Enzymatic approaches to the preparation of chiral epichlorohydrin
Huo-Xi Jin
* and
Xiao-Kun OuYang
School of Food Science and Pharmaceutics, Zhejiang Ocean University, Zhoushan 316022, P. R. China. E-mail: jinhuoxi@163.com; xkouyang@zjou.edu.cn; Tel: +86-18768082687 Tel: +86-15957094799
First published on 23rd October 2015
Abstract
Enantiomerically pure epichlorohydrin is a key chiral synthon in the preparation of 4-chloro-3-hydroxybutyrate, pheromones, L-carnitine, and β-adrenergic blockers. Various methods are known for obtaining the enantiomerically pure epoxides, including chemical and enzymatic approaches, but a clear understanding of the synthesis process in case of chiral epichlorohydrin is unavailable. This review gives an overview of the enzymatic approaches for preparation of the chiral epichlorohydrin, highlighting the synthetic routes using haloalcohol dehalogenase and epoxide hydrolase as biocatalysts.
Introduction
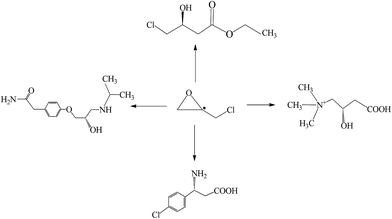
Enantiomerically pure epoxides are versatile building blocks and important chiral synthons in synthetic chiral chemistry due to their high reactivity and directable regioselectivity.1,2 As one of several promising epoxides, enantiopure epichlorohydrin has been widely used to prepare many biologically active compounds, including (S)-4-chloro-3-hydroxybutyrate,3 β-adrenergic blockers,4 baclofen,5 and L-carnitine6 (Fig. 1). Various methods including chemical and biologic are known for obtaining the chiral epichlorohydrin. They can be divided in three general preparation strategies: asymmetric synthesis, use of the chiral precursor, and kinetic resolution of racemate.The first strategy is the production of chiral epichlorohydrin, starting from a prochiral compound. For example, 3-chloropropene and dichloropropanol could be converted into chiral epichlorohydrin by external asymmetric induction using the chiral catalyst-peroxidase and haloalcohol dehalogenase, respectively. This method is the commercially most attractive due to the high theoretical yield with 100% of the enantiopure epichlorohydrin.
A chiral precursor for synthesis of the chiral epichlorohydrin is the enantiopure 2,3-dichloro-1-propanol, which can be converted into chiral epichlorohydrin by chemical or biological methods. However, the industrial application prospect of this method is not optimistic since the enantiopure 2,3-dichloro-1-propanol is expensive.
The third strategy is the kinetic resolution of racemic epichlorohydrin, which is based on the differences in reaction rate of the enantiomers. In the reaction mixture, one enantiomer of epichlorohydrin is remained in enantiomerically pure form when the other enantiomer has been entirely converted by chemo-catalyst or biocatalyst.7–9 As two of the biocatalysts used in preparation of chiral epichlorohydrin, the haloalcohol dehalogenase and epoxide hydrolase have great potential for development due to their rich resources, as well as efficient catalytic specificity and environmental friendly. However, a drawback to this method is that the maximum yield is only 50% of the total amount of the racemate.
Although a number of papers have been published on the preparation of chiral epichlorohydrin, review that specifically focuses on the enzymatic synthesis of chiral epichlorohydrin have not been reported so far. In this paper, we focus on introducing the synthesis of chiral epichlorohydrin by biotransformation reactions, including by direct epoxidation of alkenes using peroxidase, by enantioselective ring closure of dichloropropanol using haloalcohol dehalogenases, by enantioselective ring opening of racemic epichlorohydrin using haloalcohol dehalogenases, and by enantioselective kinetic resolution of racemic epichlorohydrin using epoxide hydrolases.
Direct epoxidation of alkenes by peroxidase
Chloroperoxidase, a versatile heme-peroxidase, is able to catalyze a variety of different reactions due to its unique active site structure, such as halogenation, epoxidation, peroxidation, sulfoxidation, and hydroxylation.10,11 More importantly, this enzyme has the broad substrate adaptability, and shows enantioselectivity for epoxidation of alkenes and hydroxylation of alkynes.12,13 A number of epoxides produced from alkenes by chloroperoxidase catalysis can be used as chiral synthons for preparation of chiral drug. Hager et al. had investigated the substrate specificity of chloroperoxidase from Caldariomyces fumago for preparation of chiral epoxides.14 The results indicated that chloroperoxidase showed the high activity for alkenes with chain lengths of less than ten carbon atoms, and methallyl alkenes and styrenes can function as good substrates.Chiral epichlorohydrin would be produced by direct epoxidation of 3-chloropropene using chloroperoxidase (Fig. 2). This method can obtain the 100% theoretical yield of chiral epichlorohydrin, but the low enantioselectivity, as well as inactivation of this enzyme at high concentration of H2O2, had greatly limited its development and application.15 Wu et al. used t-butyl hydroperoxide as O2 donor in order to eliminate the inhibition, the (R)-epichlorohydrin with 97.1% enantiomeric excess (e.e.) and 88.8% yield was obtained by asymmetric epoxidation of 3-chloropropene using chloroperoxidase from Caldariomyces fumago in homogenous phosphate buffer/ionic liquid mixtures.16 In this reaction system, the imidazole ionic liquids as co-solvent remarkably increased the yield of (R)-epichlorohydrin.
