Fabrication of AS1411 aptamer functionalized Gd2O3-based molecular magnetic resonance imaging (mMRI) nanoprobe for renal carcinoma cell imaging†
Yue Daiab,
Aiping Zhangb,
Jia Youb,
Jingjing Li*ab,
Huiting Xuab and
Kai Xu*b
aDepartment of Radiology, Affiliated Hospital of Xuzhou Medical College, Xuzhou 221006, China
bSchool of Medical Imaging, Xuzhou Medical College, Xuzhou 221004, China. E-mail: xkpaper@163.com
First published on 7th September 2015
Abstract
Magnetic resonance imaging (MRI) as a noninvasive diagnostic technology with high spatial resolution has been widely used in clinics. However, the relatively low sensitivity is the main shortcoming of this technology. To address this issue, we would like to develop a molecular MRI nanoprobe for the sensitive and specific MRI of renal carcinoma cells with BSA-Gd2O3 nanoparticles as MRI contrast agents, mesoporous silica nanoparticles (mSiO2 NPs) as nanocarriers and the AS1411 aptamer as a targeting molecule. To achieve this aim, BSA-Gd2O3 NPs were assembled onto mSiO2 NPs with the help of anionic polyelectrolyte, sodium polystyrene sulfonate (PSS), and cationic polyelectrolyte, poly dimethyl diallyl ammonium chloride (PDDA) layer by layer. Such successful assembly was confirmed by transmission electron microscopy (TEM), FT-IR spectroscopy, zeta-potential analysis, hydrodynamic diameter determination and gel electrophoresis. After assembly, the mSiO2/PSS/PDDA/BSA-Gd2O3 nanoprobe presented a larger longitudinal relaxivity (r1) (26.1 s−1 mM−1 Gd) than BSA-Gd2O3 NPs (11.8 s−1 mM−1 Gd) and commercially used Gd–DTPA (3.87 s−1 mM−1 Gd). Additionally, with the AS1411 aptamer as a targeting molecule, our fabricated mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobes could recognize clear cell renal carcinoma cells (ccRCC) specifically by MRI in vitro.
Introduction
Magnetic resonance imaging (MRI), which provides inherent soft-tissue contrast, high spatial resolution and lack of ionizing radiation is thought to be one of the best strategies used in clinical diagnosis. However, the relatively low sensitivity is the major limitation of MRI technology. In order to improve the visibility of internal body structures, various MRI contrast agents (CAs) have been introduced by shortening the relaxation parameters of water.1 As the representation of positive MRI CAs, chelated gadolinium compounds such as Gd–DTPA and Gd–DOTA are widely used in clinic to improve the contrast between normal and diseased tissues. However, it should be mentioned that the limited contrast enhancement ability, short blood circulation time and non-specificity still hampered their further applications. Particularly, with the demand of the development of molecular magnetic resonance imaging (mMRI), MRI contrast agents with better proton relaxivity and easy functionalization ability are needed urgently.2 In recent years, with the development of nanotechnology, nanoparticle-based positive MRI CAs have been paid more and more attentions due to their easy design and functionalization. Gd2O3 nanoparticles (Gd2O3 NPs) as positive MRI CA have emerged to present larger T1 relaxivitiy, good biocompatibility and easy conjugation with other biomolecules or imaging agents for mMRI and multimodal molecular imaging.3–5 Thus, in this study, we would like to introduce BSA-Gd2O3 NPs as MRI CA to fabricate AS1411 aptamer functionalized mSiO2/PSS/PDDA/BSA-Gd2O3 mMRI nanoprobe to achieve better MRI contrast enhancement and specific tumor cell targeting. Mesoporous silica nanoparticles (mSiO2 NPs) as nanocarriers have attracted great interest since they exhibit low cytotoxicity and excellent chemical stability and their surface can be easily modified.6,7 In terms of biocompatibility, silica is accepted as “Generally Recognized As Safe” (GRAS) by the United States Food and Drug Administration (FDA).8 Furthermore, dye-doped silica nanoparticles, called Cornell dots (C dots), have received approval from the FDA for the first Investigational NewDrug (IND) application for targeted molecular imaging of cancer.9 Decuzzi's group has successfully confined gadolinium in the pores of mSiO2 NPs to improve the relaxivity.10 In this study, we choose mSiO2 NPs as the nanocarrier to load more BSA-Gd2O3 NPs through layer by layer approach with the help of poly(diallyldimethylammonium chloride) (PDDA) and poly(4-styrenesulfonic acid) (PSS). AS1411 aptamer was employed as targeting molecules due to its selective binding to nucleolin11,12 and internalized into a variety of cancer cell lines including renal, breast, and other adenocarcinoma cell lines.13–15 To confirm the specific MRI ability to tumor cells, renal cell carcinoma (RCC) were chosen as model. RCC accounts for approximately 90% of all renal malignancies.16 The main subtype of RCC is clear cell RCC (ccRCC, approximately 70%).Results and discussion
