Role of pentose phosphate pathway in lipid accumulation of oleaginous fungus Mucor circinelloides†
Lina Zhaoa,
Xin Tanga,
Xiao Luana,
Haiqin Chenab,
Yong Q. Chenab,
Wei Chenab,
Yuanda Song*a and
Colin Ratledgec
aState Key Laboratory of Food Science and Technology, School of Food Science and Technology, Jiangnan University, 1800 Lihu Avenue, Wuxi, Jiangsu 214122, P. R. China. E-mail: ysong@jiangnan.edu.cn; Fax: +86 510 85197130; Tel: +86 510 85197130
bSynergistic Innovation Center for Food Safety and Nutrition, Wuxi, P. R. China
cDepartment of Biological Sciences, University of Hull, Hull, UK
First published on 2nd November 2015
Abstract
Lipid biosynthesis in oleaginous fungi requires acetyl-CoA, as the essential precursor of fatty acids, and a supply of reducing power, NADPH. Malic enzyme provides the majority of NADPH needed for lipid biosynthesis in most oleaginous fungi, while the reactions of the pentose phosphate pathway (PPP) provide additional NADPH. We have therefore overexpressed the genes coding for glucose 6-phosphate dehydrogenase (g6pd) and 6-phosphogluconate dehydrogenase (6pgd) from the PPP in the oleaginous fungus Mucor circinelloides to analyze its effect on lipid accumulation. The results showed that overexpressing g6pd or 6pgd increased the lipid content of cell dry weight by 20–30% compared to the control strain, and with higher G6PD and 6PGD activities and higher mRNA levels in the overexpressing strains. The results suggest that G6PD and 6PGD are important NADPH providers and play a key role on lipid accumulation in oleaginous fungus M. circinelloides.
1. Introduction
Microbial lipids have gained interest because some contain high amounts of polyunsaturated fatty acids (PUFAs) such as arachidonic acid (AA), eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA) and γ-linolenic acid (GLA) that have nutritional benefits. Additionally, microbial oils are being considered as potential sources of biofuels. Oleaginous microorganisms, such as yeasts, Yarrowia lipolytica1 and Rhodosporidium toruloides,2 bacteria Rhodococcus opacus,3 microalgae, Aurantiochytrium limacinum4 and Schizochytrium sp.,5 and filamentous fungi, Mortierella alpina6 and Mucor circinelloides7 can accumulate more than 20% (w/w) lipid of their cell dry weight (CDW), some can accumulate more than 80% lipid but some of them will only accumulate lipid as low as 20% (w/w, CDW). Although the mechanism of lipid accumulation is broadly similar in most of these oleaginous microorganisms,8 the reasons why the final lipid contents of the cells varies so greatly is still not clear. Evidently, however, the maximum lipid content of a cell must be under genetic control otherwise all oleaginous microorganisms would accumulate the same levels of lipid in their cells.Lipid biosynthesis in oleaginous eukaryotic microorganisms is a cytosolic event: the two key events are the provision of acetyl-CoA, as the essential precursor of fatty acids, and the provision of NADPH.9 NADPH is vital as the source of reducing power for fatty acid synthesis and is generated in many, but not all, oleaginous microorganisms by NADP+-dependent malic enzyme (ME), that converts malate to pyruvate. It has been suggested to be the rate-limiting step for fatty acid biosynthesis in oleaginous fungi.10,11 