Enantioselective ring-closure of dichloropropanol by haloalcohol dehalogenase
Haloalcohol dehalogenases catalyse the conversion of halohydrins into their corresponding epoxides by intramolecular nucleophilic displacement of a halogen, as well as the reverse reaction.17–21 Halohydrin dehalogenase, halohydrin epoxidase or hydrogen-halide lyase are alternative names for the haloalcohol dehalogenase.22 Haloalcohol dehalogenase can be found in several organisms, such as Flavobacterium sp.,23 Agrobacterium radiobacter,24 Arthrobacter sp.,25,26 Pseudomonas sp.,27 Corynebacterium sp.,28 Alcaligenes sp.,29 Agrobacterium tumefaciens,30 Agromyces mediolanus,31 and so on. Most haloalcohol dehalogenases genes were cloned and sequenced.24,30–32 They were divided in three general types: HheA, HheB, and HheC due to the sequence homology. The haloalcohol dehalogenases in the same type are very close to each other with an 88.7–98.3% homology; while it was only 18.9–33.5% between different groups.22 Recently, the structures and mechanism of HheA from Arthrobacter strain AD2 and HheC from Agrobacterium radiobacter AD1 had been reported,33–36 but there is still no report on the structure information of HheB. These three haloalcohol dehalogenases have great difference in substrate specificity. HheA and HheB have the higher catalytic activity for long-chain halohydrin, while HheC has the higher catalytic activity for short-chain halohydrin, and high enantioselectivity for different aromatic or aliphatic compounds.The enantioselectivity of ring-closure reactions of halohydrin catalysed by haloalcohol dehalogenases makes them promising biocatalysts for the preparation of chiral epoxides. Both 1,3-dichloro-2-propanol and 2,3-dichloro-1-propanol are the direct precursors for production of chiral epichlorohydrin by haloalcohol dehalogenases (Fig. 3). However, most haloalcohol dehalogenases which can catalyse the ring closure of 1,3-dichloro-2-propanol display low activity or no activity for 2,3-dichloro-1-propanol, only a few haloalcohol dehalogenases exhibit an excellent activity for 2,3-dichloro-1-propanol (Table 1).
 | ||
| Fig. 3 Synthesis of chiral epichlorohydrin from dichloropropanol catalyzed by haloalcohol dehalogenases. | ||
| Microbial strains | Relative activitiesa (%) |
|---|---|
| a The activity of each haloalcohol dehalogenase for 1,3-dichloro-2-propanol was defined as 100%, and the activity of each haloalcohol dehalogenase for 2,3-dichloro-1-propanol is relative to that for 1,3-dichloro-2-propanol. | |
| Arthrobacter sp. AD2 (ref. 17) | 0 |
| Arthrobacter erithii H10a (ref. 37) | 0 |
| Agromyces mediolanus31 | 0 |
| Corynebacterium sp. N-1074 (ref. 38 and 39) | 0.089 |
| Pseudomonas sp. OS-K-29 (ref. 27) | 9.8 |
| Arthrobacter sp. PY1 (ref. 26) | 10 |
| Agrobacterium radiobacter AD1(ref. 25) | 25 |
| Agrobacterium tumefaciens (ref. 30) | 28.3 |
| Agrobacterium sp. NHG3 (ref. 40) | 34 |
| Alcaligenes sp. DS-K-S38 (ref. 29) | 47 |
The different haloalcohol dehalogenases often exhibit significant differences in enantioselectivity for the ring closure of 1,3-dichloro-2-propanol. The haloalcohol dehalogenases HheA from Arthrobacter strain AD2 and Corynebacterium sp. revealed no enantioselectivity, while haloalcohol dehalogenases from Agromyces mediolanus and Agrobacterium radiobacter AD1 displayed low enantioselectivity.31,41 The haloalcohol dehalogenases HheB from Corynebacterium sp. yielded (S)-epichlorohydrin with 90% e.e. in the initial stage of the reaction.42 However, the enantiomerical purity of the formed epichlorohydrin continuously decreased during the reaction. This phenomenon was also observed with the haloalcohol dehalogenases from Arthrobacter erithii H10a and Agrobacterium radiobacter AD1.37,43 The prochiral 1,3-dichloro-2-propanol was initially converted to (R)-epichlorohydrin with 89% e.e. by the haloalcohol dehalogenases from Arthrobacter erithii H10a, but it decreased upon prolonged incubation.37 Jin et al. obtained (S)-epichlorohydrin with 60% e.e. in the beginning of the reaction by haloalcohol dehalogenase HheC from Agrobacterium radiobacter AD1, but the e.e. decreased to almost zero after 20 min.43 The above phenomenon could be explained by the enzyme-catalysed racemisation of epichlorohydrin via the reverse reaction. In the presence of Cl−, the preferentially formed epichlorohydrin was also preferentially converted into 1,3-dichloro-2-propanol by haloalcohol dehalogenases, resulting in decline of the enantiomerical purity. The racemization depends on the reaction equilibrium, which is related to the type of halogen substituent. The equilibrium tends to form the halohydrin for the chloro-substituted alcohols, and follows the order: Cl− > Br− > I−.44 Therefore, a way to obtain the chiral epichlorohydrin by ring closure of dichloropropanol using haloalcohol dehalogenase is timely removal of Cl− in the reaction mixture, which will be a magnet for new investigation.
