Spirochromone-chalcone conjugates as antitubercular agents: synthesis, bio evaluation and molecular modeling studies†
Abstract
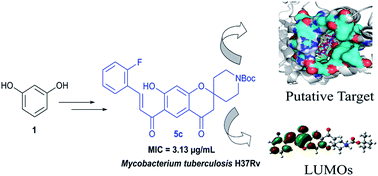
A new series of spirochromone annulated chalcone conjugates were synthesized and evaluated for their antitubercular activity against Mycobacterium tuberculosis H37Rv strain. These compounds were subjected to molecular modeling studies using docking and chemoinformatics based approaches. The docking simulations were performed against a range of known receptors for chalcone derived compounds to reveal MTB phosphotyrosine phosphatase B [MtbPtpB] protein as the most probable target based on the high binding affinity scores. Five compounds exhibit significant inhibition, showing minimum inhibitory concentration values i.e. MIC values ranging from 3.13–12.5 μg mL−1. Further analysis of the synthesized compounds with known and in-house developed chemoinformatics tools unequivocally established their potential as anti-tubercular compounds. QSAR modeling revealed a quantitative relationship between biological activities and frontier molecular orbital energies of synthesized compounds. The predictive model can be employed further for virtual screening of new compounds in this series.

 Please wait while we load your content...
Please wait while we load your content...