Understanding the structure–activity relationship between quercetin and naringenin: in vitro
Bao Tu,
Zhi-Juan Liu,
Zhi-Feng Chen,
Yu Ouyang and
Yan-Jun Hu*
Hubei Collaborative Innovation Center for Rare Metal Chemistry, Hubei Key Laboratory of Pollutant Analysis & Reuse Technology, Department of Chemistry, Hubei Normal University, Huangshi 435002, PR China. E-mail: yjhu@263.net; Fax: +86-714-6573832; Tel: +86-714-6515602
First published on 10th December 2015
Abstract
Quercetin and naringenin are polyphenolic flavonoids obtained from plant sources. A comparative study of their structure–activity relationships (SAR) has been carried out by multiple methods. Spectroscopy experiments show that quercetin intercalates into the double helix of deoxyribonucleic acid (DNA), while naringenin displays a groove binding mode. Molecular docking studies visually display different binding modes, interaction forces and binding regions while electrochemistry experiments indicate the main factor that effects the DNA–drug interaction during the electrochemical process. Antioxidant assay was performed to find out the difference in their anti-oxidative ability. The results obtained from all these experimentations illustrate that the presence of a double bond in ring A of quercetin is very important to maintain its structural planarity, which influences its binding mode, the main interaction force and the control step in electrochemistry experiments. Quercetin showed better antioxidant properties as compared to naringenin which may be attributed to the presence of a greater number of hydroxyl groups. These findings give an understanding of their structure–activity relationships which may be helpful in the design of analogs of these flavonoids and their application in drug and food industries.
1. Introduction
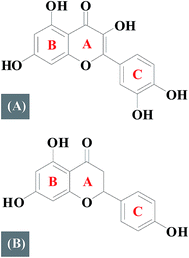
Flavonoids are polyphenolic compounds which can be classified into several subclasses according to their basic chemical structure: flavones, flavonols, flavanones, flavanonols, and flavanols.1,2 Quercetin (Fig. 1A) is a dietary flavonoid that belongs to a flavonol subfamily, possessing three phenolic rings (A, B and C) and five hydroxyl groups which has drawn increased attention due to its potential as a chemo-preventing agent which significantly reduces cancer risk and has been reported to intervene with anti-apoptotic proteins of the Bcl-2 family.3,4 Naringenin (Fig. 1B) is a major flavanone present in grapefruit which has three phenolic rings and three hydroxyl groups and is associated with antioxidative, anti-hyperlipidemic activities, and inhibits HCV production.5–7In recent years, studies about the SAR have aroused people's concern.8–10 Differences in structure may responsible for the different pharmacological properties. Therefore, investigating SAR is significant for designing and understanding numerous biological activities of flavonoids based drugs. DNA, is perhaps the most studied biopolymer to date because of its importance in controlling the heredity of life by its base sequence.11,12 Interactions about small molecules with DNA are of great interest from the perspective of medicinal or clinical research.13 A lot of effort have been made by various researchers to discover novel therapeutic agents.14–16 However, most of them has just investigated their binding modes only.
In this study, quercetin and naringenin belonging to different subfamily have been selected for a comparative study of their interaction with DNA and to investigate their binding mode, binding constants, the main binding force, the impact on the structure of DNA and the influence of different factors on these systems. Furthermore, antioxidative potential of these flavonoids was compared. This study has a colossal significance for not only in the development of new flavonoid based therapeutic agents that target DNA, but also beneficial in the development of antioxidants that can be used in drug and food industries.
2. Materials and methods
2.1. Materials
Calf thymus DNA and 2,2-diphenyl-1-picrylhydrazyl were obtained from Sigma-Aldrich (St. Louis, MO); quercetin and naringenin were obtained from Chengdu MUST Bio-Technology Co., Ltd. (Chengdu, China); total antioxidant capacity determination kit was obtained from Nanjing Jiancheng Bioengineering Institute (Nanjing, China); Na2HPO4–NaH2PO4 (PBS) had a purity of no less than 99.5% and all other chemicals used were analytical pure and used as such as purchased without further purification, except for the DNA solution, which was dissolved in buffer solution for one day in advance before use. The purity of DNA was verified by monitoring the ratio of absorbance at 260/280 nm (A260/A280). The concentration of DNA stock solution was determined according to the absorbance at 260 nm by using an extinction coefficient of 6600 mol L−1 cm−1.17 Appropriate blanks were run under the same condition and subtracted from the sample spectra.2.2. Measurement of UV-visible spectra