The enantiomerical purity of the formed epichlorohydrin was low by ring closure of 1,3-dichloro-2-propanol or 2,3-dichloro-1-propanol using haloalcohol dehalogenase due to the reverse reaction. Promisingly, some haloalcohol dehalogenases show a good enantioselectivity for kinetic resolution of racemic 2,3-dichloro-1-propanol, remaining the single enantiomer with high enantiomerical purity.29,45,46 The chiral epichlorohydrin is prepared from the remaining 2,3-dichloro-1-propanol with treatment of aqueous NaOH (Fig. 4). However, the kinetic resolution of the racemic 2,3-dichloro-1-propanol using haloalcohol dehalogenase was inhibited by the formed epichlorohydrin. Therefore, it is necessary to remove the formed epichlorohydrin instantly. A lack of accumulation of the epichlorohydrin would improve significantly the e.e. and yield of the remaining 2,3-dichloro-1-propanol.
 | ||
| Fig. 4 Synthesis of chiral epichlorohydrin by preparation of chiral 2,3-dichloro-1-propanol using haloalcohol dehalogenase. | ||
Kasai and co-workers obtained (R)- and (S)-2,3-dichloro-1-propanol with 100% e.e. from the racemate by resting cells of Alcaligenes sp. and Pseudomonas sp., respectively, both of which contained haloalcohol dehalogenase and epoxide hydrolase. However, the e.e. of (S)- or (R)-epichlorohydrin did not reach 100% by treating the (R)- or (S)-2,3-dichloro-1-propanol with aqueous NaOH. This results was probably attributed to isomerization of chiral epichlorohydrin caused by Payne rearrangement under alkaline conditions.29,45 The e.e. of (S)-2,3-dichloro-1-propanol was 96% by kinetic resolution of its racemate with haloalcohol dehalogenase from A. radiobacter AD1, while it was increased to >99% with addition of the excess epoxide hydrolase.46 The formed epichlorohydrin was immediately hydrolysed into 3-chloro-1,2-propanediol by epoxide hydrolase, resulting in high e.e. of the remaining 2,3-dichloro-1-propanol by removing the inhibition of epichlorohydrin.
Although the high enantiomerical purity of epichlorohydrin could be obtained by preparation of chiral 2,3-dichloro-1-propanol with haloalcohol dehalogenase, it is difficult for industrial applications due to the two drawbacks as follows: (1) the price of chiral 2,3-dichloro-1-propanol is higher than chiral epichlorohydrin; (2) most haloalcohol dehalogenases have no or low activities for 2,3-dichloro-1-propanol as described in Table 1.
Enantioselective ring opening of epichlorohydrin by haloalcohol dehalogenases
Haloalcohol dehalogenase is known to be a versatile biocatalyst based on the fact that it catalyse the enantioselective ring closure of vicinal halo-alcohols to epoxides, as well as the reverse reaction in the presence of nucleophiles. The enantioselective ring-opening of epoxides catalysed by haloalcohol dehalogenases have been widely used for synthesis of enantiomerically pure β-substituted alcohols and epoxides.47,48 Haloalcohol dehalogenase can accept many kind of nucleophiles, not only the halogen ions, but also some small negatively charged ions like N3−, NO2−, CN−, SCN− and OCN− in the ring opening reaction.49 It was reported that haloalcohol dehalogenase HheC from A. radiobacter AD1 is the most selective among the three different haloalcohol dehalogenases (HheA, HheB, and HheC) by describing the substrate specificity and enantioselectivity.48 The activity, enantioselectivity, and enantiopreference were associated with the enzyme, nucleophiles, and the substrate structure. For example, the (R)-epichlorohydrin was preferentially converted by HheA from Arthrobacter erithii H10a in the presence of Cl−, while (S)-epichlorohydrin in the presence of Br−.37 In the presence of N3−, NO2−, or CN−, the HheC showed high enantioselectivity for the ring opening of epoxides with high β-regioselectivity. NO2− is the most interesting and complex one among these nucleophiles because both oxygen and nitrogen atoms can attack the carbon atoms of epoxides, yielding two different products (Fig. 5).50 The formed nitrite ester is chemically unstable, especially at low pH, and spontaneously hydrolyses to the diol. In this way, haloalcohol dehalogenases function as an epoxide