H2O2 (pH = 13) and 1 μM substance concentration. | ||
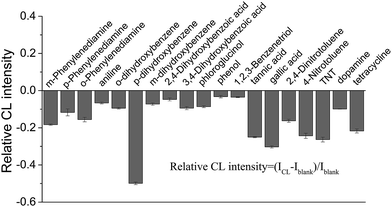 | ||
| Fig. 5 Effect of phenols, anilines and nitrotoluenes on CL reaction between the ABEI-Au colloid and H2O2. Reaction conditions: 100 μL 0.15 M H2O2 (pH = 13) and 1 μM substance concentration. | ||
CL enhancement and inhibition by various organic compounds for luminol CL reactions have been documented.28–30 It is known that the substances that facilitate the formation of oxygen-related radicals, such as OH˙, O2˙−, CO3˙−, CO4˙−, lead to CL enhancement, while the substances that eliminate these radicals result in CL inhibition. In the present study, some of the tested substances cause CL enhancement and others CL inhibition, which follow the similar roles in earlier studies.22–24,26 We considered that the proposed label-free CL method might be applicable to measure the binding constant of specific interactions and the concentration of targets that lead to a change in CL intensity, which is explored below.
Measurement of dissociation constant of DNA aptamer-binding target and concentration of target inhibiting CL signal
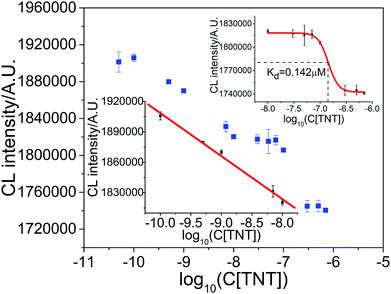
To investigate whether or not the CL strategy is applicable for the simultaneous measurement of aptamer–target binding and target concentration when the target inhibits CL intensity, three types of model targets, TNT, dopamine and tetracycline, and corresponding aptamers were tested.As shown in Fig. 6, the CL signal intensity decreased linearly with the logarithm of TNT concentration at lower TNT concentrations over the range of 1.0 × 10−10 M–1.0 × 10−8 M. The linear response of the logarithm of TNT concentration can be fitted by the equation ICL = 1.4840 × 106 − 4.2454 × 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C with a correlation coefficient of R = 0.9964, where ICL is the CL integrated intensity in 10 s, and C is the concentration of target TNT. The limit of detection (LOD), at a signal to noise ratio of three (S/N = 3), was 0.080 nM. The relative standard deviation (RSD) of seven replicate detections of TNT at 1.0 × 10−9 M (n = 7) was 0.11%, which indicates the good repeatability of the proposed strategy. When the TNT concentration was greater than 1.0 × 10−8 M, the CL intensity deviated from the linear relationship. The binding ratio of aptamer for TNT was estimated to be less than 2.3% and could be ignored when the TNT concentration was below 7.0 nM (ESI, Section S1†). Based on the principle used in the measurement of dissociation constants,31–35 a sigmoid fitted curve from 1.0 × 10−8 to 7.0 × 10−7 M was obtained, as shown in Fig. 6 (upper inset). From the sigmoid fitted curve, a dissociation constant of 0.142 μM for aptamer binding TNT was obtained, which is comparable with the value of 0.0710 μM from the literature.17,36,37
C with a correlation coefficient of R = 0.9964, where ICL is the CL integrated intensity in 10 s, and C is the concentration of target TNT. The limit of detection (LOD), at a signal to noise ratio of three (S/N = 3), was 0.080 nM. The relative standard deviation (RSD) of seven replicate detections of TNT at 1.0 × 10−9 M (n = 7) was 0.11%, which indicates the good repeatability of the proposed strategy. When the TNT concentration was greater than 1.0 × 10−8 M, the CL intensity deviated from the linear relationship. The binding ratio of aptamer for TNT was estimated to be less than 2.3% and could be ignored when the TNT concentration was below 7.0 nM (ESI, Section S1†). Based on the principle used in the measurement of dissociation constants,31–35 a sigmoid fitted curve from 1.0 × 10−8 to 7.0 × 10−7 M was obtained, as shown in Fig. 6 (upper inset). From the sigmoid fitted curve, a dissociation constant of 0.142 μM for aptamer binding TNT was obtained, which is comparable with the value of 0.0710 μM from the literature.17,36,37
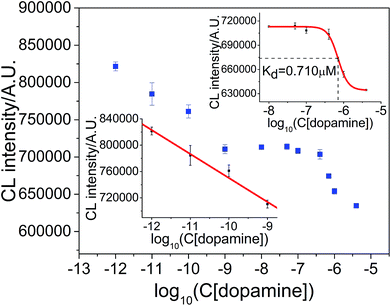
The result for the measurement of aptamer–dopamine dissociation constant and dopamine concentration is shown in Fig. 7. The regression equation is ICL = 3.8602 × 105 − 3.6418 × 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C, where ICL is the CL integrated intensity in 10 s, and C is the concentration of target dopamine, with a correlation coefficient of R = 0.9945. The linear range was 1.0 × 10−12–1.0 × 10−9 M, LOD was 0.90 pM (S/N = 3) and RSD at 1.0 × 10−10 M dopamine (n = 7) was 2.52%. The CL intensity deviated from linearity when the dopamine concentration was greater than 1.0 × 10−9 M. The curve from 1.0 × 10−9 M to 4.0 × 10−6 M was applied for sigmoid fitting and the dissociation constant was calculated to be 0.710 μM (the constant obtained in the literature is 0.700 μM (ref. 18 and 38)).