Preparation and characterization of mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobe
Gd-based chelates as MRI contrast agents have been widely used in clinic to improve the sensitivity of MRI diagnosis. However, the chelation reduced the unpaired electrons of Gd3+ greatly, resulting in a limited proton relaxivity. Gd2O3 nanoparticles with high number of gadolinium atoms were emerged to address this issue.17 In this study, BSA-Gd2O3 NPs were employed as MRI contrast agent, which were prepared according to a previous report.18 In order to obtain the best MRI behavior, BSA-Gd2O3 NPs were further deposited onto mesoporous SiO2 (mSiO2) surface through layer by layer (LBL) assembly with the help of polyelectrolytes, PDDA and PSS. Driven by the electrostatic force, a uniform monolayer of negatively charged PSS and positively charged PDDA as well as negatively charged BSA-Gd2O3 NPs were alternatively adsorbed onto the positively charged NH2–mSiO2 NPs (Scheme 1). This assembly process was monitored by the determination of the changes of zeta potentials and hydrodynamic diameter. As shown in Fig. 1A, the potential value of mSiO2 NPs was −10 mV. After the amino group functionalization, the value was 22.6 mV. With PSS and PDDA assembly, the potential values were changed to −40.9 mV and 42.8 mV, respectively. The zeta-potential of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex was −1.97 mV, which might come from the negatively charged BSA. Additionally, the hydrodynamic diameter was increased accordingly after each step of assembly (Fig. 1B). They were 202.8 nm for mSiO2, 238.8 nm for mSiO2/PSS, 275.6 nm for mSiO2/PSS/PDDA, 284.3 nm for mSiO2/PSS/PDDA/BSA-Gd2O3 and 299.1 nm for mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411, indicating the successful assembly. For the fabrication of specific nanoprobe, the obtained mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex was conjugated with AS1411 aptamer through the covalent coupling between the carboxyl group of aptamer and amino group of nanocomplex. Such conjugation was confirmed by FT-IR absorption spectrum and gel electrophoresis. The emerging absorption peak at 1620 cm−1 was ascribed to acrylamide vibration, which could not be observed in the mixture of mSiO2/PSS/PDDA/BSA-Gd2O3 and AS1411 aptamer (Fig. 1C). For gel electrophoresis (Fig. 1D), mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex (Lane 4) and mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobe (Lane 3) stayed in the well because of their relatively large size. The stronger band intensity of mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 (Lane 3) than mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex (Lane 4) was ascribed to the conjugated AS1411 aptamer. The size and morphology of mSiO2 NPs and mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex were further characterized by transmission electron microscopy (TEM), shown in Fig. 2. Before LBL assembly and BSA-Gd2O3 NPs loading, mSiO2 exhibited uniformly ordered pores (Fig. 2A). After the assembly of polyelectrolytes and BSA-Gd2O3 NPs, however, the porous structure became weaker (Fig. 2B), indicating the successful assembly of BSA-Gd2O3 NPs on the surface of mSiO2 NPs. The size of mSiO2 NPs was increased from 74.01 nm to 85.24 nm after assembly. | ||
| Scheme 1 Schematic illustration of the fabrication process of mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobes. | ||
MRI behavior of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex
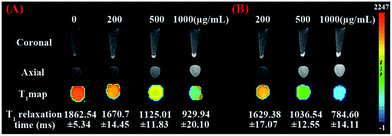