The UV-visible (UV-vis) absorbance spectrum was recorded at room temperature using a UV9000 spectrophotometer (Shanghai, China) equipped with 1.0 cm quartz cell. Appropriate blanks corresponding to PBS buffer were subtracted to correct the background.2.3. Measurement of fluorescence spectra
All fluorescence spectra were recorded using a LS-55 spectrofluorimeter (Perkin Elmer, USA) equipped with 1.0 cm quartz cells and a thermostat bath. The width of the excitation slit was set to 10 nm and the emission slit to 4 nm. Appropriate blanks corresponding to the buffer were subtracted to correct the fluorescence background.2.4. Molecular docking study
Molecular docking was performed by using a Surflex Dock program available in Sybyl-X 2.1.1 software. The crystal structure of DNA was obtained from the Protein Data Bank (http://www.pdb.org/, PDB ID: 1Z3F and 1VZK). The protomol was generated in ligand mode with the threshold kept at 0.50 and the bloat 0. The structures of quercetin and naringenin were drawn using the structure drawing tool available in Sybyl-X 2.1.1 package and the energy of molecules was minimized using a tripos force field and charges were computed using a Gasteiger–Huckel method. The lowest binding energy conformer was searched out of 20 kind conformers for each docking simulation and the resultant one was used for further analysis. Ring flexibility was considered and other parameters were determined through a number of attempts during docking.2.5. Measurement of circular dichroism spectra
Circular dichroism (CD) spectra were recorded on a JASCO J-810 spectropolarimeter using a rectangular quartz cuvette of path length of 1 cm. The reported CD spectra are an average of three successive scans with a scan rate of 50 nm per minute and an appropriately corrected baseline.2.6. Electrochemistry experiments
Electrochemical experiments were carried out using a CHI 660E electrochemical workstation (Chenhua Instrumental Company, Shanghai) equipped with a three-electrode system at the room temperature (298 K). DNA (1.0 × 10−4 mol L−1) was selectively immobilized on Au electrode as per the process reported in the literature.10 The DNA/Au electrode (2 mm diameter) served as a working electrode, a platinum wire as a counter electrode, and the Ag/AgCl electrode as a reference electrode. Electrochemical impedance spectroscopy was performed with a frequency range from 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 0.1 Hz, a dc potential of +0.24 V, and ac amplitude of 5 mV. 5 mL of electrolyte contains 5 mmol L−1 [Fe(CN)6]3−/4− and 1 mmol L−1 KCl (pH 7.4) and all the electrodes were immersed in electrolyte for 10 min before experimentation. Throughout the experiments, the scan rate was kept at 0.1 V s−1 while for different scan rate experiments, scan rate was changed from 0.05 to 0.5 V s−1 and other conditions remained same. The experiments were reproducible.
000 to 0.1 Hz, a dc potential of +0.24 V, and ac amplitude of 5 mV. 5 mL of electrolyte contains 5 mmol L−1 [Fe(CN)6]3−/4− and 1 mmol L−1 KCl (pH 7.4) and all the electrodes were immersed in electrolyte for 10 min before experimentation. Throughout the experiments, the scan rate was kept at 0.1 V s−1 while for different scan rate experiments, scan rate was changed from 0.05 to 0.5 V s−1 and other conditions remained same. The experiments were reproducible.
2.7. Measurement of anti-oxidative activity
Total antioxidant capacity determination experiment was carried out using a kit and the tests were in accordance with the relevant steps. For the experiment, different concentration (0.1 μg mL−1 and 0.5 μg mL−1) of quercetin, naringenin and vitamin C (standard) was combined with 3 mL of reagent solution. The absorbance of the solution was measured at 520 nm using a UV-visible spectrophotometer against blank after cooling to the room temperature. During the total reducing power determination experiment, the reaction mixture containing 10–50 μL 2 mg mL−1 quercetin, naringenin and vitamin C in phosphate buffer (0.2 M, pH 6.6), was incubated with K3Fe(CN)6 (1% w/v) at 50 °C for 30 min. The reaction was terminated by the addition of trichloroacetic acid solution (10% w/v) and the mixture was centrifuged (6000 rpm, 20 °C) for 10 min. The supernatant was mixed with distilled water and ferric chloride (0.1% w/v) solution, incubated for another 10 min until the formation of ferrous ion (Fe2+) and absorbance was measured at 700 nm. 0.08 mmol L−1 DPPH stock solution was prepared before this experiment and all samples were dissolved in ethanol (0–20 μg mL−1). Pure ethanol was used as a blank. Whole reaction was carried out at the room temperature and away from light. For each of the above assays, the tests were run in triplicate by varying the concentration of the complexes.3. Results and discussion
3.1. UV-vis spectroscopy
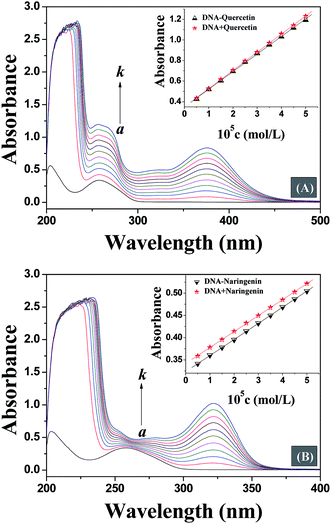