However, a recent report has demonstrated that it is not the only bottleneck for lipid accumulation in filamentous fungi.12 While, the contribution of NADPH-generating pathways to lipid accumulation in oleaginous microorganisms has been theoretically analyzed by Ratledge, who demonstrated that ME plays an important role in the majority of species, but cannot provide all the NADPH that is required for lipid biosynthesis; the reactions of pentose phosphate pathway (PPP), glucose 6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD) appear to be the most likely route to provide the rest NADPH for lipid biosynthesis.9 Chen reported that G6PD is the major contributor to the NADPH pool during lipid accumulation under nitrogen starvation in M. alpina.6 Our previous work demonstrated that M. circinelloides WJ11 can produce lipid up to 36% of CDW, which is about twice that of M. circinelloides CBS 277.49 that has no more than 15% of CDW as lipid. The activities of G6PD and 6PGD in strain WJ11 were greater than in CBS 277.49.7 13C-metabolic flux analysis of lipid accumulation in the oleaginous fungus M. circinelloides has been performed in our laboratory, and showed that the high oleaginous strain WJ11 had a higher flux of carbon through the PPP compared with strain CBS 277.49, suggesting that the PPP plays an important role to provide NADPH for lipid biosynthesis.13
Therefore, it is necessary to investigative the role of the PPP during lipid biosynthesis in oleaginous fungi. The filamentous fungus, M. circinelloides, is of interest because it produces high levels of polyunsaturated fatty acid, GLA. Indeed, its oil was the first commercially produced single cell oil.14 M. circinelloides CBS 277.49 has emerged as one of the model of oleaginous filamentous fungi because of the availability of its genome sequence (http://genome.jgi.doe.gov/Mucci2/Mucci2.home.html) as well as well-developed tools for its genetic engineering.15 Therefore, in this study, we used this strain as a model organism to study the effect of the genes, g6pd and 6pgd, of the PPP on lipid biosynthesis.
2. Materials and methods
2.1 Strains, growth and transformation conditions
Mucor circinelloides f. lusitanicus CBS 277.49 was used as the source of the gene g6pd and 6pgd. The uridine auxotroph, pleu-MU402, was used as recipient strain in transformation experiments to overexpress g6pd and 6pgd. MU402 (ref. 16) is auxotrophic strain for leucine and uridine, plasmid pLEU4 that contains the wild-type allele of leuA gene,17 transformed into MU402 generated to strain pleu-MU402. Cultures were grown at 30 °C in YPG18 or MMC media.16 Media were supplemented with uridine (200 μg mL−1) when required. The pH was adjusted to 4.5 for mycelial and colonial growth. Transformation was carried out as described previously.15Strains Mc-G6PD (g6pd-overexpression), Mc-6PGD (6pgd-overexpression) and Mc-1552 (as the control, pMAT1552 was transformed into strain pleu-MU402) were initiated by inoculation of 2.5 × 105 spores per mL into 200 mL modified K & R medium19 containing 80 g glucose per L and held in 1 L flasks equipped with baffles to improve aeration. The cultures were incubated for 72 h at 30 °C with shaking at 150 rpm. Escherichia coli strain Top 10 was used for all cloning experiments.