C, where ICL is the CL integrated intensity in 10 s, and C is the concentration of target dopamine, with a correlation coefficient of R = 0.9945. The linear range was 1.0 × 10−12–1.0 × 10−9 M, LOD was 0.90 pM (S/N = 3) and RSD at 1.0 × 10−10 M dopamine (n = 7) was 2.52%. The CL intensity deviated from linearity when the dopamine concentration was greater than 1.0 × 10−9 M. The curve from 1.0 × 10−9 M to 4.0 × 10−6 M was applied for sigmoid fitting and the dissociation constant was calculated to be 0.710 μM (the constant obtained in the literature is 0.700 μM (ref. 18 and 38)).
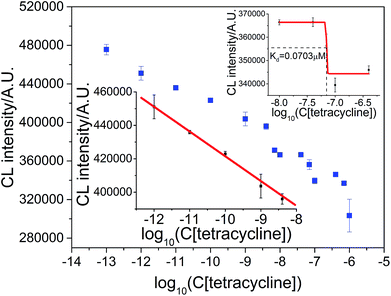
The result for the measurement of the aptamer–tetracycline dissociation constant and tetracycline concentration is shown in Fig. 8. The CL intensity was a linear function of the logarithm of tetracycline concentration in the range of 1.0 × 10−12 M–4.0 × 10−9 M with the regression equation ICL = 2.6581 × 105 − 1.5547 × 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (R = 0.9925), as shown in Fig. 8 (lower inset). The detection limit was (S/N = 3) 0.20 pM, and the RSD at 1.0 × 10−10 M tetracycline was 0.25% (n = 7). The dissociation constant of the tetracycline–aptamer was calculated to be 0.0703 μM, which is comparable to 0.0636 μM measured by Javed and coworkers.4,19,39
C (R = 0.9925), as shown in Fig. 8 (lower inset). The detection limit was (S/N = 3) 0.20 pM, and the RSD at 1.0 × 10−10 M tetracycline was 0.25% (n = 7). The dissociation constant of the tetracycline–aptamer was calculated to be 0.0703 μM, which is comparable to 0.0636 μM measured by Javed and coworkers.4,19,39
These results demonstrate that the proposed CL strategy is suitable for multiple types of targets that can inhibit ABEI-Au colloid initiated CL.
Measurement of dissociation constant of antibody–antigen complex and concentration of antigen
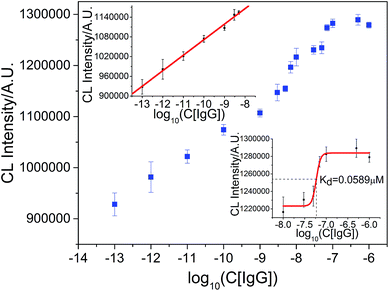
To investigate whether or not the CL strategy could be applicable for the measurement of the dissociation constant of an antibody–antigen complex and antigen concentration in a single detection procedure, goat-anti-human IgG and hIgG were chosen as the model. As shown in Fig. 9, the linear response of the logarithm of hIgG concentration could be fitted by the equation ICL = 1.5440 × 106 + 4.7244 × 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C in the range of 1.0 × 1 0−13 M–5.0 × 10−9 M (R = 0.9976). The detection limit was (S/N = 3) 0.092 pM and RSD at 1.0 × 10−11 M hIgG was 2.9% (n = 7). The curve from 1.0 × 10−8 M to 1.0 × 10−6 M was applied for sigmoid fitting and the dissociation constant was calculated to be 0.0589 μM. These results demonstrate that the proposed CL strategy is also suitable for the simultaneous measurement of the binding constant between an antigen and antibody and concentration of the antigen.