As mMRI nanoprobe, the MRI behavior of our fabricated mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex was evaluated. With different amounts of BSA-Gd2O3 NPs, the T1 relaxation times of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex varied. As shown in Fig. 3, with the increase of BSA-Gd2O3 NP amount, the T1 relaxation time of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex was decreased gradually. But when the amount of BSA-Gd2O3 NPs was up to 8.48 μmol, the signal intensity reached a critical level and the T1 relaxation time changed slowly with the increase of BSA-Gd2O3 amount. Thus, 8.48 μmol was chosen for the fabrication of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex in the following experiments. It should be noted that the existence of PDDA and PSS favored more BSA-Gd2O3 NPs loaded on the surface of mSiO2 NPs. Positively charged NH2–mSiO2 NPs could adsorb negatively charged BSA-Gd2O3 NPs directly. However, mSiO2/BSA-Gd2O3 displayed weaker MRI signal than mSiO2/PSS/PDDA/BSA-Gd2O3 at the same concentrations of mSiO2 and BSA-Gd2O3 NPs (Fig. S1†). The loading amounts of BSA-Gd2O3 NPs were determined by ICP-MS with 11.6 μmol Gd/g mSiO2 for mSiO2/BSA-Gd2O3 and 49.6 μmol Gd/g mSiO2 for mSiO2/PSS/PDDA/BSA-Gd2O3, coming from the stronger positively charged mSiO2/PSS/PDDA. To further evaluate the ability of our fabricated nanocomplex as mMRI nanoprobe, the relaxivity values of Gd–DTPA, BSA-Gd2O3 NPs, and mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex were determined and compared by measuring longitudinal proton relaxation time (T1) as a function of Gd concentration. As shown in Fig. 4, the r1 value of BSA-Gd2O3 was 11.8 s−1 mM−1 Gd, which was 3 times that of the commercial MRI contrast agents, Gd–DTPA (r1 = 3.87 s−1 mM−1 Gd). More importantly, the relaxivity of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex was further increased to 26.1 s−1 mM−1 Gd. Such boost relaxivity might come from the increased molecular size after assembly. Theoretically, the proton relaxivity of Gd(III) compound is determined by the equation r1 = Cqμeff2τcr−6, in which C is a constant, q is the number of inner sphere water molecules, μeff is the effective magnetic moment, τc is the molecular correlation time, and r is the Gd⋯H (H2O) distance.19–21 The molecular correlation time τc is determined by the following parameters: rotational correlation time τr, the electronic correlation time τs, and the proton residence time τm, as expressed in the equation τc−1 = τr−1 + τs−1 + τm−1. To obtain higher r1, improvement of τr and τm values is commonly considered. Changing molecular size is one of the possible approach to increase τr. Gd chelates conjugated with polymers, dendrimers, or biomacromolecules presented an increased r1.22–25 Protein-bound Gd–DTPA has relaxivities approaching 20 mM−1 s−1, compared to 4 mM−1 s−1 for Gd–DTPA alone. In our case, r1 was increased gradually with the assembly of nanocomplex, from 11.8 s−1 mM−1 Gd of BSA-Gd2O3, to 16.23 s−1 mM−1 Gd of mSiO2/BSA-Gd2O3 (Fig. S2†), and 26.1 s−1 mM−1 Gd of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex, which favored them for the fabrication of mMRI nanoprobe and their biomedical applications. | ||
| Fig. 3 (A) T1-weighted MR images and T1-map images of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex prepared with different amount of BSA-Gd2O3 NPs. (B) The corresponding T1 relaxation time. | ||
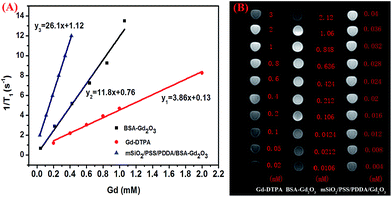 | ||
| Fig. 4 r1 relaxivity curves (A) and T1-weighted MR images (B) of Gd–DTPA, BSA-Gd2O3 nanoparticles and mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex with various Gd concentrations. | ||
In vitro cytotoxicity
The safety assessment of nanoparticles is a vital step before their clinical applications. To evaluate the cell toxicity of the nanoprobes, MTT assay were performed to look for the potentially safe concentrations for the following targeting experiments. mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex with seven different concentrations, ranging from 100 to 1000 μg mL−1, were incubated with 786-0 cells and normal human umbilical vein endothelial cells for 24 h, respectively. As shown in Fig. 5, mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex displayed good biocompatibility, and no significant cytotoxicity was observed on 786-0 renal carcinoma cells or normal human umbilical vein endothelial cells even under a high concentration of 1000 μg mL−1, indicating their excellent biocompatibility as mMRI nanoprobe.In vitro MR imaging
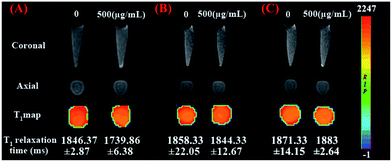