UV-vis absorption spectroscopy is a simple, widely used and one of the most effective methods in detecting the interaction of small molecules with DNA.18 In general, the interaction of small molecules with DNA and the formation of a new complex lead to changes in UV-vis spectra.19 On interaction with small molecules, any destabilisation of secondary structure of DNA leads to hyperchromism, while hypochromism originates from the stabilization of DNA secondary structure either by electrostatic effects or intercalation of small molecules.20,21 Thus, in order to provide evidence for the possibility of binding mode of quercetin and naringenin to DNA, spectroscopic titration of a solution of DNA with flavonoids has been performed. Fig. 2 shows that the maximum absorption peak of DNA at 260 nm exhibited a proportional increase with an increase in the concentration of flavonoids studied without any noticeable shift, but an non-ignorable difference was noted which shows that the increase in degree of absorption at 260 nm is almost equal to at 378 nm for quercetin, while for naringenin, a smaller increase in absorption was observed at 260 nm than at 320 nm. These results indicate that the interaction mode between DNA and small molecules is different for quercetin and naringenin. In order to remove ambiguity, the same concentration of drug was used to study the interaction further. The insets in Fig. 2 show that the absorption value of the simple sum of free DNA and free drug was slightly greater than the measured value of drug–DNA complex, which means that a weak hypochromic effect existed. However, when the UV absorption spectra of quercetin and naringenin were compared, large differences in “so-called hypochromic effect” were noted. The ordinate value for DNA–quercetin system shifted from 0.4 to 1.2, while for DNA–naringenin system it changed from 0.35 to 0.55. The change in ordinate value is very small for DNA–naringenin system as compared to DNA–quercetin system. Furthermore, the changes in the slope of the illustrations were also noted. For DNA–quercetin system, the slope was in the order of DNA + quercetin (0.18) > DNA–quercetin (0.17) which means that a weak hypochromic effect existed. As the drug concentration increased gradually, the difference between the slopes also increased leading to enhancement in hypochromic effect. From these data it can be concluded that an intercalation binding actually existed between DNA and quercetin. However in case of DNA–naringenin system, the slope was equal which suggests that the binding mode of naringenin with DNA may be groove binding. This observation was also supported by subsequent experiments.3.2. Fluorescence competitive displacement assays
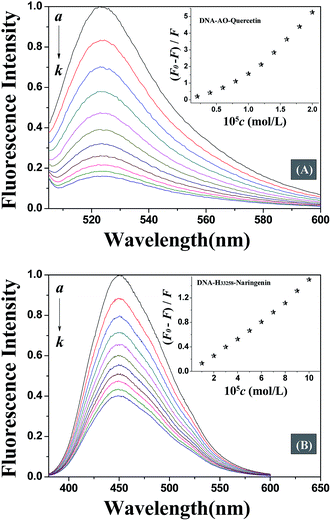
Fluorescence spectroscopy is one of the most widely exploited techniques to study the DNA–drug interaction. Due to weak endogenous fluorescence intensity of DNA and studied flavonoids, competitive displacement assay was carried out in this study to investigate DNA–flavonoid interaction. Competitive displacement assays was performed using fluorescent dye. Any small molecule that displaces the dye already bound to DNA will interact with DNA in the same mode as the displaced dye.22Acridine orange (AO) is a classic DNA intercalating dye that is extensively used as a fluorescence probe. The fluorescence of AO increases when it gets intercalated between DNA base pairs. Any molecule that binds to DNA via the same mode as AO, will displace AO from DNA helix and result in a decrease in the fluorescence intensity of DNA–AO system. Fig. 3A displays that the fluorescence intensity decreased with the addition of quercetin which means that quercetin intercalated into the double helix of DNA, displaced the location of AO bound to DNA. However, the fluorescence intensity was not affected by addition of naringenin which suggests a non-intercalation binding mode. In order to further confirm the binding mode of DNA–naringenin system, another fluorescence probe Hoechst 33258 (H33258), a typical groove type probe was used.23 Fluorescence intensity of H33258 increased tremendously while binding to DNA, however, with the addition of naringenin, the fluorescence intensity was decreased indicating that naringenin interacts with DNA by groove binding (Fig. 3B). These experiments show interesting results in terms of differences in the binding mode of flavonoids with DNA.
An investigation of structure–activity relationships of quercetin and naringenin suggests that a double bond in ring A (Fig. 1) is responsible for the different binding mode with DNA. The presence of a double bond in ring A of quercetin plays an important role in maintaining the planarity of the molecule where as in naringenin, a single bond is easy to rotate but difficult to maintain a planar structure which is a necessary requirement for intercalation into the double helix of DNA.