hydrolases for the ring opening of epoxides in the presence of NO2−.A promising route of preparing the chiral epichlorohydrin is enantioselective ring opening of its racemate using haloalcohol dehalogenases in nucleophiles-mediated (Fig. 6). The HheC revealed the higher enantioselectivity than HheA and HheB for the epichlorohydrin.51 Spelberg et al. indicated that pH of the reaction system had significant influences on the reaction rate and enantioselectivity in the ring opening of epichlorohydrin by HheC and N3−, both of which decreased sharply as pH increase from 5.5 to 8.5. However, the reaction rate of the ring closure of 1,3-dichloro-2-propanol catalysed by HheC increased as pH increase within this pH range.52 Therefore, (R)-epichlorohydrin with e.e. >99% was obtained from its racemate by HheC and N3− at pH 4.5. This was attributed to no racemisation because of very low rate of ring-closure at this pH. In addition, the (R)-epichlorohydrin with 99% e.e. and 41% yield had been successfully prepared using NO2− as the nucleophile at the low pH (pH 5.0) in our studies.43
 | ||
| Fig. 6 Synthesis of chiral epichlorohydrin by enantioselective ring opening of its racemate using haloalcohol dehalogenases. | ||
As described above, it was known that the HheC had a low enantioselectivity in the ring closure of 1,3-dichloro-2-propanol and a high enantioselectivity in the ring opening of epichlorohydrin using the N3− or NO2− as the nucleophile. Furthermore, the optimal reaction pH of ring closure and ring opening showed a huge difference. Consequently, it was possible to obtain the chiral epichlorohydrin from 1,3-dichloro-2-propanol using HheC by adjusting the pH. In our studies, the chiral epichlorohydrin was formed by addition of excess NO2− and adjustment of pH from 8.0 to 5.0 after the ring closure of 1,3-dichloro-2-propanol with HheC.43 In addition, Assis et al. had reported another method without adjustment of the reaction pH for preparation of the chiral epichlorohydrin from 1,3-dichloro-2-propanol. Consequently, the (R)-epichlorohydrin with >95% e.e. was obtained by addition of excess Br− using HheA from Arthrobacter erithii H10a.37 This was attributed to the fact that the epichlorohydrin formed preferentially in the ring closure of 1,3-dichloro-2-propanol was (R)-isomer, while (S)-isomer was preferentially converted in the ring opening of epichlorohydrin with Br− as nucleophile.
Enantioselective hydrolysis of epichlorohydrin by epoxide hydrolases
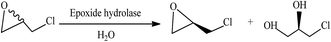
Epoxide hydrolases, which catalyze the hydrolysis of epoxides to yield the corresponding diols, have been widely used in preparation of the chiral epoxides and vicinal diols.53–55 Epoxide hydrolase activity has been found in animals, plants, and microorganisms.56–59 The epoxide hydrolases from microorganisms prompted an increased interest in biocatalytic applications due to the excellent enantioselectivity and those be easily obtained in large amounts.60,61The epoxide hydrolase shows great difference in activity and enantioselectivity based on the structure of epoxides. A correct combination of epoxide hydrolases and substrates resulted in various substituted chiral epoxides and diols. A number of epoxide hydrolases display the high enantioselectivity for kinetic resolution of disubstituted or polysubstituted epoxides because of the steric effect.62,63 It was showed that the epoxide hydrolases from bacteria have almost absolute enantioselectivity for the epoxides with disubstituent on the chiral centre, namely priority hydrolysis of (S)-enantiomer. The level of enantioselectivity was related to the type of the two substituents.64 The benzyl carbon atom of aromatic epoxides is conducive to be attacked by nucleophilic groups, resulting in that the epoxide hydrolases with enantioselective hydrolysis of this range of epoxides are relatively common in microorganism.65,66 A very extensive study showed that the chiral recognition for the mono-substituted epoxides by some epoxide hydrolases was difficult. This was caused by the regional freedom of nucleophilic attack due to the teeny steric hindrance of this range of epoxides.
