Leaf-inspired artificial microvascular networks (LIAMN) for three-dimensional cell culture†
Abstract
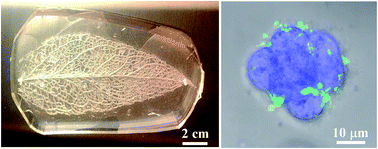
Construction of vascular architecture capable of distributing oxygen and nutrients is critically important for 3D cell culture. Here, we demonstrate leaf-inspired artificial microvascular networks (LIAMN) that serve as an effective perfusion system for 3D cell culture in hydrogel constructs. Because cell-containing hydrogel constructs with desired thickness can be constructed using embedded LIAMN layers, our strategy paves a new avenue to engineering effective vascular transport system for 3D cell culture and the development of functional artificial organs.

 Please wait while we load your content...
Please wait while we load your content...