3.3. Quenching mechanism
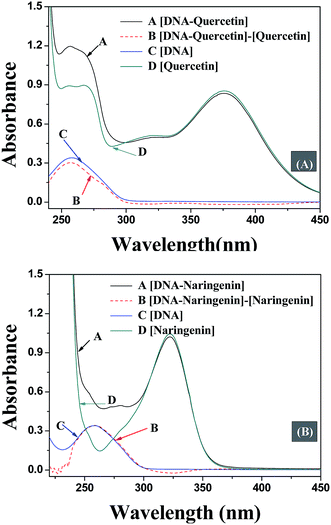
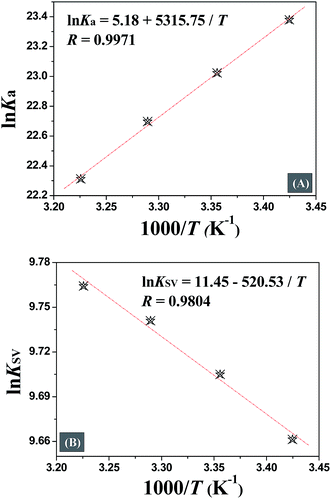
The quenching mechanism followed was either static or dynamic mode. Static quenching usually results from the formation of a complex between quencher and fluorophore in the ground state, while for dynamic quenching the fluorophore and quencher get in touch with each other during the transient existence of the excited state.19 So, the measurement of absorbance spectra is an easy way to determine the quenching mechanism. Fig. 4A and B show that when quercetin binds to DNA, the ultraviolet absorption spectrum of DNA changes (B and C), whereas for naringenin no such change was observed. These information indicating that the quenching mechanism is static for DNA–quercetin and dynamic for DNA–naringenin. Another way of easily distinguishing the quenching mechanism is by following their temperature dependence.24 For dynamic quenching, high temperature results in a quicker diffusion that leads to an increase in quenching constant. However, the stability of the complex is reduced with an increase in temperature in case of static quenching, resulting in decreased quenching constant. The decrease in fluorescence intensity is usually described by the well-known Stern–Volmer equation:
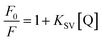 | (1) |
Table 1 shows that the quenching constants is inversely correlated with temperature for quercetin indicating that the fluorescence quenching mechanism of the DNA–quercetin is initiated by compound formation, whereas for naringenin, quenching constants increases with an increase in temperature suggesting a dynamic quenching.
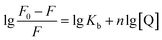 | (2) |
| T (K) | Quercetin | Naringenin | ||||||
|---|---|---|---|---|---|---|---|---|
| 10−4 KSV (L mol−1) | 10−4 Kb (L mol−1) | n | ΔH (kJ mol−1) | ΔG (kJ mol−1) | ΔS (J mol−1 K−1) | 10−4 KSV (L mol−1) | Ea (kJ mol−1) | |
| 292 | 26.59 | 14.24 | 1.60 | −12.90 | −25.49 | 43.10 | 1.57 | 4.33 |
| 298 | 17.18 | 9.97 | 1.48 | −25.75 | 1.64 | |||
| 304 | 10.95 | 7.20 | 1.35 | −26.01 | 1.70 | |||
| 310 | 6.77 | 3.10 | 1.29 | −26.27 | 1.74 | |||
Double-logarithm equation was brought in to get the binding constants Kb and n stand for the binding constant and binding number, respectively.
In Table 1, a decreasing trend of Kb with increasing temperature was in accordance with KSV's dependence on temperature as discussed earlier for quercetin.
The interaction forces between small molecules and DNA may include electrostatic interactions, multiple hydrogen bonds, van der Waals interactions, hydrophobic and steric contacts within the antibody-binding site, etc.24 When the enthalpy change (ΔH) does not vary significantly within the temperature range studied, its value and entropy change (ΔS) could be evaluated from the van't Hoff equation:
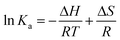 | (3) |
| ΔG = ΔH − TΔS | (4) |
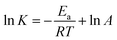 | (5) |
Eqn (4) was introduced to evaluate the free energy change for quercetin while Arrhenius equation was used for naringenin system, where A is the pre-exponential, Ea is the apparent activation energy and R is the gas constant.
Fig. 5 displays the van't Hoff and Arrhenius plots of quercetin and naringenin, respectively. Table 1 shows negative enthalpy change (ΔH) and positive entropy change when quercetin binding to DNA. The negative sign of free energy change means that the binding process for quercetin to DNA is spontaneous. Fig. 5B indicates that the apparent activation energy (Ea) of naringenin system remained almost constant at different temperatures whereas the quenching constants increased with an increase in temperature indicating that it is an intensified quenching process, which stands for the endothermic process (ΔH > 0), while, spontaneous process means the negative sign of free energy change (ΔG < 0), so the entropy change must be positive (ΔS > 0). According to Ross,25 electrostatic force is the main force for quercetin system, while hydrophobic force is responsible for naringenin's binding to DNA. This difference is mainly due to diversity in their structure, five hydroxyl groups connected at quercetin create higher electron density surrounding the molecule which is convenient for intercalating into the double helix of DNA while keeping away from the phosphoric acid skeleton, whereas the single bond in ring A may cause partly collapsing, leading three hydroxyl group outside form a hydrophobic hydrosphere. Several hydroxyl groups present in quercetin and naringenin could form intermolecular hydrogen bonds that help in strengthening the binding force in the DNA–drug complex.