The specific cellular targeting of mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 mMRI nanoprobe was evaluated by MRI. AS1411 aptamer as the target molecules could selectively bind to nucleolin, which overexpressed in a variety of cancer cell lines, including renal, breast, and other adenocarcinoma cell lines.13–15,26–28 The 786-0 renal carcinoma cells were incubated with mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex and mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 mMRI nanoprobes, respectively. To obtain the best signal-to-noise value, 200, 500, and 1000 μg mL−1 nanoprobes were compared. As shown in Fig. 6B, the higher concentration of mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobe, the stronger signal intensity from 786-0 cells was observed. Furthermore, the presence of AS1411 aptamer could facilitate more mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobe to bind with 786-0 cells and present a brighter MRI signal at all these three concentrations. But when the concentration of nanoprobe reached 1000 μg mL−1, the non-specificity adsorption obviously existed. Thus, 500 μg mL−1 mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobe was chosen for the specific in vitro MRI finally. To further confirm such AS1411 aptamer-based specific targeting, NIH-3T3 cells and EA.hy926 cells were introduced as control and treated with 500 μg mL−1 mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobe, respectively. As shown in Fig. 7A and B, no significant MRI signal could be observed from these two control cell lines, indicating the specific MRI ability of our fabricated nanoprobe to ccRCC in vitro. Additionally, when 786-0 cells were pretreated with AS1411 aptamer before the incubation with 500 μg mL−1 mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobe, the MRI signal was weakened obviously (Fig. 7C). The binding blocking with AS1411 aptamer to nucleolin inhibited the following cellular binding with mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobe, indicating the specific MRI signal came from AS1411 aptamer.Experimental
Bovine serum albumin (BSA), (3-aminopropyl)triethoxysilane (APTES) and (3-aminopropyl) tetraethylorthosilicate (TEOS) were obtained from Solarbio (China) and Aladdin (USA). Gd(NO3)3·6H2O and cetyl-trimethylammonium bromide (CTAB) were purchased from Sinopharm Chemical Reagent Co. Ltd (Shanghai, China). Sodium polystyrene sulfonate (PSS), poly dimethyl diallyl ammonium chloride (PDDA), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC·HCl) and N-hydroxysuccinimide (NHS) were purchased from Sigma-Aldrich (USA). Dimethyl sulfoxide (DMSO) was bought from PIERCE (USA). DNA oligos were synthesized and purified by Shanghai Sangon Biotechnology Co. Ltd (Shanghai, China). All chemicals involved in this work were analytical grade. All aqueous solutions were prepared with ultrapure water (≥18 MΩ, Milli-Q, Millipore). The DNA sequence was listed as follows.COOH-AS1411![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5′–COOH–C6-GGTGGTGGTGGTTGTGGTGGTGGTGG-3′. 5′–COOH–C6-GGTGGTGGTGGTTGTGGTGGTGGTGG-3′. |
Apparatus and characterization
The size and morphology of our nanoparticles were observed by transmission electron microscopic (TEM) (TECNAI G2, USA). FT-IR spectra were obtained from the infrared absorption spectroscopy (Bruker, Germany). Zeta potentials and hydrodynamic diameters were determined by Nano ZS90 (Malvern, England). Gel imaging was obtained by gel Dox™ EZ Imager (BIO-RAD, USA). The absorbances for MTT assay were determined by a microplate reader (Multiskon MK3, USA) at 490 nm. The determination of gadolinium content was performed with inductively coupled plasma-mass spectrometry (ICPMS) (Optima 5300DV, PerkinElmer, USA). MRI scanning was performed on 3.0 T human magnetic resonance scanner (GE Signa, USA).Cells and cell culture
The 786-0 renal carcinoma cells, NIH-3T3 mouse fibroblast cells and EA.hy926 normal human umbilical vein endothelial cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The 786-0 renal carcinoma cells were propagated in a 10% FBS containing RPMI 1640 medium supplemented with penicillin (100 mg mL−1), and streptomycin (100 mg mL−1). NIH-3T3 cells and normal human umbilical vein endothelial cells were cultured in 10% FBS-containing DMEM medium (Gibco, Grand Island, NY) supplemented with penicillin (100 mg mL−1), and streptomycin (100 mg mL−1). All cells were grown in a humidified incubator (Thermo, USA) at 37 °C under 5% CO2 atmosphere.Preparation of mesoporous silica nanoparticles