Hydrolytic kinetic resolution is an effective way for synthesis of chiral epichlorohydrin (Fig. 7). However, epichlorohydrin is a kind of the mono-substituted and small molecule epoxides, most epoxide hydrolases revealed the low enantioselectivity for it. As early as 1991, Weijers et al. had reported that the strain Nocardia H8 by enantioselective degradation gave (R)-epichlorohydrin in high enantiomerical purity (>98% e.e.) from racemic mixtures, but the yield was only 19%.67 Choi and his partner successfully obtained (S)-epichlorohydrin from its racemate using an Aspergillus niger with epoxide hydrolase activity, the yield was <5% in the aqueous system but 20% in the organic system by reducing the spontaneous chemical hydrolysis of epichlorohydrin.68,69
More enantioselective epoxide hydrolases from microorganisms were screened and purified in the 21st century, and the genes were also cloned and expressed.70–72 Kim et al. performed the hydrolysis of 50 mM (R,S)-epichlorohydrin using an recombinant epoxide hydrolase from the Rhodotorula glutinis, yielding enantiopure (R)-epichlorohydrin with 26% yield.73 Lee also prepared the chiral epichlorohydrin using a recombinant epoxide hydrolase, and finally (R)-epichlorohydrin with 99% e.e and 28.5% yield at 20 mM of the racemate was obtained.74 The epoxide hydrolase from Novosphingobium aromaticivorans can enantioselectively hydrolyze the racemic epichlorohydrin at 500 mM substrate concentration, but producing (S)-epichlorohydrin in a yield of only 11.9%.75 It was indicated that these processes were not suitable for industrial production because of the low substrate concentration or yield. With the rapid development of genetic engineering and bioinformatics, it is possible to obtain novel epoxide hydrolase with the higher yield of chiral epichlorohydrin by directed evolution and sequential analysis. Mutant S4 of epoxide hydrolase from A. radiobacter with 20-fold higher enantioselectivity for epichlorohydrin was obtained by error-prone PCR and DNA shuffling.76 The yield of (R)-epichlorohydrin with >99% e.e. was over 40% by kinetic resolution of 25.6 mM racemate using this mutant.77
The group of professor Zheng from Zhejiang University of Technology performed a very extensive research for preparation of chiral epichlorohydrin using epoxide hydrolases.77–81 The genes of epoxide hydrolases from A. radiobacter, Agromyces mediolanus, Rhodococcus sp., and Rhodosporidium toruloides were cloned and expressed in Escherichia coli.53,77,78,80 Table 2 shows the characteristics of epoxide hydrolases from different microorganisms towards epichlorohydrin.77,78,80 The results indicated that both the enantiopure (R)- and (S)-epichlorohydrin were obtained from (R,S)-epichlorohydrin using the corresponding epoxide hydrolases. The epoxide hydrolase from A. radiobacter exhibited the excellent property for the high yield and reaction rate. The (S)-epichlorohydrin was preferentially hydrolyzed due to the lower Km, but the (R)-epichlorohydrin would be hydrolyzed with a much faster compared to (S)-epichlorohydrin because of the higher Vm of (R)-epichlorohydrin when the (S)-epichlorohydrin was completely converted.77 In addition, the writer also performed the enantioselective hydrolysis of racemic epichlorohydrin using whole cells of Aspergillus niger ZJB-09173 in cyclohexane. The results showed that the water content had significant influence on the e.e. and yield of (S)-epichlorohydrin. The substrate inhibition, rather than product inhibition was observed in this process. The substrate concentration was markedly increased by continuous feeding of substrate for reducing the substrate inhibition.81 In another research, both substrate and product inhibition were observed in kinetic resolution of epichlorohydrin using the A. radiobacter epoxide hydrolase. The (R)-epichlorohydrin with a high yield (>27%) and e.e. (>98%) was obtained from over 500 mM substrate concentration in two-phase system by intermittent feeding of the substrate, laying the foundations for its application on the industrial scale.77
| Microorganism | Isomer | % e.e. | Yield (%) | VmS (μmol min−1 mg−1) | VmR (μmol min−1 mg−1) | KmS (mM) | KmR (mM) | |
|---|---|---|---|---|---|---|---|---|
| a VmS, KmS, VmR, KmR represent the Vmax and Km for (S)- and (R)-epichlorohydrin, respectively. | ||||||||
| A. radiobacter | R | >99 | 42.7 | 27.8 | 62.5 | 5.3 | 42.5 | |
| A. mediolanus | S | >99 | 21.5 | 7.9 | 35.6 | 161 | 56.6 | |
| R. toruloides | R | 100 | 18 | — | — | — | — | |
The haloalcohol dehalogenase catalyses the ring closure of 1,3-dichloro-2-propanol to yield epichlorohydrin with low enantioselectivity, but the epoxide hydrolase catalyses enantioselective hydrolysis of epichlorohydrin to remain a single enantiomer. Accordingly, it is believed that there is a good potential for production of chiral epichlorohydrin from 1,3-dichloro-2-propanol by combination of these two enzymes. The reaction was performed in a specially designed reactor by two-step biocatalysis.82 The reaction mixtures in the first reactor flowed into the second reactor after the ring closure reaction of 1,3-dichloro-2-propanol, but the immobilized haloalcohol dehalogenase was intercepted in the first reactor in order to avoid the racemization of chiral epichlorohydrin in the second step. The formed epichlorohydrin was hydrolyzed with high enantioselectivity by epoxide hydrolase in the second reactor, and finally the (R)-epichlorohydrin with >99% e.e. was successfully obtained. This research offered a potential method to produce the chiral epichlorohydrin from 1,3-dichloro-2-propanol.
Concluding remarks
Enantiomerically pure epichlorohydrin is a valuable chiral intermediate for synthesis of chiral pharmaceuticals. Currently, chiral epichlorohydrin is mainly produced by the chemical methods. Kinetic resolution of racemic epichlorohydrin by chemical catalyst salen-Co revealed the excellent enantioselectivity with >99% e.e. and >45% yield. However, the salen-Co catalyst is expensive and pollutes the environment. It is necessary to find an economical, environmentally friendly, and efficient process of producing the chiral epichlorohydrin. Enzymatic synthesis is qualified for these requirements and considered as a promising alternative method. A lot of haloalcohol dehalogenase and epoxide hydrolase have been screened and applied for preparation of chiral epichlorohydrin, but it is still a long way for their applications in industrial production. The further studies would focus mainly on improvement of substrate concentration and yield by genetic engineering and protein engineering technologies.Acknowledgements
This work is supported by the Scientific Research Foundation of Zhejiang Ocean University (No. Q1420).References
- W. J. Choi, Appl. Microbiol. Biotechnol., 2009, 84, 239–247 CrossRef CAS PubMed.
