Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of a visible nanothermometer with a highly emissive 2′-O-methylated guanosine analogue†
Seigi
Yamamoto
a,
Soyoung
Park
*a and
Hiroshi
Sugiyama
*ab
aDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-oiwakecho, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: oleesy@kuchem.kyoto-u.ac.jp; hs@kuchem.kyoto-u.ac.jp
bInstitute for Integrated Cell-Material Sciences (iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto 606-8501, Japan
First published on 27th November 2015
Abstract
We have synthesized a fluorescent base analogue, 2-aminothieno[3,4-d]pyrimidine based G-mimic deoxyribonucleoside, 2′-OMe-thG, and investigated its photophysical properties and DNA incorporation. The 2′ methoxy group of 2′-OMe-thG effectively induces the Z-form DNA. Finally we have constructed a visible nanothermometer based on the B–Z transition of DNA using 2′-OMe-thG.
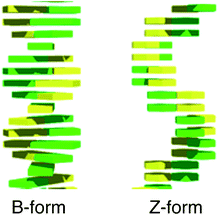
The development of nanodevices and molecular machines such as nanorobots and molecular switches has been a very active research area in the field of nanotechnology. A variety of nanomaterials such as gold nanoparticles, graphene oxide, and mesoporous silica have been investigated, and are now widely utilized to develop nanodevices. DNA is an outstanding natural building block with which to construct two- and three-dimensional nanostructures because of its unique complementarity, chemical stability, and dynamic conformational changes by external stimuli.1–5 Besides natural nucleobases (A, T, C, G), with the rapid advancements in chemical biology, multifarious functionalized nucleobase analogues have been developed, and these artificial genetic alphabet characters widen the sphere of DNA technology applications.6–10 Previously, Tor and coworkers have developed isomorphic fluorescent RNA nucleosides derived from thieno[3,4-d]-pyrimidine and demonstrated their attractive photophysical properties including visible light emission and a high quantum yield.11 Recently, we have synthesized a highly emissive deoxyguanosine analogue, thdG, as a fluorescent probe and found that thdG fluorescence could be applied to visualize B to Z transitions of DNA based on different π-stacking of B- and Z-DNA (Fig. 1).12 In this context, we have focused the nucleotide modifications to enhance the utility of thdG and synthesized a highly emissive 2′-O-methylated guanosine analogue, 2′-OMe-thG, and investigated its photophysical properties as a fluorescent base analogue.
2′-O-Methyl-modification of oligonucleotides is a well-known strategy used to increase the binding affinity of oligonucleotides for their target and to enhance the thermal stability of the resulting duplex structure. This methylation also has the advantage that the 2′-O-methyl substituent inhibits the hydrolysis of oligonucleotides in vivo.13–18 Herein, we report the development of a visible nanothermometer by a combination of highly emissive 2′-O-methylated guanosine analogue, 2′-OMe-thG, and distinctive B–Z transition of DNA.
The synthesis of 2′-OMe-thG (3) was achieved based on reported procedures for thdG nucleosides and 2′-O-methyl-modified oligonucleotides (Scheme 1).11a,12a,19 The protected 2′-O-methylated ribonucleoside (2) was obtained through Friedel–Crafts C-glycosylation between thienoguanine (1) and an acylated sugar derivative. This coupling mainly afforded a β-anomer in 83% yield. The benzoyl-protecting groups were removed in a methanolic base and the N,N-dimethylformamidine group was introduced for protection of the purine amino group (4). Subsequently, the 5′-hydroxyl group was protected with the dimethoxytrityl ether (DMTr) and the desired β-anomer (5) could be isolated in 75% yield. The configuration at the C-1 carbons of the β-anomer was confirmed by 1D and 2D (NOESY) 1H NMR experiments (see ESI†).