2.2 Plasmids construction
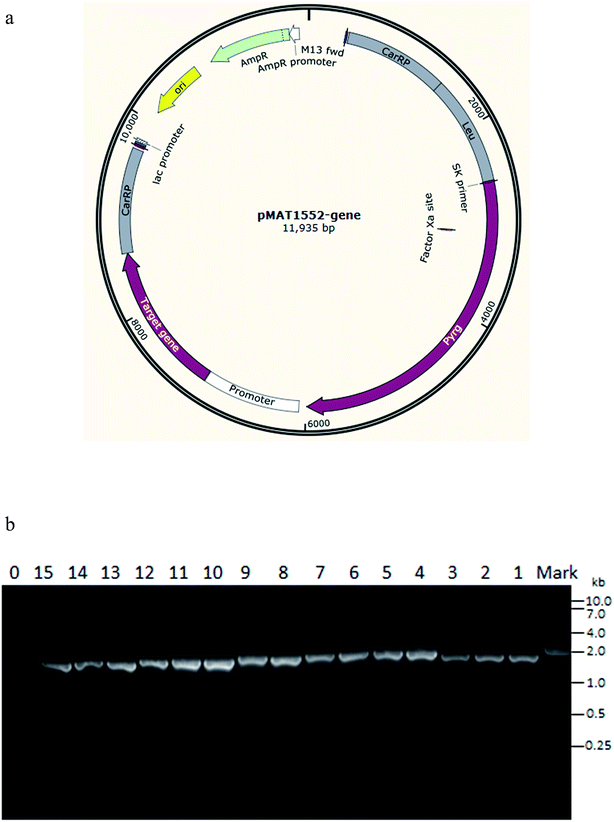
Plasmid, pMAT1552, that contains the M. circinelloides pyrG gene surrounded up- and down-stream by 1 kb of CarRP sequences, was used for construction of g6pd-overexpressing and 6pgd-overexpressing plasmids. The g6pd and 6pgd genes were isolated by PCR amplification from the genome of M. circinelloides CBS 277.49 with corresponding primers g6pd1-F/R, g6pd2-F/R, g6pd3-F/R, 6pgd1-F/R, 6pgd2-F/R (Table S1†). These contain 30 bp homologous sequences of both sides of XhoI restriction sites in pMAT1552. The PCR fragment was then cloned into plasmid pMAT1552 using XhoI to generate plasmids pMAT1552-g6pd1, pMAT1552-g6pd2, pMAT1552-g6pd3, pMAT1552-6pgd1 and pMAT1552-6pgd2 (Fig. 1a) (One step cloning kit, Takara).2.3 Genome DNA preparation and RT-qPCR analysis
For DNA extraction, M. circinelloides was grown for 3 days in K & R medium at 30 °C with shaking at 150 rpm. The mycelium was harvested by filtration through a Buchner funnel under reduced pressure and then washed three times with distilled water. Genomic DNA was extracted using the DNA Quick Plant System kit (Tiangen Biotech Co., Ltd).For reverse transcription-quantitative PCR (RT-qPCR) analysis, Mc-G6PD, Mc-6PGD and Mc-1552 strains were grown in 1 L baffled flasks with modified K & R medium, and the mycelium harvested at 24 h, 48 h and 72 h. Total RNA of M. circinelloides was isolated with Trizol after grinding under liquid N2 and reverse-transcribed using the Prime ScriptRT Reagent kit (Takara) according to the manufacturer's instructions. RT-qPCR was performed using corresponding primers g6pd1-qF/qR and 6pgd2-qF/qR (Table S1†) on CFX Connect Real-Time System (Bio-Rad) with iTaq™ Universal SYBR Green PCR Supermix (Bio-Rad) according to the manufacturer's instructions. The mRNA expression levels of g6pd and 6pgd were normalized to levels of 18S rRNA mRNA and the results were expressed as relative expression levels. The date were quantified by the method of 2−ΔΔCt.
2.4 Determination of cell dry weight and lipid content
The mycelium were harvested and washed as before, then frozen at −80 °C. The frozen biomass was lyophilized and the CDW was determined gravimetrically. Lipids were extracted from approx. 20 mg lyophilized biomass with chloroform/methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).20 Pentadecanoic acid (15
1, v/v).20 Pentadecanoic acid (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) was added as an internal standard. Fatty acid methyl esters were formed using 10% (w/w) HCl/methanol and analyzed by GC with a DB-Waxetr column (30 m by 0.32 mm; film thickness, 0.25 μm; Shimadzu Co., Ltd, Japan). The program was as follows: 120 °C for 3 min, increased to 190 °C at 5 °C per min, then raised to 220 °C at 4 °C per min, and held for 20 min. N2 was the carrier gas at a constant flow of 3 mL per min.
0) was added as an internal standard. Fatty acid methyl esters were formed using 10% (w/w) HCl/methanol and analyzed by GC with a DB-Waxetr column (30 m by 0.32 mm; film thickness, 0.25 μm; Shimadzu Co., Ltd, Japan). The program was as follows: 120 °C for 3 min, increased to 190 °C at 5 °C per min, then raised to 220 °C at 4 °C per min, and held for 20 min. N2 was the carrier gas at a constant flow of 3 mL per min.