mSiO2 NPs were synthesized according to literature procedures with some modifications.29 Briefly, cetyl-trimethylammonium bromide (CTAB, 1.0 g) and NaOH (aqueous) (2.00 M, 3.50 mL) were dissolved in 480 mL of double distilled water and stirred at 80 °C. Subsequently, triethoxysilane (5.00 mL) was added dropwise to the solution, and the mixture was allowed to stir for 2 h at 80 °C. The resultant white precipitate was isolated by centrifugation and washed with ethanol for three times. In order to remove the structure-directing agent of CTAB, the production was refluxed in a solution composed of methanol (80 mL) and HCl (37%, 1 mL) for 20 h. After washing with ethanol three times, mSiO2 NPs were obtained by drying under 60 °C.Synthesis of NH2–mSiO2 NPs
For the easy conjugation with target molecules, amino group was further modified on the surface of mSiO2 NPs. 300 mg mSiO2 NPs were suspended in 20 mL anhydrous toluene inside a round-bottom flask, and an excess of APTES (0.3 mL) was added. The solution was stirred at 50 °C under nitrogen for 4 h. Then it was centrifuged, washed with ethanol for three times, and dried at 60 °C to obtain NH2–mSiO2 NPs.Preparation of BSA-Gd2O3 NPs
BSA-Gd2O3 NPs were synthesized according to the literature with some modifications.18 1.25 g of BSA was dissolved in 45 mL of ultrapure water. Then, 5 mL of 50 mM Gd(NO3)3 was added to the above solution slowly under vigorous stirring. After the introduction of 5 mL of 2 M NaOH 5 min later, the mixture was allowed to react under vigorous stirring at 37 °C for 12 h. Finally, the prepared BSA-Gd2O3 was dialyzed against ultrapure water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, v/v) to remove excess precursors.
1000, v/v) to remove excess precursors.
Inductively coupled plasma-mass spectrometry (ICP-MS) analysis
The concentration of gadolinium in BSA-Gd2O3 nanoparticles was determined by ICP-MS analysis (Optima 5300DV, PerkinElmer, USA). 500 μL 10-time concentrated BSA-Gd2O3 NPs were mixed with 500 μL 14 M HNO3. After heated for 30 min at 80 °C, 1 mL of the above solution was diluted with 25 mL 5% HNO3 for ICP-MS analysis. The sample preparation of mSiO2/BSA-Gd2O3 and mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex for ICP-MS analysis were similar with BSA-Gd2O3 NPs.Fabrication of mSiO2/PSS/PDDA/Gd2O3-AS1411 mMRI nanoprobe
To confirm the role of PDDA and PSS for more BSA-Gd2O3 NP loading, mSiO2/BSA-Gd2O3 nanocomplex were synthesized first. Briefly, 10 mg NH2–mSiO2 NPs were dispersed in 2 mL BSA-Gd2O3 (4.24 μmol Gd) solution and sonicated for 20 min. After centrifugation and washed with water for three time, the mSiO2/BSA-Gd2O3 nanocomplex were dispersed in 2 mL H2O for further use. For the preparation of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex, 10 mg NH2–SiO2 NPs were dispersed in 2 mL PSS solution (2 mg mL−1, 0.2 M NaCl) and sonicated for 30 min, and excess PSS was removed by centrifugation and wash with water. Then, mSiO2/PSS was suspended in 2 mL PDDA solution (2 mg mL−1, 0.2 M NaCl) and sonicated for 30 min, and excess PDDA was removed by centrifugation and wash with water. Finally, mSiO2/PSS/PDDA was dispersed in different amounts BSA-Gd2O3 solution (1.016 μmol, 2.12 μmol, 4.24 μmol, 6.36 μmol, 8.48 μmol, 10.6 μmol, 12.72 μmol, 14.84 μmol Gd) and sonicated for 30 min. After centrifugation and washed with water for three times, mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex were dispersed in 2 mL H2O for MRI scanning. AS1411 aptamer was finally functionalized onto the surface of mSiO2 NPs by the covalent coupling between amino group of mSiO2 and carboxyl group modified at the 5′ end of AS1411 aptamer with the help of EDC and NHS. 100 μM COOH-AS1411 (50 μL) were mixed with EDC (100 μL, 10 mg mL−1) in 300 μL PBS (10 mM, pH 7.4) and incubated at 37 °C for 15 min to active carboxyl group. Then, NHS (100 μL, 10 mg mL−1) and mSiO2/PSS/PDDA/BSA-Gd2O3 (500 μL, 5.0 mg mL−1) were added into the mixture and reacted at 37 °C for 2 h. The unreacted bio-molecules were removed by two centrifugation/washing cycles. Then the mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 nanoprobes were dispersed in 1 mL pH 7.4 PBS (10 mM) for further use.Gel electrophoresis analysis
12% polyacryilamide gel was employed for the characterization of AS1411 aptamer attached to mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex. Electrophoresis was carried out at 100 V for 1 h at room temperature. Low range DNA ladder was used as the size marker. After separation, the gel was stained with ethidium bromide and imaged using the fluorescence gel imaging system.Relaxivity calculation of BSA-Gd2O3 NPs, Gd-DTPA, mSiO2/BSA-Gd2O3 and mSiO2/PSS/PDDA/BSA-Gd2O3 nanocompex