C in the range of 1.0 × 1 0−13 M–5.0 × 10−9 M (R = 0.9976). The detection limit was (S/N = 3) 0.092 pM and RSD at 1.0 × 10−11 M hIgG was 2.9% (n = 7). The curve from 1.0 × 10−8 M to 1.0 × 10−6 M was applied for sigmoid fitting and the dissociation constant was calculated to be 0.0589 μM. These results demonstrate that the proposed CL strategy is also suitable for the simultaneous measurement of the binding constant between an antigen and antibody and concentration of the antigen.
Measurement of dissociation constant of double-strand DNA hybrid and concentration of specific DNA sequence
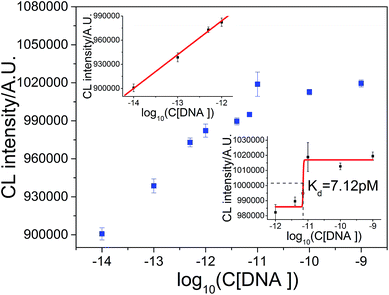
The DNA hybridization reaction was also explored by taking DNA from TB and its complementary chain as an example. As shown in Fig. 10, the linear response of the logarithm of DNA concentration could be fitted by the equation ICL = 1.4878 × 106 + 4.1982 × 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C in the range from 1.0 × 10−14 M to 1.0 × 10−12 M (R = 0.9981). The detection limit was (S/N = 3) 4.1 fM and the RSD at 1.0 × 10−12 M DNA was 2.6% (n = 7). The dissociation constant between TB DNA and its complementary sequence was 7.12 pM, as shown in Fig. 10 (lower inset). This result demonstrates that the interaction between DNA sequences could also be studied by this method. In addition, it also demonstrates that this CL method could be applicable for the measurement of a wide range of dissociation constants.
C in the range from 1.0 × 10−14 M to 1.0 × 10−12 M (R = 0.9981). The detection limit was (S/N = 3) 4.1 fM and the RSD at 1.0 × 10−12 M DNA was 2.6% (n = 7). The dissociation constant between TB DNA and its complementary sequence was 7.12 pM, as shown in Fig. 10 (lower inset). This result demonstrates that the interaction between DNA sequences could also be studied by this method. In addition, it also demonstrates that this CL method could be applicable for the measurement of a wide range of dissociation constants.
Measurement of dissociation constant of protein-binding small molecules and concentration of small molecule
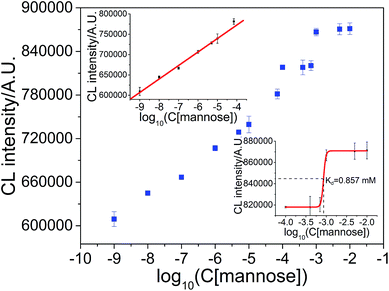
The applicability of this method for the measurement of the dissociation constant of protein-binding small molecules was examined by taking concanavalin A and mannose as the model. The interaction between concanavalin A and mannose is much weaker than those we studied above. Surprisingly, this CL method was still valid. As shown in Fig. 11 (upper inset), the CL intensity was a linear function of the logarithm of mannose concentration in the range from 1.0 × 10−9 M to 1.0 × 10−4 M with the regression equation ICL = 9.0548 × 105 + 3.3181 × 104logC (R = 0.9918). The detection limit was (S/N = 3) 0.93 nM and the RSD at 1.0 × 10−6 M mannose was 0.52% (n = 7). As shown in Fig. 11 (lower inset), the dissociation constant of concanavalin A-mannose was 0.857 mM, which is comparable to that measured in the literature (0.450 mM).40–43Conclusion
In this study, it has been found that some substances, including bases, sugars, amino acids and proteins, could enhance the CL reaction between the ABEI-Au colloid and H2O2, whereas other substances, including anilines, phenols and nitrotoluenes, could inhibit the CL reaction. This study also demonstrated that the proposed label-free CL strategy could be used to determine the binding constant between a DNA aptamer and target and the concentration of the target that could not only enhance but also inhibit the CL signal. Moreover, the CL strategy is also