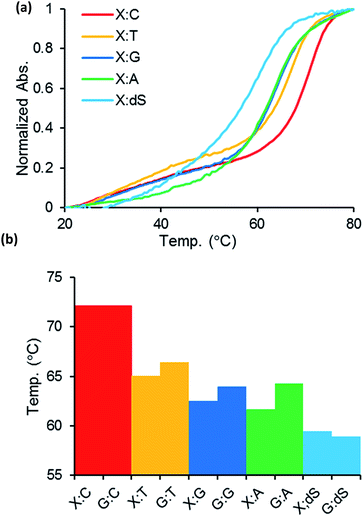
To evaluate oligonucleotides containing the 2′-O-methylated guanosine analogue, the phosphoramidite 2′-OMe-thG (6) was synthesized and incorporated into the center of 18-mer DNA oligonucleotides of 5′-d(CGTCCGTCXTACGCACGC)-3′, where X = 2′-OMe-thG, by automated solid-phase synthesis. The complementary strands of ODN1 containing matched or mismatched bases and the corresponding native DNA duplexes with G were also prepared. The 2′-OMe-thG–C base pair showed almost identical thermal stability (Tm = 72.1 °C) compared with native duplex DNA with a G–C base pair (Tm = 72.1 °C), as shown in Fig. 2. The complementary strands containing mismatched bases (ODN4–7) decreased the melting temperature compared with one obtained using the complementary stands containing dG. The thermodynamic stability and base pairing selectivity indicate that 2′-OMe-thG could replace a G base in the strand without structural disruption. The photophysical properties of 2′-OMe-thG monomer were also investigated (see ESI†). The fluorescence of 2′-OMe-thG ribonucleoside (1) shows absorption at 320 nm and visible emission at 457 nm with a high quantum yield of 0.652 under neutral conditions in water.
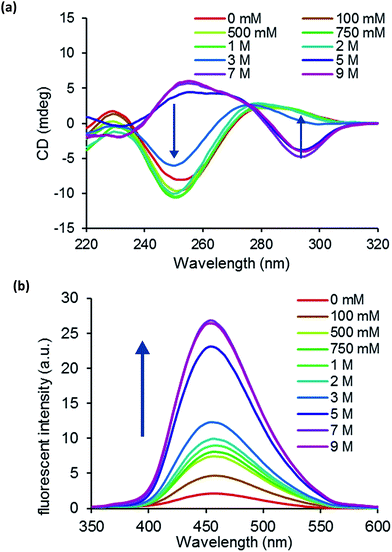
We prepared a self-complementary decamer 5′-d(CGCXCGCGCG)-3′ (ODN8), where X = 2′-OMe-thG, and examined its conformation using circular dichroism (CD) spectra. Fig. 3a shows the CD spectra of ODN8 in various concentrations of NaClO4 at 5 °C. At 1–3 M NaClO4, a negative Cotton effect at around 250 nm and a positive Cotton effect at around 285 nm were observed; ODN8 maintained the B-form. When we increased the NaClO4 concentrations from 5 to 9 M, we observed a positive Cotton effect at around 260 nm and a negative Cotton effect at around 290 nm; 2′-OMe-thG-containing decamer duplex converted to Z conformation. In a recent study, we demonstrated that the B–Z transition could be visualized by the fluorescence intensity of thdG.12 Although thdG indicated the comparable resemblance to the native dG nucleosides regarding thermodynamic stability and base pairing selectivity, the B–Z transition became more difficult when thdG was incorporated as a replacement for a dG nucleotide. Because thdG favors the anti-conformation to stabilize B-form DNA, 8-methylguanine (m8G) was additionally introduced into DNA sequences as a Z-stabilizing unit.20 Fortunately, the obtained results indicate that the oligonucleotide possessing 2′-OMe-thG could convert Z conformation without the aid of a Z-DNA inducer. Subsequently, we observed the change in fluorescent intensity by B–Z transition of ODN8. As shown in Fig. 3b, the fluorescence of ODN8 increased significantly with increasing NaClO4 concentration. This result indicated that strong fluorescence enhancement was observed in Z-DNA compared with B-DNA.
