A microRNAs–mRNAs network involved in the control of graphene oxide toxicity in Caenorhabditis elegans†
Yunli Zhao,
Qiuli Wu and
Dayong Wang*
Medical School, Southeast University, Nanjing 210009, China. E-mail: dayong_w@yahoo.com
First published on 15th October 2015
Abstract
A previous study has suggested that microRNAs (miRNAs) are involved in the control of the toxicity of graphene oxide (GO) in the in vivo Caenorhabditis elegans assay system. However, it is still unclear for miRNAs–mRNAs networks functioning to regulate GO toxicity. In the present study, we first employed a HiSeq 2000 sequencing technique to examine dysregulated mRNAs in GO-exposed nematodes and identified 970 up-regulated and 995 down-regulated mRNAs. Analysis of both gene ontology and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway implied that these dysregulated mRNAs mediated many important biological processes. Some dysregulated genes encode the JNK signaling pathway, which was proven to be involved in the control of GO toxicity. In the JNK signaling pathway, JKK-1 and MEK-1 function in the same pathway as JNK-1 to regulate GO toxicity. Moreover, we raised a miRNAs–mRNAs network, which at least potentially explained molecular basis for the roles of oxidative stress, intestinal barrier, and defecation behavior in regulating GO toxicity. The established link between miRNAs and mRNAs provides the key basis for further elucidating the molecular mechanisms of GO toxicity in organisms.
Introduction
Graphene oxide (GO), a member of graphene family, is a type of two-dimensional (2D) carbon engineered nanomaterial (ENM) and has a single or few layers of sp2-bonded carbon atoms. Due to its unique size, shape, and physicochemical properties, GO is potentially used as a material for biomedical applications.1,2 However, previous in vitro and in vivo studies have suggested the adverse effects of GO on both human health and environmental organisms.3–10 Oxidative stress is considered as one of the crucial cellular mechanisms for GO toxicity.8 Besides the cellular mechanisms, Qu et al. (2013) have indicated that the toll-like receptor (TLR4) may be the predominant molecular mechanism for GO-induced macrophagic necrosis.11 More recently, the dysregulated genes were identified in HepG2 cells exposed to GO.12,13 Global microRNAs (miRNAs) expression has been further investigated in GO-exposed GLC-82 pulmonary adenocarcinoma cells with GO induced dysregulated miRNAs found to be involved in the induction of cell death by activating both the death receptor pathway and mitochondrial pathway.7Caenorhabditis elegans is a typical animal model because of its properties of short lifespan, ease of manipulation, low cost, and well-described genetic background.14 Moreover, C. elegans enables both the assessment of toxicity at the organism level and analysis of the molecular responses (e.g. stress responses) to chemical exposure.15,16 C. elegans is considered as an excellent test organism for the toxicological study of stresses or toxicants.15,16 C. elegans has been used for both the assessment of toxicity and the toxicological study of carbon-based ENMs, including carbon nanotubes (CNTs) and several members of the graphene family.17–22 With the aid of lifespan, brood size, locomotion behavior, reactive oxygen species (ROS) production, and intestinal autofluorescence as toxicity assessment endpoints, previous studies have demonstrated that exposure to GO can cause several aspects of toxicity in nematodes.23–25 The distribution or translocation pattern of GO has also been examined in nematodes. GO could be translocated in both the primary targeted organs such as intestine and the secondary targeted organs such as reproductive organs.23 In nematodes, oxidative stress, permeability of intestinal barrier, and defecation behavior have been suggested to play crucial roles in regulating GO toxicity.24–26
In C. elegans, our recent study identified a series of miRNAs, whose dysregulated expression patterns were observed in GO-exposed nematodes.27 miRNAs are a large class of short non-coding RNAs and usually act to post-transcriptionally inhibit gene expression.28 However, it is still unclear for the possible miRNAs–mRNAs network involved in the control of GO toxicity in nematodes. Thus, in the present study, we first investigated the global mRNA expression patterns in GO-exposed nematodes using a HiSeq 2000 sequencing technique. In C. elegans, the c-Jun N-terminal kinase (JNK) signaling pathway mainly contains the members MEK-1, JKK-1, and JNK-1 and is involved in the control of stress response or response to heavy metals.29,30 Among the dysregulated genes induced by GO exposure, we further investigated the role of the JNK signaling pathway in the control of GO toxicity. Moreover, we examined the possible miRNAs–mRNAs network functioning to regulate the GO toxicity in nematodes. The raised miRNAs–mRNAs network in the current study will be helpful for a deeper understanding of the underlying molecular mechanism for GO toxicity in organisms.