applicable for the measurement of the binding constant between a peptide aptamer and target, antibody and antigen, protein and small molecules and DNA hybridization and target concentration. The dissociation constant could be measured from the millimole to picomole level and the concentration of target, including TNT, dopamine, tetracycline, hIgG, TB DNA and mannose, could be detected sensitively with the detection limit of 0.93 nM–4.1 fM. Compared with the previously reported label-free methods based on specific interactions, the LOD for TNT is comparable with that mentioned in the literature. The LOD for dopamine, tetracycline and TB DNA is 1–3 orders of magnitude lower than those mentioned in the literature; however, the LOD for hIgG is higher than that in the literature (Table S1†). Therefore, the CL strategy is a general and sensitive strategy for the measurement of the binding constant of specific interactions and the concentration of targets that can lead to a change in CL intensity from the reaction of the ABEI-Au colloid with H2O2. This provides a rapid, simple and effective method to study specific interactions between aptamers and targets, antibody and antigen, protein and small molecules and DNA hybridization as well as target concentration, which shows great potential in fundamental research as well as in applications such as clinical diagnosis, therapy, drug design, bioassays, environmental monitoring and food safety assessment.Acknowledgements
The support of this research by the National Natural Science Foundation of P. R. China (Grant Nos. 21173201, 21075115, 20625517 and 20573101), and the Fundamental Research Funds for the Central Universities (Grant No. WK2060190007) is gratefully acknowledged.References
- M. Jing and M. T. Bowser, Anal. Chim. Acta, 2011, 686, 9–18 CrossRef PubMed.
- Y. Li, H. J. Lee and R. M. Corn, Nucleic Acids Res., 2006, 34, 6416–6424 CrossRef CAS PubMed.
- S. Jaouen, L. de Koning, C. Gaillard, E. Muselíková-Polanská, M. Štros and F. Strauss, J. Mol. Biol., 2005, 353, 822–837 CrossRef PubMed.
- J. H. Niazi, S. J. Lee and M. B. Gu, Bioorg. Med. Chem., 2008, 16, 7245–7253 CrossRef CAS PubMed.
- D. S. Hage and S. A. Tweed, J. Chromatogr. B: Biomed. Sci. Appl., 1997, 699, 499–525 CrossRef.
- D. E. Huizenga and J. W. Szostak, Biochemistry, 1995, 34, 656–665 CrossRef PubMed.
- M. Müller, J. E. Weigand, O. Weichenrieder and B. Suess, Nucleic Acids Res., 2006, 34, 2607–2617 CrossRef PubMed.
- J. A. Cruz-Aguado and G. Penner, J. Agric. Food Chem., 2008, 56, 10456–10461 CrossRef CAS PubMed.
- D. K. Mandal, N. Kishore and C. F. Brewer, Biochemistry, 1994, 33, 1149–1156 CrossRef CAS PubMed.
- L. G. Fägerstam, Å. Frostell-Karlsson, R. Karlsson, B. Persson and I. Rönnberg, J. Chromatogr. A, 1992, 597, 397–410 CrossRef.
- V. C. Ozalp, Analyst, 2011, 136, 5046–5050 RSC.
- Y. Chai, D. Tian and H. Cui, Anal. Chim. Acta, 2012, 715, 86–92 CrossRef CAS PubMed.
- X. Yu, Y. Chai, J. Jiang and H. Cui, J. Photochem. Photobiol., A, 2012, 241, 45–51 CrossRef CAS.
- F. Li and H. Cui, Biosens. Bioelectron., 2013, 39, 261–267 CrossRef CAS PubMed.
- F. Li, Y. Yu, H. Cui, D. Yang and Z. Bian, Analyst, 2013, 138, 1844–1850 RSC.
- S. Li, D. Chen, Q. Zhou, W. Wang, L. Gao, J. Jiang, H. Liang, Y. Liu, G. Liang and H. Cui, Anal. Chem., 2014, 86, 5559–5566 CrossRef CAS PubMed.
- Y. Yu, Q. Cao, M. Zhou and H. Cui, Biosens. Bioelectron., 2013, 43, 137–142 CrossRef CAS PubMed.
- R. Walsh and M. C. deRosa, Biochem. Biophys. Res. Commun., 2009, 388, 732–735 CrossRef CAS PubMed.
