Chemistry of heterocyclic-2-thiones: in situ generated 1-(4,5-dihydro-3-alkyl-imidazolidin-2-yl)-3-alkyl-imidazolidine-2-thione and triiodide form novel mixed valent CuI–II/CuII complexes†
Abstract
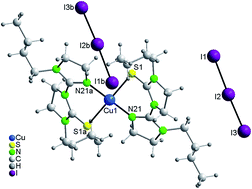
Equimolar reaction of copper(I) iodide with N-methyl-imidazolidine-2-thione (imdzSH-Me) in acetonitrile under aerobic conditions involved C–S rupture and formation of C–N bond between two heterocyclic rings giving rise to the formation of 1-(4,5-dihydro-3-methyl-imidazolidin-2-yl)-3-methyl-imidazolidine-2-thione (L-NMe) which has yielded black prismatic crystals of mixed valent trinuclear complex, [CuI2CuII(κ2-N,S-L-NMe)(κ1N,μ-S-L-NMe)(μ-I)2(κ1-I)2] 1 along with formation of CuSO4·5H2O. A similar reaction of N-n-propyl-imidazolidine-2-thione (imdzSH-Prn) with copper(I) iodide has formed white {Cu(μ-S-imdzSH-Prn)2Cu(μ-I)2}n 2 and brown [Cu3(κ1N,μ-S-L-NPrn)2(μ-I)2(κ1-I3)2]n 3 polymers. The N-n-propyl-imidazolidine-2-thione ligand is unchanged in 2, but it changed into 1-(4,5-dihydro-3-propyl-imidazolidin-2-yl)-3-propyl-imidazolidine-2-thione (L-NPrn) in polymer 3. Finally, in reaction of copper(I) iodide with N-n-butyl-imidazolidine-2-thione (imdzSH-Bun), the thio-ligand changed into 1-(4,5-dihydro-3-butyl-imidazolidin-2-yl)-3-butyl-imidazolidine-2-thione (L-NBun) which coordinated to the metal centre and formed black compound [Cu(κ2-N,S-L-NBun)2](I3)2 4. The formation of in situ generated new ligands, L-NR (R = Me, Prn and Bun) coordinated in 1, 3 and 4 occurs in the same manner involving C–S rupture, fusion of two N-alkyl-imidazolidine-2-thione rings and formation of new C–N bonds. Compounds 1–4 have been characterized with the help of analytical data, IR spectroscopy, X-ray crystallography and ESI-mass studies.

 Please wait while we load your content...
Please wait while we load your content...