3.4. Molecular docking study
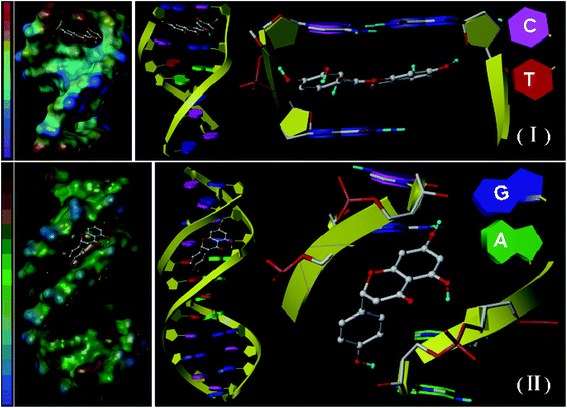
Molecular docking can offer a visual representation of the binding characteristics of small ligands to DNA, which is useful in complementing and substantiating experimental results.19,24 The docking experiment in this study was performed to obtain an insight of the preferred orientation of the quercetin and naringenin at DNA. Fig. 6 showed that quercetin intercalated into double helix of DNA and the binding domain is concentrated on C–G enrichment region. The deeper electrostatic surface color means that electrostatic force is existed and three hydrogen bonds were formed which is in consistent with the aforementioned spectrophotometry experiments. In case of naringenin, the perfect binding mode is groove binding differ from quercetin which mainly due to the larger plane angle (77.26° vs. 41.83°). The deeper hydrophobic surface color means the existence of hydrophobic force, hydrogen bonds is also existed and naringenin prefers to binding between two different types of base pairs.3.5. Effect of degeneration conditions
Chemical denaturants like guanidine HCl (GuHCl) and urea are frequently used to destabilize the double stranded DNA helix.19 Such denaturants are exploited to analyze the binding mode of quercetin and naringenin with DNA in this study. The results in Table 2 show that the addition of both denaturing agents decreases quenching constants for both the two systems which is attributed due to unwinding of double helix. It was also found that the denaturation of DNA helix by GuHCl and urea was almost equal, but their effect on quercetin system was larger as compared to naringenin system which may be due to the release of intercalated quercetin into the solution leading to an alteration in the fluorescence behavior.| System | Chemical denaturation | Thermal denaturation | Effect of pH | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blank | GuHCL | Urea | 25 | 60 | 80 | 100 | 4.2 | 6.2 | 7.2 | 8.2 | 10.2 | ||
| Quercetin | 10−5 KSV (L mol−1) | 1.76 | 1.49 | 1.39 | 1.78 | 2.12 | 2.82 | 3.69 | 0.11 | 3.44 | 42.93 | 12.24 | 1.06 |
| 10−5 Ka (L mol−1) | 1.10 | 1.00 | 0.96 | 1.26 | 1.36 | 1.67 | 1.80 | 0.13 | 1.78 | 18.45 | 6.17 | 1.23 | |
| Naringenin | 10−4 KSV (L mol−1) | 1.71 | 1.65 | 1.67 | 1.47 | 1.57 | 1.71 | 1.87 | 0.39 | 0.40 | 1.04 | 1.67 | 1.84 |
Thermal denaturation experiment is performed to elucidate the binding mode of small molecules with DNA.24 By increasing the temperature of solution, the DNA double helix is denatured and single stranded regions are generated. The results reported in Table 2 indicate that with an increase in temperature, quenching constant increases for naringenin which means that the increasing temperature is accompanied with the depolarization of double-stranded DNA leading to the creation of an conducive environment for binding with DNA at groove area. An interesting phenomenon was observed in relation to the effect of temperature on quenching constants and binding constants for quercetin which was shown in the order of 100 > 80 > 60 > 25 °C. In general, higher temperature is not conducive for the binding of a molecule into the double helix of DNA, however, the unusual phenomena seen in case of quercetin may be due to the existence of electrostatic force and hydrogen bonding in the system.
pH plays an important role in a variety of chemical and biological processes. The stability of DNA double-stranded structure is very closely related to the environmental pH.26 The maximum emission intensity of DNA–quercetin complex dropped significantly at pH 7.2. The quenching and binding constants for quercetin in strong acid and alkaline solution are much lower suggesting that the interaction is strongest at physiological conditions, and easier effected by acidic solution than alkaline solution suggests the existence of electrostatic force when quercetin binding to DNA (Table 2). In case of naringenin, the quenching constant increased with an increase in pH value which means that alkaline condition favors its binding with DNA, which could be due to the existence of hydrophobic force.