- M. Kotik, V. Tepanek, M. Grulich, P. Kyslík and A. Archelas, J. Mol. Catal. B: Enzym., 2010, 65, 41–48 CrossRef CAS.
- N. Kasai, T. Suzuki and Y. Furukawa, Chirality, 1998, 10, 682–692 CrossRef CAS.
- K. Kitaori, Y. Takehira, Y. Furukawa, H. Yoshimoto and J. Otera, Pharm. Bull., 1997, 45, 412–414 CrossRef CAS.
- S. Shuto, N. Shibuya, S. Yamada, T. Ohkura, R. Kimura and A. Matsuda, Chem. Pharm. Bull., 1999, 47, 1188–1192 CrossRef CAS PubMed.
- M. M. Kabat, A. R. Daniewski and W. Burger, Tetrahedron: Asymmetry, 1997, 8, 2663–2665 CrossRef CAS.
- X. Zheng, C. W. Jones and M. Weck, Chem.–Eur. J., 2006, 12, 576–583 CrossRef PubMed.
- C. S. Gill, K. Venkatasubbaiah, N. T. S. Phan, M. Weck and C. W. Jones, Chemistry, 2008, 14, 7306–7313 CrossRef CAS PubMed.
- Z. P. Zhao, M. S. Li, J. Y. Zhang, H. N. Li, P. P. Zhu and W. F. Liu, Ind. Eng. Chem. Res., 2012, 51, 9531–9539 CrossRef CAS.
- R. L. Osborne, M. K. Coggins, T. James and J. H. Dawson, J. Am. Chem. Soc., 2007, 129, 14838–14839 CrossRef CAS PubMed.
- J. Aburto, J. Correa-Basurto and E. Torres, Arch. Biochem. Biophys., 2008, 480, 33–40 CrossRef CAS PubMed.
- V. M. Dembitsky, Tetrahedron, 2003, 59, 4701–4720 CrossRef CAS.
- S. K. Karmee, C. Roosen, C. Kohlmann, S. Lütz, L. Greiner and W. Leitner, Green Chem., 2009, 11, 1052–1055 RSC.
- L. P. Hager, F. J. Lakner and A. Basavapathruni, J. Mol. Catal. B: Enzym., 1998, 5, 95–101 CrossRef CAS.
- J. B. Park and D. S. Clark, Biotechnol. Bioeng., 2006, 93, 1190–1195 CrossRef CAS PubMed.
- J. Wu, C. Liu, Y. Jiang, M. Hu, S. Li and Q. Zhai, Catal. Commun., 2010, 11, 727–731 CrossRef CAS.
- A. J. van den Wijngaard, P. T. Reuvekamp and D. B. Janssen, J. Bacteriol., 1991, 1, 124–129 Search PubMed.
- A. Archelas and R. Furstoss, Annu. Rev. Microbiol., 1997, 51, 491–525 CrossRef CAS PubMed.
- K. Zheng and L. Tang, J. Chem. Ind. Eng., 2008, 59, 2971–2977 CAS.
- T. Koudelakova, S. Bidmanova, P. Dvorak, A. Pavelka, R. Chaloupkova, Z. Prokop and J. Damborsky, Biotechnol. J., 2013, 8, 32–45 CrossRef CAS PubMed.
- S. Marcus, R. J. Floor, H. Bernhard, B. Michael, P. A. Jekel, H. J. Wijma, B. W. Dijkstra and D. B. Janssen, ChemBioChem, 2013, 14, 870–881 CrossRef PubMed.
- Z. Y. You, Z. Q. Liu and Y. G. Zheng, Appl. Microbiol. Biotechnol., 2013, 97, 9–21 CrossRef CAS PubMed.
- C. E. Castro and E. W. Bartnicki, Biochemistry, 1968, 7, 3213–3218 CrossRef CAS PubMed.
- J. E. van Hylckama Vlieg, L. Tang, J. H. Lutjie Spelberg, T. Smilda, G. J. Poelarends and T. Bosma, et al., J. Bacteriol., 2001, 183, 5058–5066 CrossRef CAS PubMed.
- A. J. Van Den Wijngaard, D. B. Janssen and B. Witholt, J. Gen. Microbiol., 1989, 135, 2199–2208 CAS.
- R. Yonetani, H. Ikatsu, C. Miyake-Nakayama, E. Fujiwara, Y. Maehara, S. I. Miyoshi, H. Matsuoka and S. Shinoda, J. Health Sci., 2004, 50, 605–612 CrossRef CAS.
- N. Kasai, K. Tsujimura, K. Unoura and T. Suzuki, Agric. Biol. Chem., 1990, 54, 3185–3190 CrossRef CAS.
- T. Nakamura, F. Yu, W. Mizunashi and I. Watanabe, Agric. Biol. Chem., 1991, 55, 1931–1933 CrossRef CAS.