Previously, we have found that B–Z transition can be controlled by temperature and high salt conditions, and demonstrated a DNA-based switching device that responds to thermal stimuli.21 Based on previous studies and the photophysical properties of 2′-OMe-thG, we devised a visible nanothermometer. To test this concept, the 2′-OMe-thG-containing oligonucleotide 5′-d(CGCGCXCGCGCGCG)-3′ (ODN9), where X = 2′-OMe-thG, was prepared and conformational changes by temperature were investigated in 3.5 M NaClO4. At 5 °C, ODN9 predominantly converted to Z-DNA because of its lower entropy. As the temperature increased from 5 °C to 40 °C, the proportion of B-DNA gradually increased (Fig. 4a and c). We ascertained that the equilibrium between B-DNA and Z-DNA of ODN9 could be controlled by temperature. Therefore, the fluorescence of ODN9 was observed at different temperatures in 3.5 M NaClO4. To our delight, the fluorescence intensity of ODN9 changed depending on the temperature in conjunction with its B–Z transition. As shown in Fig. 3b, very strong fluorescence enhancement was observed at 5 °C, whereas the fluorescence intensity of ODN9 decreased significantly at 40 °C. The proportions of Z-DNA, B-DNA, and single-strand DNA are shown in the ESI (Fig. S6†). To examine the utility of 2′-OMe-thG-containing oligonucleotide as a nanothermometer, we monitored the fluorescence of ODN9 under a repetitive temperature cycle between low temperature (5 °C) and high temperature (40 °C). Consequently, a reproducible fluorescence of ODN9 was observed according to the change in the proportion of Z- and B-DNA (Fig. 4c). As shown in Fig. 4d, the distinguishable blue emission of the nanothermometer at 5 °C was visible to the naked eye.
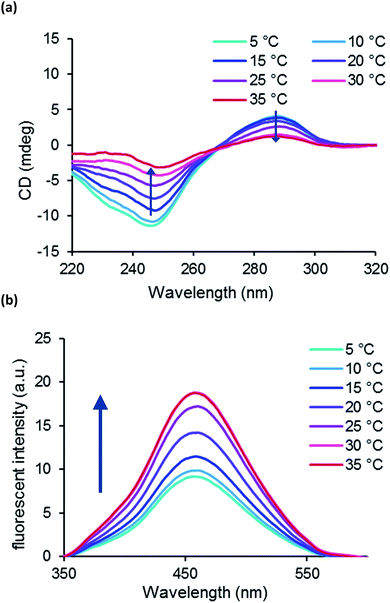
For further investigation of the devised nano-thermometer we prepared two 2′-OMe-thG-containing oligonucleotide 5′-d(CGCXCXCGCG)-3′ (ODN10), where X = 2′-OMe-thG, and evaluated it by CD spectra and fluorescent spectra measurements. Although ODN10 did not indicate dramatic B–Z transition in the range of 0 M to 5 M NaClO4, fluorescent intensity strongly increased with increasing NaClO4 concentration from 7 M to 9 M (Fig. S7†). Therefore, CD and emission spectra of ODN10 were measured in 7 M NaClO4 at various temperatures. Interestingly, ODN10 indicated that the population of Z-form increased by increasing temperature from 5 °C to 35 °C and the emission increased in line with the proportion of Z conformation (Fig. 5); the thermal response of ODN10 was in inverse to that of ODN9. It is known that Z-RNA is the more stable at higher temperature.21b,22 This result suggests that four 2′-OMe group in duplex ODN10 may induce RNA-like thermal conformational change and we can reverse the response to the same stimuli by control the incorporation of 2′-OMe-thG.