3.6. Circular dichroism experiments
The conformational changes in DNA associated with a small molecule binding can be studied efficiently by CD spectroscopy during the drug–DNA interactions.27 The CD spectra of the DNA duplex displayed a canonical B-form conformation characterized by a positive band at 273–280 nm due to base stacking and a negative band at 245 nm due to right handed helicity.28 CD spectra show insignificant change in case of minor groove binding and electrostatic binding, whereas the intercalative binding make influences both the positive and negative bands.29 As illustrated in Fig. 7A, it reveals that both bands of the CD spectrum of DNA are changed obvious with increasing concentration of quercetin, indicating that the binding of the quercetin with DNA disturb the stacking of the bases. Fig. 7B indicates that the binding of naringenin with DNA does not disturb the stacking of the bases. This observation optimistically rules out intercalation binding implying that naringenin binds to the host DNA through groove binding which is in consistent with the results obtained from the previous spectrum experiment. The main reason for this difference may be due to the absence of double bond in ring A of naringenin.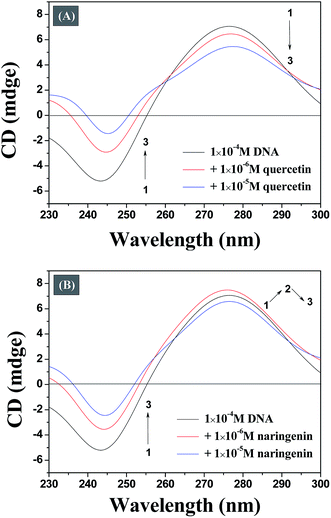 | ||
| Fig. 7 Representative CD spectrum of DNA resulting from the interaction of quercetin (A) and naringenin (B). | ||
3.7. Cyclic voltammetry experiments (CV)
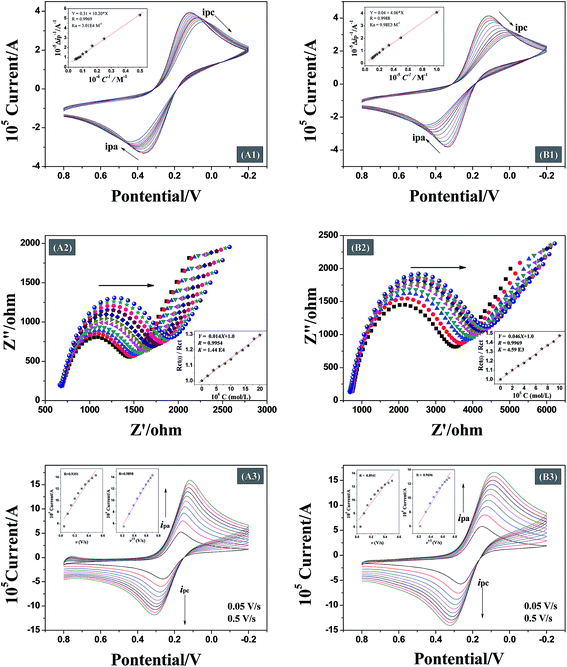
Analysis of the changes induced in the electrochemical signal by DNA–drug interaction can provide evidences for the interaction.24 With a gradual increase in both quercetin and naringenin concentration, the values of peak current ip (Fig. 8) showed an obvious decrease and was accompanied with a bigger difference in peak potential, indicating that the binding of flavonoids with DNA prevents the process of oxidation and reduction on the modified electrode surface. Meanwhile, the change in the peak current can be used to calculate the binding constant with the help of following equation (the inset in Fig. 8):
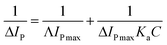 | (6) |
3.8. Electrochemical impedance spectroscopy (EIS)
Being able to detect interfacial properties at the electrode surface, EIS has become an increasingly versatile and efficient method for highly sensitive observation of antibody–antigen interactions.10 In this study, EIS was used to characterize the interfacial resistance change during the binding process of quercetin and naringenin with DNA. Prominent increases in diameter of the semicircle in the Nyquist plots were found (Fig. 8) which was in consistent with aforementioned CV results (Fig. 8). The binding equilibrium constant was calculated (the inset in Fig. 8) as per the eqn (7) mentioned below.
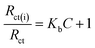 | (7) |
Both CV and EIS results are shown in Table 3. The results suggest higher affinity of quercetin towards DNA, which may comes from the different binding mode, the formed DNA–quercetin complex could increase the possibility of touch between them.
| System | CV | EIS | ESR | |
|---|---|---|---|---|
| 10−4 Ka (L mol−1) | 10−4 Kb (L mol−1) | v–i | v1/2–i | |
| DNA–quercetin | 3.01 | 1.44 | 0.9351 | 0.9898 |
| DNA–naringenin | 0.99 | 0.46 | 0.8941 | 0.9696 |
3.9. The effect of scan rate (ESR)
The binding between DNA modified electrode and drug can be controlled by a diffusion process or an adsorption process. In this study, the scan rate is a changeable factor (from 0.05 V s−1 to 0.5 V s−1) for investigating the control step in the binding process of flavonoids with DNA.30 When there is a good linear relationship between peak current and scan rate, adsorption process is the main controlling step. On the contrary, a good linear relationship between square root of the scan rate and peak current indicates that the binding is controlled by a diffusion process. Fig. 8 shows that the peak current increases with an increase in the scan rate. The insets show linearity of the scan rate (v) or square root of the scan rate (v1/2) along with peak current. The data shown in Table 3 indicate that the binding between DNA modified Au electrode and quercetin is controlled by a diffusion process, however, the two linearity for naringenin means that the binding process is controlled by both diffusion and adsorption processes. The reason for both the flavonoid systems that are controlled by diffusion process is attributed due to the presence of hydroxyl groups which are adverse to diffusion process. The presence or absence of a double bond in ring-A may be responsible for a controlled adsorption process for flavonoids. In case of naringenin, the presence of a single bond makes it rotate to another un-planar structure easily. For DNA adsorption on the surface of Au electrode, better the planarity of the molecule, easier is the adsorption.3.10. Antioxidant activities measurement
Polyphenols like phenolics, flavonoids and phenolic acids are considered to be the major natural antioxidants capable of preventing cellular damage generated by oxidative stress.31 In order to explore the relationship between structure and antioxidative potential of flavonoids, three different antioxidant assays were used viz., total antioxidant, total reducing power and DPPH radical scavenging capacity. Vitamin C (ascorbic acid), a major antioxidants found in food, is frequently used as a reference antioxidant while evaluating antioxidant properties of other compounds.32
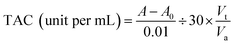 | (8) |
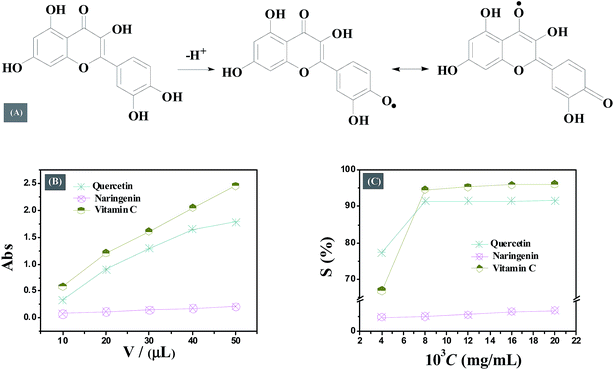
The results reported in Table 4 indicate that the total antioxidant capacity of flavonoids is much smaller than vitamin C. Quercetin shows stronger antioxidative properties than naringenin within the concentration range studied. The difference in antioxidant properties of quercetin and naringenin is accounted due to their structural diversity. Adjacent hydroxyl groups in ring C in quercetin can be transformed into a half quinone type free radical, which may form intramolecular hydrogen bond with ortho phenolic hydroxyl to gain the stability; or the half quinone type free radical form the benzoquinone structure by resonance stabilization resulting in the accumulation of higher electron density on ortho oxygen leading to decrease in inner energy and enhancing the reaction between phenol hydroxyl and oxygen free radical (Fig. 9A).