- N. Kasai, K. Tsujimura, K. Unoura and T. Suzuki, J. Ind. Microbiol., 1992, 10, 37–43 CrossRef CAS.
- Z. Q. Liu, A. C. Gao, Y. J. Wang, Y. G. Zheng and Y. C. Shen, J. Ind. Microbiol. Biotechnol., 2014, 41, 1145–1158 CrossRef CAS PubMed.
- X. Feng, Z. Q. Liu, N. W. Wan and Y. G. Zheng, Appl. Biochem. Biotechnol., 2014, 174, 352–364 CrossRef PubMed.
- F. Yu, T. Nakamura, W. Mizunashi and I. Watanabe, Biosci., Biotechnol., Biochem., 1994, 58, 1451–1457 CrossRef CAS PubMed.
- R. M. de Jong, J. J. Tiesinga, H. J. Rozeboom, K. H. Kalk, L. Tang, D. B. Janssen and B. W. Dijkstra, EMBO J., 2003, 22, 4933–4944 CrossRef CAS PubMed.
- R. M. de Jong, H. J. Rozeboom, K. H. Kalk, L. Tang, D. B. Janssen and B. W. Dijkstra, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2002, 58, 176–178 CrossRef.
- R. M. D. Jong, J. J. W. Tiesinga, A. Villa, L. Tang, D. B. Janssen and B. W. Dijkstra, J. Am. Chem. Soc., 2005, 127, 13338–13343 CrossRef PubMed.
- R. M. D. Jong, K. H. Kalk, T. Lixia, D. B. Janssen and B. W. Dijkstra, J. Bacteriol., 2006, 188, 4051–4056 CrossRef PubMed.
- H. M. S. Assis, A. T. Bull and D. J. Hardman, Enzyme Microb. Technol., 1998, 22, 545–551 CrossRef CAS.
- T. Nakamura, T. Nagasawa, F. Yu, I. Watanabe and H. Yamada, J. Bacteriol., 1992, 174, 7613–7619 CAS.
- T. Nakamura, F. Yu, W. Mizunashi and I. Watanabe, Appl. Environ. Microbiol., 1993, 59, 227–230 CAS.
- A. J. Effendi, S. D. Greenaway and B. N. Dancer, Appl. Environ. Microbiol., 2000, 66, 2882–2887 CrossRef CAS PubMed.
- S. P. Zou, E. H. Du, Z. C. Hu and Y. G. Zheng, Biotechnol. Lett., 2013, 35, 937–942 CrossRef CAS PubMed.
- T. Nakamura, Appl. Environ. Microbiol., 1994, 60, 1297–1301 CAS.
- H. X. Jin, Z. C. Hu, Z. Q. Liu and Y. G. Zheng, Biotechnol. Appl. Biochem., 2012, 59, 170–177 CrossRef CAS PubMed.
- D. B. Janssen, E. M. Majeri, G. Hasnaoui, B. Hauer and J. H. Lutje Spelberg, Biochem. Soc. Trans., 2006, 34, 291–295 CrossRef CAS PubMed.
- N. Kasai, K. Tsujimura, K. Unoura and T. Suzuki, J. Ind. Microbiol., 1992, 9, 97–101 CrossRef CAS.
- J. H. Lutje Spelberg, J. E. T. van Hylckama Vlieg, T. Bosma, R. M. Kellogg and D. B. Janssen, Tetrahedron: Asymmetry, 1999, 10, 2863–2870 CrossRef.
- G. Hasnaoui-Dijoux, M. Majeric Elenkov, J. H. Lutje Spelberg, B. Hauer and D. B. Janssen, ChemBioChem, 2008, 9, 1048–1051 CrossRef CAS PubMed.
- M. M. Elenkov, B. Hauer and D. B. Janssen, Adv. Synth. Catal., 2006, 348, 579–585 CrossRef CAS.
- J. H. Lutje Spelberg, L. Tang, M. V. Gelder, R. M. Kellogg and D. B. Janssen, Tetrahedron: Asymmetry, 2002, 13, 1083–1089 CrossRef CAS.
- G. Hasnaoui, J. H. Lutje Spelberg, E. D. Vries, L. Tang, B. Hauer and D. B. Janssen, Tetrahedron: Asymmetry, 2005, 16, 1685–1692 CrossRef CAS.
- T. Nakamura, T. Nagasawa, F. Yu, I. Watanabe and H. Yamada, Tetrahedron, 1994, 50, 11821–11826 CrossRef CAS.
- J. H. Lutje Spelberg, L. Tang, R. M. Kellogg and D. B. Janssen, Tetrahedron: Asymmetry, 2004, 15, 1095–1102 CrossRef CAS.
- Z. Q. Liu, Y. Li, Y. Y. Xu, L. F. Ping and Y. G. Zheng, Appl. Microbiol. Biotechnol., 2007, 74, 99–106 CrossRef CAS PubMed.
- A. Archelas and R. Furstoss, Curr. Opin. Chem. Biol., 2001, 5, 112–119 CrossRef CAS PubMed.
- H. Jin, Q. Wang and Z. Y. Li, Chin. J. Chem., 2001, 19, 272–275 CrossRef CAS.