2.5 Determination of enzyme activity
The activities of G6PD and 6PGD were determined as described previously.7 The mycelium was harvested and washed three times with distilled water, then frozen and ground under liquid N2 and then resuspended in 100 mM KH2PO4/KOH, pH 7.5 extraction buffer containing 20% (w/v) glycerol, 1 mM benzamidine·HCl and 1 mM DTT. The suspensions were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min at 4 °C and then the supernatants were used for enzyme analysis. Protein concentrations were determined according to Bradford's method using bovine serum albumin as standard. Enzyme activity was measured by continuous spectrophotometric assays at 340 nm, the units of enzyme activity was defined as the amount of enzyme required to produce 1 mol NADPH per min.
000 × g for 10 min at 4 °C and then the supernatants were used for enzyme analysis. Protein concentrations were determined according to Bradford's method using bovine serum albumin as standard. Enzyme activity was measured by continuous spectrophotometric assays at 340 nm, the units of enzyme activity was defined as the amount of enzyme required to produce 1 mol NADPH per min.
2.6 Statistical analysis
A statistical analysis of the data was carried out using SPSS 16.0 for Windows (SPSS Inc., Chicago, IL). The mean values and the standard error of the mean were calculated from the data obtained from two independent experiments. The differences between the means of the test were evaluated by Student's t-test and P < 0.05 was considered as significantly different.3. Results
3.1 Generation of g6pd-overexpressing and 6pgd-overexpressing strains of M. circinelloides by genetic engineering
Based on the genomic data of M. circinelloides CBS 277.49, we found three genes encoding for G6PD and two genes encoding for 6PGD. The genes ID145983 (1476 bp), ID189960 (1539 bp) and ID186010 (1530 bp) probably code for G6PD and, therefore, they were named as g6pd1, g6pd2, and g6pd3. The genes ID90680 (1455 bp) and ID107657 (1452) probably code for 6PGD and were named as 6pgd1 and 6pgd2, respectively.To overexpress the target gene, the plasmid pMAT1552-gene that contains the target gene coding region under the control of the strong zrt1 promoter of M. circinelloides12 was generated (Fig. 1a). In addition to the target gene, the plasmid contains the pyrG gene (coding for uridine) as a selectable marker and flanking sequences corresponding to regions surrounding the carotenogenic CarRP gene to allow its chromosomal integration by homologous recombination. The target gene-overexpressing plasmids, pMAT1552-g6pd1, pMAT1552-g6pd2, pMAT1552-g6pd3, pMAT1552-6pgd1 and pMAT1552-6pgd2, plus the empty plasmid pMAT1552 were transformed into the uridine auxotrophic strain, pleu-MU402, and selection of the colonies were carried out as described by Rodríguez-Frómeta.12 For each overexpression strain, three transformants were selected, named Mc-G6PD1-1, Mc-G6PD1-2, Mc-G6PD1-3; Mc-G6PD2-1, Mc-G6PD2-2, Mc-G6PD2-3; Mc-G6PD3-1, Mc-G6PD3-2, Mc-G6PD3-3; Mc-6PGD1-1, Mc-6PGD1-2, Mc-6PGD1-3; Mc-6PGD2-1, Mc-6PGD2-2, and Mc-6PGD2-3 plus the control transformant that was named Mc-1552.
Integration of the target genes into the genome of overexpression transformants, were confirmed by PCR analysis. Amplification was carried out using a primer pair (1552-F/R) that amplified the target gene and 100 bp sequences of the plasmid pMAT1552. The PCR product fragments for each transformants were 1576 bp, 1639 bp, 1630 bp, 1555 bp, and 1552 bp, as expected, in these corresponding transformants genome, respectively, whereas 100 bp fragment was amplified in the control strain Mc-1552 (Fig. 1b). PCR amplification results confirmed that the target genes had been integrated into the genome of the fungus in these transformants.