MRI behavior test of BSA-Gd2O3 NPs was performed with 3.0 T human magnetic resonance scanner (GE Signa, USA). Various concentrations of BSA-Gd2O3 NPs solution were prepared before MRI scanning, which varied from 0.0106 mM to 2.12 mM with a volume of 600 μL. The following parameters were adopted in data acquisition.5 ① T1 weighted images: echo time (TE) = 16.5 ms, repetition time (TR) = 425 ms, field of view (FOV) = 14 cm × 14 cm, matrix = 384 × 256, slice thickness = 2.0 mm, spacing = 1.5 mm; ② T1-map images: TE = 7.4 ms, TR = 200–800 ms, FOV = 14 cm × 14 cm, matrix = 384 × 256, slice thickness = 2.0 mm, spacing = 1.5 mm. Quantitative T1 relaxation maps were reconstructed from datasets using function software at a workstation (ADW 4.2). The signal intensity of the samples was measured, and the T1 values were calculated accordingly. MRI scannings of Gd–DTPA and mSiO2/PSS/PDDA/BSA-Gd2O3 nanoprobe with different amount were carried out in the same way. The relaxivity values of BSA-Gd2O3 NPs, Gd-DTPA, mSiO2/BSA-Gd2O3 and mSiO2/PSS/PDDA/BSA-Gd2O3 nanoprobe were determined by measuring longitudinal proton relaxation time (T1) as a function of Gd concentration.MTT assay
786-0 cells and EA.hy926 normal human umbilical vein endothelial cells were cultured on 96-well plates at a density of 104 cells each well. After 24 h incubation, the medium was substituted with 100 μL of fresh medium containing different concentrations of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex (0, 100, 200, 400, 500, 600, 800, 1000 μg mL−1). After 24 h incubation, the medium was removed, and fresh medium (100 μL) containing MTT (20 μL, 5 mg mL−1) was added into each well. Four hours later, the culture medium was carefully removed, and 100 mL dimethyl sulfoxide (DMSO) was added to each well to dissolve the formazan crystals for 10 min. The absorbance at 490 nm was measured by microplate reader (Multiskon MK3, USA).In vitro specific targeting of mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 mMRI nanoprobe to 786-0 cells
The 786-0 cells were seeded into 6-well plates at a density of 105 cell per well (2 mL) and cultured for 24 h in a humidified incubator at 37 °C under 5% CO2 atmosphere, respectively. Then, the culture media was removed and the cells were washed with PBS twice. Afterward, 500 μL different concentrations of mSiO2/PSS/PDDA/Gd2O3-AS1411 (200, 500, 1000 μg mL−1) and mSiO2/PSS/PDDA/Gd2O3 (200, 500, 1000 μg mL−1) were added into the well respectively. After 1 h incubation at 37 °C, the free nanoprobes were removed and the cells were washed and lysed by trypsin. The cells that harvested by centrifugation at 1000 rpm for 10 min were fixed by 500 μL paraformaldehyde solution and kept at 4 °C for MRI scanning. The untreated cells that incubated with culture medium were taken as control. For the T1 relaxation time determination, the harvested cells were dispersed in 300 μL of 1% agarose for axial MRI scanning as well. For control experiment, NIH-3T3 cells and EA.hy926 normal human umbilical vein endothelial cells were seeded as 786-0 cells. After incubated with 0 and 500 μg mL−1 mSiO2/PSS/PDDA/Gd2O3-AS1411, the cells were harvested and imaged on MRI scanner.Conclusions
In summary, we fabricated a mSiO2/PSS/PDDA/BSA-Gd2O3-AS1411 mMRI nanoprobe for the specific MRI of renal carcinoma cells by layer by layer approach. The present of polyelectrolytes, PDDA and PSS increased the loading amount of BSA-Gd2O3 NPs on the surface of mSiO2 and improved the longitudinal relaxivity r1 of mSiO2/PSS/PDDA/BSA-Gd2O3 nanocomplex significantly. With the help of AS1411 aptamer specific targeting to nucleolin, the fabricated nanoprobes could recognize clear cell renal carcinoma cells sensitively and specifically in vitro.Acknowledgements
This work was supported by National Natural Science Foundation of China (21305120, 81470075), Natural Science Foundation of Jiangsu Province (BK20130211), and Natural Science Fund for Colleges and Universities in Jiangsu Province (13KJB150036).Notes and references
- C. Lu, J. Li, K. Xu, C. Yang, J. Wang, C. Han and X. Liu, Fabrication of mAb G250-SPIO molecular magnetic resonance imaging nanoprobe for the specific detection of renal cell carcinoma in vitro, PLoS One, 2014, 9(7), e101898–101905 Search PubMed.