Results and discussion
The properties of prepared GO
Based on an assay using transmission electron microscopy (TEM, JEM-200CX, JEOL, Japan), the sizes of most of the GO in K-medium after sonication (40 kHz, 100 W, 30 min) were in the range of 40–60 nm (Fig. 1a). The GO aggregation size was 248 ± 78 nm (Fig. 1a). Based on an assay using atomic force microscopy (AFM, SPM-9600, Shimadzu, Japan), the prepared GO was sheet-like and the thickness of GO was approximately 1.0 nm in topographic height, corresponding to approximately one layer (Fig. 1b). The zeta potential of GO (10 mg L−1) in K-medium was −22.9 ± 1.4 mV. | ||
| Fig. 1 The properties of GO. (a) TEM image showing the GO distribution and size distribution in K-medium after sonication. (b) AFM results of GO before sonication. | ||
Sequence assembly
Using the Illumina HiSeq™ 2000 technique, the transcriptomes from control and GO exposure groups were sequenced. In C. elegans, L1-larvae are more sensitive than L4-larvae or young adults to the toxicity of ENMs.16 A previous study has suggested that GO exposure from L1-larvae to adult day-1 caused adverse effects on the function of both the primary and secondary targeted organs in nematodes.24 In this study, GO (10 mg L−1) was exposed from L1-larvae to adult day-1. The raw sequence data containing adaptor fragments were filtered to obtain clean reads data. The GC percentages for the transcriptomes of the control and GO exposure groups are shown in Fig. 2a. The total read of the transcripts for the control group was 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 159
159![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 711 and the total read of the transcripts for the GO exposure group was 20
711 and the total read of the transcripts for the GO exposure group was 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 776
776![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 091. Among these reads, 99.11% of the total reads of transcripts for the control group and 98.63% of the total reads of transcripts for the GO exposure group could be mapped. The number of mapped bases for the control group was 1
091. Among these reads, 99.11% of the total reads of transcripts for the control group and 98.63% of the total reads of transcripts for the GO exposure group could be mapped. The number of mapped bases for the control group was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130
130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 145
145![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 261 and the number of mapped bases for the GO exposure group was 1
261 and the number of mapped bases for the GO exposure group was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 059
059![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 580
580![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 641. The length distribution of sequences of the protein coding region prediction (CDS) is shown in Fig. 2b. Among the CDS, approximately 14% was less than 1000 bp in length, 29% was between 1000 and 2000 bp in length, 19% was between 2000 and 3000 bp in length, 12% was between 3000 and 4000 bp in length, 7% was between 4000 and 5000 bp in length, 5% was between 5000 and 6000 bp in length, and 15% was greater than 6000 bp in length (Fig. 2b). The distribution of CDS on the chromosomes is shown in Fig. 2c. Among the CDS, approximately 17% was distributed on chromosome I, II, or IV, 16% was distributed on chromosome III, 19% was on chromosome V, and 14% was on chromosome X (Fig. 2c).
641. The length distribution of sequences of the protein coding region prediction (CDS) is shown in Fig. 2b. Among the CDS, approximately 14% was less than 1000 bp in length, 29% was between 1000 and 2000 bp in length, 19% was between 2000 and 3000 bp in length, 12% was between 3000 and 4000 bp in length, 7% was between 4000 and 5000 bp in length, 5% was between 5000 and 6000 bp in length, and 15% was greater than 6000 bp in length (Fig. 2b). The distribution of CDS on the chromosomes is shown in Fig. 2c. Among the CDS, approximately 17% was distributed on chromosome I, II, or IV, 16% was distributed on chromosome III, 19% was on chromosome V, and 14% was on chromosome X (Fig. 2c).