3.2 Lipid accumulation in g6pd-overexpressing and 6pgd-overexpressing strains
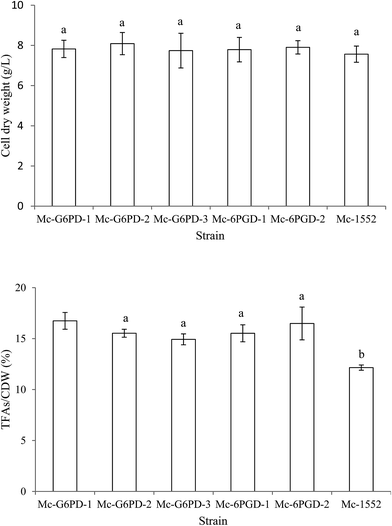
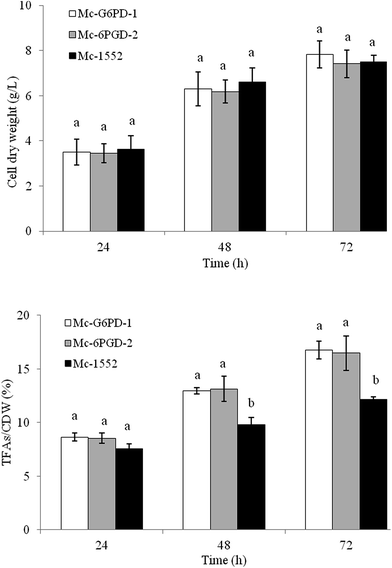
The transformants were grown in complete medium for 72 h in 1 L baffled flasks with 200 mL modified K & R medium. As our preliminary data showed that the lipid content of three transformants of each strain that was overexpressing the target gene were not significantly different (date not given), only one transformant for each mutant strain was selected for biomass and lipid accumulation analysis (renamed as Mc-G6PD-1, Mc-G6PD-2 and Mc-G6PD-3, Mc-6PGD-1 and Mc-6PGD-2). Cell dry weights and total fatty acid contents of CDW of these mutant strains are shown in Fig. 2. About 7–8 g CDW per L was achieved in these transformants; values that were not significantly different from the control strain Mc-1552 (7.6 g L−1); however, the lipid content of CDW in Mc-G6PD-1, Mc-G6PD-2, Mc-G6PD-3, Mc-6PGD-1 and Mc-6PGD-2 were 16.7%, 15.5%, 14.9%, 15.2%, 16.5% after 72 h of cultivation, respectively, which were significantly higher than the control strain Mc-1552 at 12.2% of the CDW. The results suggest that overexpressing of either g6pd or 6pgd, leads to increased lipid accumulation by 20–30% in M. circinelloides.Because the lipid content of strains Mc-G6PD-1, Mc-G6PD-2, Mc-G6PD-3, Mc-6PGD-1 and Mc-6PGD-2 were not significantly different from each other, only Mc-G6PD-1 and Mc-6PGD-2 were selected for further analysis. The cell dry weight and lipid content of CDW in strain Mc-G6PD-1, Mc-6PGD-2 and control strain Mc-1552 cultured in 1 L baffled flasks with 200 mL K & R medium at 24 h, 48 h and 72 h was analyzed. As shown in Fig. 3, the lipid contents of strains Mc-G6PD-1 and Mc-6PGD-2 were significantly higher than the control strain Mc-1552 at 48 h and 72 h of cultivation, although the CDW of these three strains was not significantly different. Conversely to the lipid content, fatty acid profile analysis showed that whether overexpressing g6pd or 6pgd had no significant effect on fatty acid composition of the strains (Table S2†).