- Y. J. Kim, Y. S. Kim, J. H. Niazi and M. B. Gu, Bioprocess Biosyst. Eng., 2010, 33, 31–37 CrossRef CAS PubMed.
- J. Jiang, Y. Chai and H. Cui, RSC Adv., 2011, 1, 247–254 RSC.
- D. Tian, H. Zhang, Y. Chai and H. Cui, Chem. Commun., 2011, 47, 4959–4961 RSC.
- S. Aoyagi, M. Yamazaki, T. Miyasaka and K. Sakai, J. Chem. Eng. Jpn., 2001, 34, 956–959 CrossRef CAS.
- H. Cui, M. Shi, R. Meng, J. Zhou, C. Lai and X. Lin, Photochem. Photobiol., 2004, 79, 233–241 CrossRef CAS PubMed.
- J. Du, Y. Li and J. Lu, Anal. Chim. Acta, 2001, 448, 79–83 CrossRef CAS.
- E. de Lamirande and C. Gagnon, Free Radicals Biol. Med., 1998, 25, 803–817 CrossRef CAS PubMed.
- Y. Jiang, H. Zhao, N. Zhu, Y. Lin, P. Yu and L. Mao, Angew. Chem., 2008, 120, 8729–8732 CrossRef.
- H. Yu, Y. He, W. Li and T. Duan, Sens. Actuators, B, 2015, 220, 516–521 CrossRef CAS.
- A. Economou, D. G. Themelis, G. Theodoridis and P. D. Tzanavaras, Anal. Chim. Acta, 2002, 463, 249–255 CrossRef CAS.
- J. Wang, H. Ye, Z. Jiang, N. Chen and J. Huang, Anal. Chim. Acta, 2004, 508, 171–176 CrossRef CAS.
- J. Zhou, H. Xu, G. Wan, C. Duan and H. Cui, Talanta, 2004, 64, 467–477 CrossRef CAS PubMed.
- T. H. Nguyen, L. J. Steinbock, H. J. Butt, M. Helm and R. Berger, J. Am. Chem. Soc., 2011, 133, 2025–2027 CrossRef CAS PubMed.
- D. Shangguan, Y. Li, Z. Tang, Z. C. Cao, H. W. Chen, P. Mallikaratchy, K. Sefah, C. J. Yang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11838–11843 CrossRef CAS PubMed.
- K. Yamana, Y. Ohtani, H. Nakano and I. Saito, Bioorg. Med. Chem. Lett., 2003, 13, 3429–3431 CrossRef CAS PubMed.
- T. Endoh, H. Funabashi, M. Mie and E. Kobatake, Anal. Chem., 2005, 77, 4308–4314 CrossRef CAS PubMed.
- W. Lee, A. Obubuafo, Y. I. Lee, L. M. Davis and S. A. Soper, J. Fluoresc., 2010, 20, 203–213 CrossRef CAS PubMed.
- J. W. Jaworski, D. Raorane, J. H. Huh, A. Majumdar and S.-W. Lee, Langmuir, 2008, 24, 4938–4943 CrossRef CAS PubMed.
- D. L. Roy, W. Yang, X. Yin, R. Y. Lai, S. H. Liou and D. J. Sellmyer, J. Appl. Phys., 2011, 109, 07E532 Search PubMed.
- H. Park and I. R. Paeng, Anal. Chim. Acta, 2011, 685, 65–73 CrossRef CAS PubMed.
- C. Berens, A. Thain and R. Schroeder, Bioorg. Med. Chem., 2001, 9, 2549–2556 CrossRef CAS PubMed.
- S. Takahashi, K. Sato and J. I. Anzai, Anal. Bioanal. Chem., 2012, 402, 1749–1758 CrossRef CAS PubMed.
- M. Farooqi, M. Saleemuddin, R. Ulber, P. Sosnitza and T. Scheper, J. Biotechnol., 1997, 55, 171–179 CrossRef CAS PubMed.
- Y. Lvov, K. Ariga, I. Ichinose and T. Kunitake, J. Chem. Soc., Chem. Commun., 1995, 22, 2313–2314 RSC.
- S. Z. Zhang, F. L. Zhao, K. A. Li and S. Y. Tong, Talanta, 2001, 54, 333–342 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1 and Table S1. See DOI: 10.1039/c5ra24928g |
| This journal is © The Royal Society of Chemistry 2016 |