- J. M. D. Carmo, A. A. D Silva, J. Morgan, Y. X. Wang, S. Munusamy and J. E. Hall, Nutr., Metab. Cardiovasc. Dis., 2012, 22, 598–604 CrossRef PubMed.
- S. Q. Huang, Y. Wang and Y. Q. Long, Chin. J. Org. Chem., 2012, 32, 877–888 CrossRef CAS.
- E. Blée and F. Schuber, Biochem. J., 1992, 282, 711–714 CrossRef.
- M. H. Jacobs, A. J. van den Wijngaard, M. Pentenga and D. B. Janssen, Eur. J. Biochem., 1991, 202, 1217–1222 CrossRef CAS PubMed.
- C. Morisseau, H. Nellaiah, A. Archelas, R. Furstoss and J. C. Baratti, Enzyme Microb. Technol., 1997, 20, 446–452 CrossRef CAS.
- Y. Liu, Q. Sha, S. Wu, J. Wang, L. Yang and W. Sun, J. Ind. Microbiol. Biotechnol., 2006, 33, 274–282 CrossRef CAS PubMed.
- U. Wandel, M. Mischitz, W. Kroutil and K. Faber, J. Chem. Soc., Perkin. Trans. 1, 1995, 7, 735–736 RSC.
- K. Faber, M. Mischitz, W. Kroutil, E. C. Roos, Q. B. Broxterman, W. J. J. van den Tweel and K. Kamphuis, Acta Chem. Scand., 1996, 50, 249–258 CrossRef CAS.
- A. Steinreiber, I. Osprian, S. Mayer, R. A. Orru and K. Faber, Eur. J. Org. Chem., 2000, 2000, 3703–3711 CrossRef.
- C. A. G. M. Weijers, Tetrahedron: Asymmetry, 1997, 8, 639–647 CrossRef CAS.
- F. Zocher, M. M. Enzelberger, U. T. Bornscheuer, B. Hauer, W. Wohlleben and R. D. Schmid, J. Biotechnol., 2000, 77, 287–292 CrossRef CAS PubMed.
- C. A. G. M. Weijers and J. A. M. D. Bont, Enzyme Microb. Technol., 1991, 13, 306–308 CrossRef CAS.
- W. J. Choi, E. C. Huh, H. J. Park, E. Y. Lee and C. Y. Choi, Biotechnol. Tech., 1998, 12, 225–228 CrossRef CAS.
- W. J. Choi, E. Y. Lee, S. J. Yoon, S. T. Yang and C. Y. Choi, J. Biosci. Bioeng., 1999, 88, 339–341 CrossRef CAS PubMed.
- E. Misawa, C. K. Kwo Chion, I. V. Archer, M. P. Woodland, N. Y. Zhou, S. F. Carter and D. A. Widdowson, Eur. J. Biochem., 1998, 253, 173–183 CAS.
- C. Morisseau, A. Archelas, C. Guitton, D. Faucher, R. Furstoss and J. C. Baratti, Eur. J. Biochem., 1999, 263, 386–395 CrossRef CAS PubMed.
- H. Visser, S. Vreugdenhil, J. A. M. D. Bont and J. C. Verdoes, Appl. Microbiol. Biotechnol., 2000, 53, 415–419 CrossRef CAS PubMed.
- H. S. Kim, J. H. Lee, S. Park and E. Y. Lee, Biotechnol. Bioprocess Eng., 2004, 9, 62–64 CrossRef CAS.
- E. Y. Lee, J. Ind. Eng. Chem., 2007, 13, 159–162 CAS.
- J. H. Woo, Y. O. Hwang, J. H. Kang, H. S. Lee, S. J. Kim and S. G. Kang, J. Biosci. Bioeng., 2010, 110, 295–297 CrossRef CAS PubMed.
- V. L. Bert, J. H. Lutje Spelberg, K. Jaap, S. Theo, M. G. Wubbolts and D. B. Janssen, Chem. Biol., 2004, 11, 981–990 CrossRef PubMed.
- H. X. Jin, Z. Q. Liu, Z. C. Hu and Y. G. Zheng, Eng. Life Sci., 2013, 13, 385–392 CrossRef CAS.
- F. Xue, Z. Q. Liu, S. P. Zou, N. W. Wan, W. Y. Zhu, Q. Zhu and Y. G. Zheng, Process Biochem., 2014, 49, 409–417 CrossRef CAS.
- Z. Q. Liu, Y. Li, L. F. Ping, Y. Y. Xu, F. J. Cui, Y. P. Xue and Y. G. Zheng, Process Biochem., 2007, 42, 889–894 CrossRef CAS.
- Z. Q. Liu, L. P. Zhang, F. Cheng, L. T. Ruan, Z. C. Hu and Y. G. Zheng, Catal. Commun., 2011, 16, 133–139 CrossRef CAS.
- H. X. Jin, Z. C. Hu and Y. G. Zheng, J. Biosci., 2012, 37, 695–702 CrossRef CAS PubMed.
- H. X. Jin, Z. Q. Liu, Z. C. Hu and Y. G. Zheng, Biochem. Eng. J., 2013, 74, 1–7 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2015 |