3.3 Activities of G6PD in g6pd-overexpressing strain and 6PGD in 6pgd-overexpressing strain
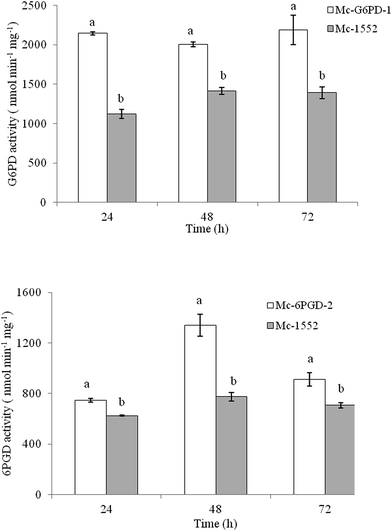
To investigate whether the enzymes coded by the integrated genes in the overexpressing strains are active or not, the activities of G6PD and 6PGD in cultures of Mc-G6PD-1 and Mc-6PGD-2 grown in 1 L baffled flasks with 200 mL modified K & R medium at 24 h, 48 h and 72 h was analyzed. As shown in Fig. 4, the activities of G6PD and 6PGD were significantly higher in Mc-G6PD-1 and Mc-6PGD-2 than the control strain. The activities of G6PD in Mc-G6PD-1 were approx. 2000 nmol min−1 mg−1 at 24 h, 48 h and 72 h, which is much higher than the control strain (about 1300 nmol min−1 mg−1) during the whole fermentation process. The activity of 6PGD in Mc-6PGD-2 was also significantly greater than the control strain, and the highest activity of 1339 nmol min−1 mg−1 was reached at 48 h. These results suggested that overexpressing of g6pd and 6pgd enhance the activities of G6PD and 6PGD in the fungus, and would provide more NADPH for lipid biosynthesis in strains Mc-G6PD-1 and Mc-6PGD-2.3.4 Expression levels of the g6pd gene in g6pd-overexpressing and the 6pgd gene in 6pgd-overexpressing strains
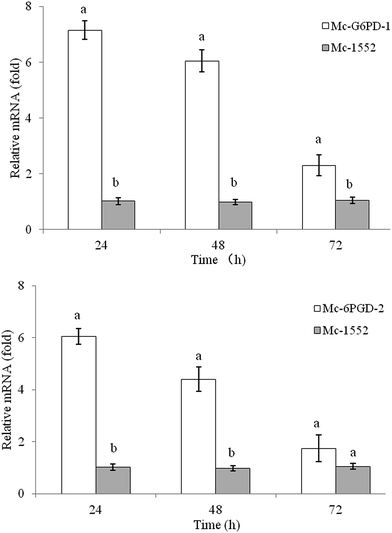
The mRNA levels of g6pd-1 and 6pgd-2 in the overexpressing strains Mc-G6PD-1 and Mc-6PGD-2 were analyzed and compared to those of the control strain Mc-1552 by RT-qPCR. As shown in Fig. 5, the levels of g6pd-1 and 6pgd-2 mRNA were notably increased in the overexpressing strains compared to the control strain, although they decreased with the incubation time. For example, the 6pgd-2 mRNA level was increased by approx. 6-fold at 24 h, 4.4-fold at 48 h, and 1.7-fold at 72 h compared to the control strain Mc-1552. The fact that the target gene mRNA was maintained at elevated levels throughout the whole culture time confirmed that the target gene is being continuously overexpressed in the transformants.4. Discussion
Lipid biosynthesis requires acetyl-CoA and the provision of NADPH. Acetyl-CoA, as the essential precursor of fatty acids, is generated in the cytosol of the cell via the cleavage of citric acid, which is initially generated in the mitochondrion, and then translocated into the cytosol. The enzyme involved is ATP: citrate lyase (ACL), which has been implicated as the key provider of acetyl-CoA, and plays an important role in lipid biosynthesis.7,8,21,22 The second critical process for fatty acid biosynthesis is the provision of reducing power in the form of NADPH. The supply of NADPH as generated by NADP+-dependent ME, that simultaneously convert malate to pyruvate, has been postulated to be the rate-limiting step for fatty acid biosynthesis in oleaginous fungi.10,11 Overexpression of ME genes from M. alpina and M. circinelloides in the strain of M. circinelloides R7B led to a 2.5-fold increase in lipid accumulation compared to the strain of R7B.23 However, Rodríguez-Frómeta discovered that leucine auxotrophy lead to a 2.5-fold increase for lipid accumulation in M. circinelloides, and the lipid content of that expression of leu gene in the strain of R7B had no difference compared to the strain of overexpression of the ME gene, the result demonstrated that ME may not be the only bottleneck for lipid accumulation in this fungus.12A recent review by Ratledge calculated the sources of NADPH through stoichiometric analysis and, suggested that ME plays an important role in the majority of species, but cannot provide all the NADPH that is required for lipid