- J. Kim, Y. Piao and T. Hyeon, Multifunctional nanostructured materials for multimodal imaging, and simultaneous imaging and therapy, Chem. Soc. Rev., 2009, 39, 372–390 RSC.
- J. Y. Park, M. J. Baek, E. S. Choi, S. Woo, J. H. Kim, T. J. Kim, J. C. Jung, K. S. Chae, Y. Chang and G. H. Lee, Paramagnetic ultrasmall gadolinium oxide nanoparticles as advanced T1 MRI contrast agent: account for large longitudinal relaxivity, optimal particle diameter, and in vivo T1 MR images, ACS Nano, 2009, 3, 3663–3669 CrossRef CAS PubMed.
- L. Faucher, M. Tremblay, J. Lagueux, Y. Gossuin and M. A. Fortin, Rapid synthesis of PEGylated ultrasmall gadolinium oxide nanoparticles for cell labeling and tracking with MRI., ACS Appl. Mater. Interfaces, 2012, 4, 4506–4515 CAS.
- J. J. Li, J. You, Y. Dai, M. L. Shi, C. P. Han and K. Xu, Gadolinium oxide nanoparticles and aptamer-functionalized silver nanoclusters-based multimodal molecular imaging nanoprobe for optical/magnetic resonance cancer cell imaging, Anal. Chem., 2014, 86, 11306–11311 CrossRef CAS PubMed.
- Y. Piao, A. Burns, J. Kim, U. Wiesner and T. Hyeon, Designed fabrication of silica-based nanostructured particle systems for nanomedicine applications, Adv. Funct. Mater., 2008, 18, 3745–3758 CrossRef CAS PubMed.
- A. Burns, H. Ow and U. Wiesner, Fluorescent core–shell silica nanoparticles: towards “Lab on a Particle” architectures for nanobiotechnology, Chem. Soc. Rev., 2006, 35, 1028–1042 RSC.
- J. E. Lee, N. Lee, T. Kim, J. Kim and T. Hyeon, Multifunctional mesoporous silica nanocomposite nanoparticles for theranostic applications, Acc Chem Res., 2011, 893–902 CrossRef CAS PubMed.
- M. Benezra, O. Penate-Medina, P. B. Zanzonico, D. Schaer, H. Ow, A. Burns, E. DeStanchina, V. Longo, E. Herz, S. Iyer, J. Wolchok, S. M. Larson, U. Wiesner and M. S. Bradbury, Multimodal silica nanoparticles are effective cancer-targeted probes in a model of human melanoma, J. Clin. Invest., 2011, 121, 2768–2780 CAS.
- J. S. Ananta, B. Godin, R. Sethi, L. Moriggi, X. Liu, R. E. Serda, R. Krishnamurthy, R. Muthupillai, R. D. Bolskar, L. Helm, M. Ferrari, L. J. Wilson and P. Decuzzi, Geometrical confinement of gadolinium-based contrast agents in nanoporous particles enhances T1 contrast, Nat. Nanotechnol., 2010, 5, 815–821 CrossRef CAS PubMed.
- Y. Teng, A. Girvan, L. Casson, W. J. Pierce, M. Qian, S. Thomas and P. Bates, AS1411 alters the localization of a complex containing protein arginine methyltransferase 5 and nucleolin, Cancer Res., 2007, 67, 10491–10500 CrossRef CAS PubMed.
- S. Soundararajan, W. Chen, E. Spicer, N. Courtenayluck and D. Fernandes, The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells, Cancer Res., 2008, 68, 2358–2365 CrossRef CAS PubMed.
- P. Bates, E. Choi and L. Nayak, G-rich oligonucleotides for cancer treatment, Methods Mol. Biol., 2009, 542, 379–392 CAS.