In conclusion, we synthesized a useful fluorescent guanine analogue, 2′-OMe-thG, and developed a visible nanothermometer using a combination of distinctive B–Z transition of DNA and robust brightness of 2′-OMe-thG in the visible region. Temperature is a critical factor influencing various biochemical transformations in living systems.23 A visible nanothermometer based on a nucleic acid may be a useful tool for the development of biocompatible nanodevices to monitor the temperature in an intracellular environment.24–29 Further investigations into the application of 2′-OMe-thG are ongoing in our laboratory.
Acknowledgements
We express sincere thanks for a Grant-in-Aid Priority Research from Japan Society for the Promotion of Science (JSPS) and the grant from the WPI program (iCeMS, Kyoto University).Notes and references
- F. C. Simmel and W. U. Dittmer, Small, 2005, 1, 284 CrossRef CAS.
- A. Condon, Nat. Rev. Genet., 2006, 7, 565 CrossRef CAS PubMed.
- A. V. Pinheiro, D. Han, W. M. Shih and H. Yan, Nat. Nanotechnol., 2011, 6, 763 CrossRef CAS PubMed.
- L. A. Yatsunyk, O. Mendoza and J.-L. Mergny, Acc. Chem. Res., 2014, 47, 1836 CrossRef CAS PubMed.
- Y. Krishnan and F. C. Simmel, Angew. Chem., Int. Ed., 2011, 50, 3124–3156 CrossRef CAS.
- R. W. Sinkeldam, N. J. Greco and Y. Tor, Chem. Rev., 2010, 110, 2579 CrossRef CAS PubMed.
- A. T. Krueger and E. T. Kool, Curr. Opin. Chem. Biol., 2007, 11, 588 CrossRef CAS PubMed.
- (a) A. A. Henry and F. E. Romesberg, Curr. Opin. Chem. Biol., 2003, 7, 727 CrossRef CAS PubMed; (b) A. M. Leconte and F. E. Romesberg, Nature, 2006, 444, 553 CrossRef CAS PubMed; (c) A. M. Leconte and F. E. Romesberg, Nat. Methods, 2006, 3, 667 CrossRef CAS PubMed.
- S. A. Benner, Acc. Chem. Res., 2004, 37, 784 CrossRef CAS.
- (a) I. Hirao, Biotechniques, 2006, 40, 711 CrossRef CAS PubMed; (b) I. Hirao and M. Kimoto, Proc. Jpn. Acad., Ser. B, 2012, 88, 345 CrossRef CAS; (c) I. Hirao, M. Kimoto and R. Yamashige, Acc. Chem. Res., 2012, 45, 2055 CrossRef CAS PubMed.
- (a) D. Shin, R. W. Sinkeldam and Y. Tor, J. Am. Chem. Soc., 2011, 133, 14912 CrossRef CAS PubMed; (b) R. W. Sinkeldam, L. S. McCoy, D. Shin and Y. Tor, Angew. Chem., Int. Ed., 2013, 52, 14026 CrossRef CAS PubMed; (c) M. Sholokh, R. Sharma, D. Shin, R. Das, O. A. Zaporozhets, Y. Tor and Y. Mély, J. Am. Chem. Soc., 2015, 137, 3185 CrossRef CAS PubMed.
- (a) S. Park, H. Otomo, L. Zheng and H. Sugiyama, Chem. Commun., 2014, 50, 1573 RSC; (b) H. Otomo, S. Park, S. Yamamoto and H. Sugiyama, RSC Adv., 2014, 4, 31341 RSC.
- P. Lubini, W. Zürcher and M. Egli, Chem. Biol., 1994, 1, 39 CrossRef CAS PubMed.
- L. L. Cummins, S. R. Owens, L. M. Risen, E. A. Lesnik, S. M. Freier, D. McGee, C. J. Guinosso and P. D. Cook, Nucleic Acids Res., 1995, 23, 2019 CrossRef CAS PubMed.
- J. Kurreck, Eur. J. Biochem., 2003, 270, 1628 CrossRef CAS PubMed.