biosynthesis. Supplementary sources of NAPDH are therefore essential. The reactions of PPP would appear to be the most likely route although there is a possibility that some NADPH might be produced by a cytosolic isocitrate dehydrogenase reaction.9 The enzymes G6PD (EC: 1.1.1.49) and 6PGD (EC: 1.1.1.44) from PPP are involved in the conversion of glucose-6-phosphate into ribulose 5-phosphate, a precursor of important molecules such as nucleic acids, and are generally considered a major source of NADPH. G6PD and 6PGD are generally considered to catalyze the following reactions with generating NADPH:
| Glucose-6-phosphate + NADP+ → 6-phospho-D-glucono-1,5-lactone + NADPH |
| 6-Phosphogluconate + NADP+ → ribulose 5-phosphate + CO2 + NADPH |
Increased flux of carbon through the PPP, and enhanced lipid accumulation in the yeast Y. lipolytica,24 the microalga Chlorella protothecoides,25 the filamentous fungus M. circinelloides13 and the plant material of maize embryos26 have been reported. These results imply that G6PD and 6PGD of the PPP could be the key supplementary sources of NADPH.
In this study, overexpressing of the genes coding for G6PD and 6PGD, increased the activities of G6PD and 6PGD, and enhanced lipid content of CDW by 20–30% in the fungus. These results indicated that G6PD and 6PGD of PPP play a vital role in lipid accumulation by providing more NADPH for lipid biosynthesis. This is in accordance with our previous findings,7 that established the activities of G6PD and 6PGD were significantly higher in a high lipid-producing strain, WJ11, than in a low lipid-producing stain, CBS 277.49, of M. circinelloides. Furthermore, metabolic flux analysis also suggested that the PPP plays an important role in providing NADPH for lipid biosynthesis in M. circinelloides.13 Hong have shown that cloning of G6PD and 6PGD into an industrially-used strain of Y. lipolytica, increased the lipid content of the yeast by 10–14%.27
In conclusion, the g6pd and 6pgd gene were overexpressed in oleaginous fungus M. circinelloides. G6PD and 6PGD activities were increased markly in strains overexpressing these genes while the levels of mRNA for g6pd and 6pgd were also increased significantly. The higher G6PD and 6PGD activities, and higher mRNA levels, resulted in the lipid content of the cells being improved by 20–30% in g6pd-overexpressing and 6pgd-overexpressing strains compared with the control strain. These results suggested that G6PD and 6PGD from the PPP is the important NADPH provider and play a vital role in lipid accumulation in oleaginous fungus M. circinelloides.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.Acknowledgements
This research was supported by the National Natural Science Foundation of China (31271812, 81071685, 21276108), the National High Technology Research and Development Program of China (2012AA022105C), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1249), the Program for New Century Excellent Talents (NCET-13-0831), and the Strategic Merieux Research Grant.References
- M. Rakicka, Z. Lazar, T. Dulermo, P. Fickers and J. M. Nicaud, Biotechnol. Biofuels, 2015, 8, 104 CrossRef PubMed.
- Z. Zhu, S. Zhang, H. Liu, H. Shen, X. Lin, F. Yang, Y. J. Zhou, G. Jin, M. Ye, H. Zou and Z. K. Zhao, Nat. Commun., 2012, 3, 1112 CrossRef PubMed.
- Z. Wei, G. Zeng, F. Huang, M. Kosa, Q. Sun, X. Meng, D. Huang and A. J. Ragauskas, Appl. Microbiol. Biotechnol., 2015, 99, 7369–7377 CrossRef CAS PubMed.
- X. Li, Y. Lin, M. Chang, Q. Jin and X. Wang, Bioresour. Technol., 2015, 181, 275–282 CrossRef CAS PubMed.