| c (μg mL−1) | TAC (unit per mL) | V (μL) | TRP (absorbance) | c (μg mL−1) | DPPH RSA (S%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | I | II | III | I | II | III | |||
| 0.1 | 9.4 | 5.8 | 23.2 | 10 | 0.33 | 0.08 | 0.58 | 4 | 77.44 | 2.30 | 66.97 |
| 0.5 | 22.4 | 11.0 | 135.5 | 20 | 0.90 | 0.12 | 1.21 | 8 | 91.47 | 2.42 | 94.63 |
| 30 | 1.29 | 0.15 | 1.61 | 12 | 91.50 | 2.72 | 95.36 | ||||
| 40 | 1.65 | 0.18 | 2.05 | 16 | 91.52 | 3.21 | 95.93 | ||||
| 50 | 1.79 | 0.21 | 2.46 | 20 | 91.59 | 3.38 | 96.11 | ||||
| S(%) = [1 − (Ai − Aj)/A0] × 100% | (9) |
Similar to the results of TRP assay, the DPPH radical scavenging activity of both quercetin and naringenin was found to be dose dependent (Fig. 9C and Table 4). Quercetin displayed high scavenging activity of 77.44% at a concentration of 4 μg mL−1 which is higher than the activity of reference antioxidant vitamin C (66.97%) whereas, with a gradual increase in drug concentration, its DPPH radical scavenging activity was found to be little less than vitamin C. This result suggests that the antioxidant properties of quercetin are closely related to concentration. On the other hand, in case of naringenin, the change in antioxidant activity was not noticeable, and scavenging activity is quite small. The number of hydroxyl groups present in the molecule may be responsible for the difference in radical scavenging activity of quercetin and naringenin, the more hydroxyl means higher electron transfer/hydrogen donating ability.
4. Conclusions
This study demonstrated that the antioxidant activities of quercetin is almost equivalent to vitamin C, and is much higher than naringenin. The high antioxidative properties of quercetin were believed due to the presence of more hydroxyl groups, at the same time, the adjacent hydroxyl groups in ring-C which may form a resonance stabilized quinone structure may be another reason. The spectroscopy experiments show that the different binding mode of quercetin and naringenin to DNA may mainly comes from the existence of the double bond in ring-A. Molecular docking was performed to obtain detailed information on the binding of flavonoids with DNA. The planarity of molecular structure is detrimental for stronger binding between flavonoids and DNA modified electrode. These study generated significant information about the structure–activity relationships which will be helpful not only for further screening and designing of potential flavonoids based drugs which target DNA but also their application in food industries.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21273065), the Research Foundation of Education Bureau of Hubei Province, China (No. B2015133), the National Undergraduate Training Program for Innovation and Entrepreneurship (No. 201510513004), and the Science and Technology Foundation for Creative Research Group of HBNU (No. T201501).References
- Z. Markovic, D. Amic, D. Milenkovic, J. M. Dimitric-Markovic and S. Markovic, Phys. Chem. Chem. Phys., 2013, 15, 7370–7378 RSC.
- A. Primikyri, G. Mazzone, C. Lekka, A. G. Tzakos, N. Russo and I. P. Gerothanassis, J. Phys. Chem. B, 2015, 119, 83–95 CrossRef CAS PubMed.
- J. Garcia-Ubasart, T. Vidal, A. L. Torres and O. J. Rojas, Biomacromolecules, 2013, 14, 1637–1644 CrossRef CAS PubMed.
- N. Scheuble, T. Geue, E. J. Windhab and P. Fischer, Biomacromolecules, 2014, 15, 3139–3145 CrossRef CAS.