- P. J. Bates, D. A. Laber, D. M. Miller, S. D. Thomas and J. O. Trent, Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer, Exp. Mol. Pathol., 2009, 86, 151–164 CrossRef CAS PubMed.
- E. M. Reyes-Reyes, Y. Teng and P. J. Bates, A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism, Cancer Res., 2010, 70, 8617–8629 CrossRef CAS PubMed.
- B. Ljungberg, S. C. Campbell, H. Y. Choi, D. Jacqmin and J. E. Lee, The epidemiology of renal cell carcinoma, Eur. Urol., 2011, 60, 615–621 CrossRef PubMed.
- J. L. Bridot, A. C. Faure, S. Laurent, C. Rivière, C. Billotey, B. Hiba, M. Janier, V. Josserand, J. L. Coll, L. V. Elst, R. Muller, S. Roux and P. Perriat, Tillement, O. Hybrid gadolinium oxide nanoparticles: multimodal contrast agents for in vivo imaging, J. Am. Chem. Soc., 2007, 129(16), 5076–5084 CrossRef CAS PubMed.
- S. K. Sun, L. X. Dong, Y. Cao, H. R. Sun and X. P. Yan, Fabrication of multifunctional Gd2O3/Au hybrid nanoprobe via a one-step approach for near-infrared fluorescence and magnetic resonance multimodal imaging in vivo, Anal. Chem., 2013, 85, 8436–8441 CrossRef CAS PubMed.
- C.-T. Yang and K.-H. Chuang, Gd(III) chelates for MRI contrast agents: from high relaxivity to “smart”, from blood pool to blood-brain barrier permeable, Med. Chem. Commun., 2012, 3, 552–565 RSC.
- J. A. Peters, J. Huskens and D. J. Raber, Lanthanide induced shifts and relaxation rate enhancements, Prog. Nucl. Magn. Reson. Spectrosc., 1996, 28, 283–350 CrossRef CAS.
- S. H. Koenig and R. D. Brown III, Field-cycling relaxometry of protein solutions and tissue: implications for MRI, Prog. Nucl. Magn. Reson. Spectrosc., 1990, 22, 487–567 CrossRef CAS.
- Z. Jaszberenyi, L. Moriggi, P. Schmidt, C. Weidensteiner, R. Kneuer, A. E. Merbach, L. Helm and E. Toth, Physicochemical and MRI characterization of Gd3+-loaded polyamidoamine and hyperbranched dendrimers, J. Biol. Inorg. Chem., 2007, 12, 406–420 CrossRef CAS PubMed.
- S. Langereis, A. Dirksen, T. M. Hackeng, M. H. P. van Genderen and E. W. Meijer, Dendrimers and magnetic resonance imaging, New J. Chem., 2007, 31, 1152–1160 RSC.
- S. Laus, A. Sour, R. Ruloff, E. Toth and A. E. Merbach, Rotational dynamics account for pH-dependent relaxivities of PAMAM dendrimeric, Gd-based potential MRI contrast agents, Chem.–Eur. J, 2005, 11, 3064–3076 CrossRef CAS PubMed.
- D. A. Fulton, M. O’Halloran, D. Parker, K. Senanayake, M. Botta and S. Aime, Efficient relaxivity enhancement in dendritic gadolinium complexes: effective motional coupling in medium molecular weight conjugates, Chem. Commun., 2005, 474–476 RSC.
- A. C. Girvan, Y. Teng, L. K. Casson, S. D. Thomas, S. Juliqer, M. W. Ball, J. B. Klein, W. M. Pierce Jr, S. S. Barve and P. J. Bates, AGRO100 inhibits activation of nuclear factorkappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin, Mol. Cancer Ther., 2006, 5, 1790–1799 CrossRef CAS PubMed.
- C. R. Ireson and L. R. Kelland, Discovery and development of anticancer aptamers, Mol. Cancer Ther., 2006, 5, 2957–2962 CrossRef CAS PubMed.
- J. W. Kotula, E. D. Pratico, X. Ming, O. Nakagawa, R. L. Juliano and B. A. Sullenger, Aptamer-mediated delivery of splice-switching oligonucleotides to the nuclei of cancer cells, Nucleic Acid Ther., 2012, 22(3), 187–195 CAS.
- K. M. Taylor, J. S. Kim, W. J. Rieter, H. An, W. Lin and W. Li, Mesoporous silica nanospheres as highly efficient MRI contrast agents, J. Am. Chem. Soc., 2008, 130, 2154–2155 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra17211j |
| This journal is © The Royal Society of Chemistry 2015 |