- X. Song, X. Zang and X. Zhang, J. Oleo Sci., 2015, 64, 197–204 CrossRef CAS PubMed.
- H. Chen, G. Hao, L. Wang, H. Wang, Z. Gu, L. Liu, H. Zhang, W. Chen and Y. Q. Chen, Sci. Rep., 2015, 5, 11247 CrossRef CAS PubMed.
- X. Tang, H. Chen, Y. Q. Chen, W. Chen, V. Garre, Y. Song and C. Ratledge, PLoS One, 2015, 10, 1–12 Search PubMed.
- C. Ratledge and J. P. Wynn, Adv. Appl. Microbiol., 2002, 51, 1–51 CrossRef CAS PubMed.
- C. Ratledge, Biotechnol. Lett., 2014, 36, 1557–1568 CrossRef CAS PubMed.
- J. P. Wynn and C. Ratledge, Microbiology, 1997, 143, 253–257 CrossRef CAS.
- J. P. Wynn, H. A. bin Abdul and C. Ratledge, Microbiology, 1999, 145, 1911–1917 CrossRef CAS PubMed.
- R. A. Rodríguez-Frómeta, A. Gutiérrez, S. Torres-Martínez and V. Garre, Appl. Microbiol. Biotechnol., 2013, 97, 3063–3072 CrossRef PubMed.
- L. Zhao, H. Zhang, L. Wang, H. Chen, Y. Q. Chen, W. Chen and Y. Song, Bioresour. Technol., 2015, 197, 23–29 CrossRef CAS PubMed.
- C. Ratledge, Biochimie, 2004, 86, 807–815 CrossRef CAS PubMed.
- S. Torres-Martínez, R. M. Ruiz-Vázquez, V. Garre, S. López-García, E. Navarro and A. Vila, Methods Mol. Biol., 2012, 898, 85–107 Search PubMed.
- F. E. Nicolás, J. P. de Haro, S. Torres-Martínez and R. M. Ruiz-Vázquez, Fungal Genet. Biol., 2007, 44, 504–516 CrossRef PubMed.
- M. I. G. Roncero, L. P. Jepsen, P. Strøman and R. van Heeswijck, Gene, 1989, 84, 335–343 CrossRef CAS PubMed.
- S. Bartnicki-García and W. J. Nickerson, J. Bacteriol., 1962, 84, 841–858 Search PubMed.
- A. Kendrick and C. Ratledge, Eur. J. Biochem., 1992, 209, 667–673 CrossRef CAS PubMed.
- J. Folch, M. Lees and G. H. S. Stanley, J. Biol. Chem., 1957, 226, 497–509 CAS.
- H. Zhang, L. Zhang, H. Chen, Y. Q. Chen, W. Chen, Y. Song and C. Ratledge, J. Biotechnol., 2014, 192, 78–84 CrossRef CAS PubMed.
- X. Y. Liu, Z. Chi, G. L. Liu, C. Madzak and Z. M. Chi, Mar. Biotechnol., 2013, 15, 26–36 CrossRef CAS PubMed.
- Y. Zhang, I. P. Adams and C. Ratledge, Microbiology, 2007, 153, 2013–2025 CrossRef CAS PubMed.
- T. M. Wasylenko, W. S. Ahn and G. Stephanopoulos, Metab. Eng., 2015, 30, 27–39 CrossRef CAS PubMed.
- W. Xiong, L. Liu, C. Wu, C. Yang and Q. Wu, Plant Physiol., 2010, 154, 1001–1011 CrossRef CAS PubMed.
- A. P. Alonso, V. L. Dale and Y. Shachar-Hill, Metab. Eng., 2010, 12, 488–497 CrossRef PubMed.
- S. P. Hong, Z. Xue and Q. Q. Zhu, US Pat., 2011/0244512, 2011.
Footnote |
| † Electronic supplementary information (ESI) available: The primers used in this study and the fatty acid composition in the overexpressing strains. See DOI: 10.1039/c5ra20364c |
| This journal is © The Royal Society of Chemistry 2015 |