- A. Grunweller, E. Wuszko, B. Bieber, R. Jahnel, V. A. Erdmann and J. Kurreck, Nucleic Acids Res., 2003, 31, 3185 CrossRef PubMed.
- T. P. Prakash, C. R. Allerson, P. Dande, T. A. Vickers, N. Sioufi, R. Jarres, B. F. Baker, E. E. Swayze, R. H. Griffey and B. Bhat, J. Med. Chem., 2005, 48, 4247 CrossRef CAS PubMed.
- H. Doessing and B. Vester, Molecules, 2011, 16, 4511 CrossRef CAS PubMed.
- (a) A. H. Haines, Tetrahedron, 1973, 29, 2807 CrossRef CAS; (b) C. Génu-Dellac, G. Gosselin and J.-L. Imbach, Tetrahedron, 1991, 216, 249 Search PubMed; (c) B. S. Ross, R. H. Springer, G. Vasquez, R. S. Andrews, P. D. Cook and O. L. Acevedo, J. Heterocycl. Chem., 1994, 31, 765 CrossRef CAS; (d) G. Parmentier, G. Schmitt, F. Dolle and B. Luu, Tetrahedron, 1994, 50, 5361 CrossRef CAS.
- (a) H. Sugiyama, K. Kawai, A. Matsunaga, K. Fujimoto, I. Saito, H. Robinson and A. H.-J. Wang, Nucleic Acids Res., 1996, 24, 1272 CrossRef CAS PubMed; (b) Y. Xu, R. Ikeda and H. Sugiyama, J. Am. Chem. Soc., 2003, 125, 13519 CrossRef CAS PubMed; (c) F. Y.-H. Chen, S. Park, H. Otomo, S. Sakashita and H. Sugiyama, Artificial DNA: PNA & XNA, 2014, 5, e28226 Search PubMed.
- (a) R. Tashiro and H. Sugiyama, Angew. Chem., Int. Ed., 2003, 42, 6018 CrossRef CAS PubMed; (b) R. Tashiro and H. Sugiyama, J. Am. Chem. Soc., 2005, 127, 2094 CrossRef CAS PubMed.
- (a) K. Hall, P. Cruz, I. Tinoco Jr, T. M. Jovin and J. H. van de Sande, Nature, 1984, 311, 584 CrossRef CAS PubMed; (b) M. K. Teng, Y. C. Liaw, G. A. van der Marel, J. H. van Boom and A. H.-J. Wang, Biochemistry, 1989, 28, 4923 CrossRef CAS PubMed; (c) P. W. Davis, R. W. Adamiak and I. Tinoco Jr, Biopolymers, 1990, 29, 109 CrossRef CAS PubMed; (d) B. A. Brown II, K. Lowenhaupt, C. M. Wilbert, E. B. Hanlon and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 13532 CrossRef PubMed.
- B. B. Lowell and B. M. Spiegelman, Nature, 2000, 404, 652 CAS.
- S. Lal, S. E. Clare and N. J. Halas, Acc. Chem. Res., 2008, 41, 1842 CrossRef CAS.
- K. M. McCabe and M. Hernandez, Pediatr. Res., 2010, 67, 469 CrossRef PubMed.
- J. Lee and N. A. Kotov, Nano Today, 2007, 2, 48 CrossRef.
- J. S. Donner, S. A. Thompson, M. P. Kreuzer, G. Baffou and R. Quidant, Nano Lett., 2012, 12, 2107 CrossRef CAS PubMed.
- F. H. Wang, D. S. Banks, A. Abu-Arish and C. Fradin, J. Am. Chem. Soc., 2007, 129, 10302 CrossRef PubMed.
- P. Leiderman, D. Huppert and N. Agmon, Biophys. J., 2006, 90, 1009 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra24756j |
| This journal is © The Royal Society of Chemistry 2015 |