- B. Tu, Y. Wang, R. Mi, Y. Ouyang and Y. J. Hu, Spectrochim. Acta, Part A, 2015, 149, 536–543 CrossRef CAS PubMed.
- A. Primikyri, M. V. Chatziathanasiadou, E. Karali, E. Kostaras, M. D. Mantzaris, E. Hatzimichael, J. S. Shin, S. W. Chi, E. Briasoulis, E. Kolettas, I. P. Gerothanassis and A. G. Tzakos, ACS Chem. Biol., 2014, 9, 2737–2741 CrossRef CAS PubMed.
- V. Ambrogi, L. Panzella, P. Persico, P. Cerruti, P. A. Lonz, C. Carfagna, L. Verotta, E. Caneva, A. Napolitano and M. Dlschia, Biomacromolecules, 2014, 15, 302–310 CrossRef CAS PubMed.
- K. B. Pandey and S. I. Rizvi, Int. J. Food Prop., 2012, 15, 702–708 CrossRef CAS.
- P. Perucca, M. Savio, O. Cazzalini, R. Mocchi, C. Maccario, S. Sommatis, D. Ferraro, R. Pizzala, L. Pretali, E. Fasani, A. Albini and L. A. Stivala, J. Photochem. Photobiol., B, 2014, 140, 57–68 CrossRef CAS.
- K. B. Pandey and S. I. Rizvi, Phytother. Res., 2010, 24, S11–S14 CrossRef PubMed.
- S. Nandi, P. Mukherjee, A. Kundu and A. K. Nandi, J. Phys. Chem. B, 2014, 118, 2649–2661 CrossRef CAS.
- S. Ghosha, P. Kundua, B. K. Paulb and N. Chattopadhyay, RSC Adv., 2014, 4, 63549–63558 RSC.
- H. D. Chen, G. Y. Chen, F. Du, Q. Q. Fu, Y. Zhao and Z. Tang, RSC Adv., 2013, 3, 16251–16254 RSC.
- B. Tu, Z. F. Chen, Z. J. Liu, R. R. Li, Y. Ouyang and Y. J. Hu, RSC Adv., 2015, 5, 73290–73300 RSC.
- A. A. Masum, M. Chakraborty, P. Pandya, U. C. Halder, M. M. Islam and S. Mukhopadhyay, J. Phys. Chem. B, 2014, 118, 13151–13161 CrossRef CAS PubMed.
- E. Sundaravadivel, S. Vedavalli, M. Kandaswamy, B. Vargheseb and P. Madankumar, RSC Adv., 2014, 4, 40763–40775 RSC.
- C. V. Kumar and E. H. Asuncion, J. Am. Chem. Soc., 1993, 115, 8547–8553 CrossRef CAS.
- L. P. Peng, S. Nagarajan, S. Rasheed and C. H. Zhou, Med. Chem. Commun., 2015, 6, 222–229 RSC.
- B. Tu, Z. F. Chen, Z. J. Liu, L. Y. Cheng and Y. J. Hu, RSC Adv., 2015, 5, 33058–33066 RSC.
- M. Rahban, A. Divsalar, A. A. Saboury and A. Golestani, J. Phys. Chem. C, 2010, 114, 5798–5803 CAS.
- S. U. Rehman, T. Sarwar, M. A. Husain, H. M. Ishqi and M. Tabish, Arch. Biochem. Biophys., 2015, 576, 49–60 CrossRef PubMed.
- R. K. Gupta, G. Sharma, R. Pandey, A. Kumar, B. Koch, P. Z. Li, Q. Xu and D. S. Pandey, Inorg. Chem., 2013, 52, 13984–13996 CrossRef CAS.
- R. Mi, B. Tu, X. T. Bai, J. Chen, Y. Ouyang and Y. J. Hu, Luminescence, 2015, 30, 1344–1351 CrossRef CAS PubMed.
- R. Mi, X. T. Bai, B. Tu and Y. J. Hu, RSC Adv., 2015, 5, 47367–47376 RSC.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS.
- X. L. Li, Y. J. Hu, H. Wang, B. Q. Yu and H. L. Yue, Biomacromolecules, 2012, 13, 873–880 CrossRef CAS PubMed.
- S. S. Mati, S. S. Roy, S. Chall, S. Bhattacharya and S. C. Bhattacharya, J. Phys. Chem. B, 2013, 117, 14655–14665 CrossRef CAS.
- A. Manna and S. Chakravorti, J. Phys. Chem. B, 2012, 116, 5226–5233 CrossRef CAS PubMed.
- B. Jana, S. Senapati, D. Ghosh, D. Bose and N. Chattopadhyay, J. Phys. Chem. B, 2012, 116, 639–645 CrossRef CAS PubMed.
- E. Huang, F. M. Zhou and L. Deng, Langmuir, 2000, 16, 3272–3280 CrossRef.
- G. Fabre, I. Bayach, K. Berka, M. Paloncyova, M. Starok, C. Rossi, J. L. Duroux, M. Otyepka and P. Trouillas, Chem. Commun., 2015, 51, 7713–7716 RSC.
- K. Saravanan and S. M. Aradhya, Food Funct., 2011, 2, 603–610 CAS.
- M. S. Blois, Nature, 1958, 181, 1199–1200 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2015 |