Transcriptomic changes in nematodes exposed to GO
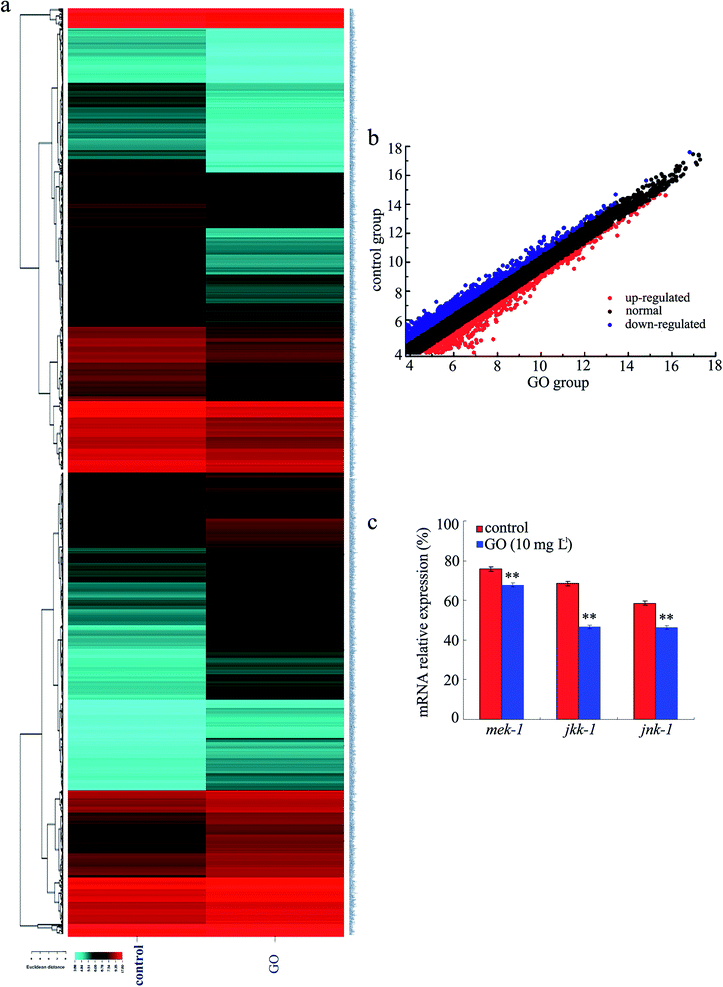
The dysregulated expression of mRNAs in GO-exposed nematodes was determined using fold change analysis and further developed for analysis based on statistical significance and the use of a two-fold change cutoff. Annotations of the differentially expressed genes were acquired by comparing the detected mRNA sequences with the databases of Genbank (Fig. 3a, Table S1†). Among the 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 752 genes examined, the transcriptional expression of 11
752 genes examined, the transcriptional expression of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 787 genes was not significantly altered upon GO exposure. In contrast, we identified a total of 1965 differentially expressed mRNAs in GO-exposed nematodes compared with the control (Fig. 3b, Table S1†). Among these 1965 mRNAs, 970 mRNAs were up-regulated, and 995 mRNAs were down-regulated (Fig. 3b, Table S1†). Thus, the expression patterns of the mRNAs were globally influenced by GO exposure in nematodes.
787 genes was not significantly altered upon GO exposure. In contrast, we identified a total of 1965 differentially expressed mRNAs in GO-exposed nematodes compared with the control (Fig. 3b, Table S1†). Among these 1965 mRNAs, 970 mRNAs were up-regulated, and 995 mRNAs were down-regulated (Fig. 3b, Table S1†). Thus, the expression patterns of the mRNAs were globally influenced by GO exposure in nematodes.
Our previous studies have demonstrated that the formation of GO toxicity may be due to the combined effects of induced oxidative stress, altered intestinal permeability, and deficit in defecation behavior in nematodes.24,31 Interestingly, in the list of dysregulated mRNAs induced by GO exposure, we found some genes associated with the control of oxidative stress, intestinal development, or defecation behavior (Table S1†). Based on the Illumina HiSeq™ 2000 sequencing data, the dysregulated genes associated with the control of oxidative stress were sod-1, sod-2, sod-3, sod-4, isp-1, gas-1, and clk-1, the dysregulated genes associated with the control of intestinal development were nhx-2, par-6, and pkc-3 and the dysregulated genes associated with the control of defecation behaviors were hlh-8, unc-93, fat-2, fat-3, unc-44, smp-1, itr-1, cab-1, mig-2, ced-10, unc-101, and egl-36 (Table S1†). The dysregulation of the genes associated with the control of oxidative stress, intestinal development and function, or defecation behavior was confirmed by our previous study based on an assay of the quantitative real-time polymerase chain reaction (qRT-PCR).24
Confirmation of the dysregulated genes in GO-exposed nematodes
Among the dysregulated genes induced by GO exposure, we further found that the transcriptional expressions of mek-1, jkk-1, and jnk-1 genes were decreased in GO-exposed nematodes compared with the control (Table S1†). We further used the qRT-PCR technique to confirm the Illumina HiSeq™ 2000 sequencing data for expression patterns of mek-1, jkk-1, and jnk-1 genes in GO (10 mg L−1) exposed nematodes. After GO exposure, the transcriptional expressions of mek-1, jkk-1, and jnk-1 genes were significantly decreased (Fig. 3c). Therefore, the qRT-PCR results for the expression patterns of genes encoding the JNK signaling pathway were similar to those from Illumina HiSeq™ 2000 sequencing data.Biological processes mediated by the dysregulated genes in GO-exposed nematodes based on a gene ontology term assay
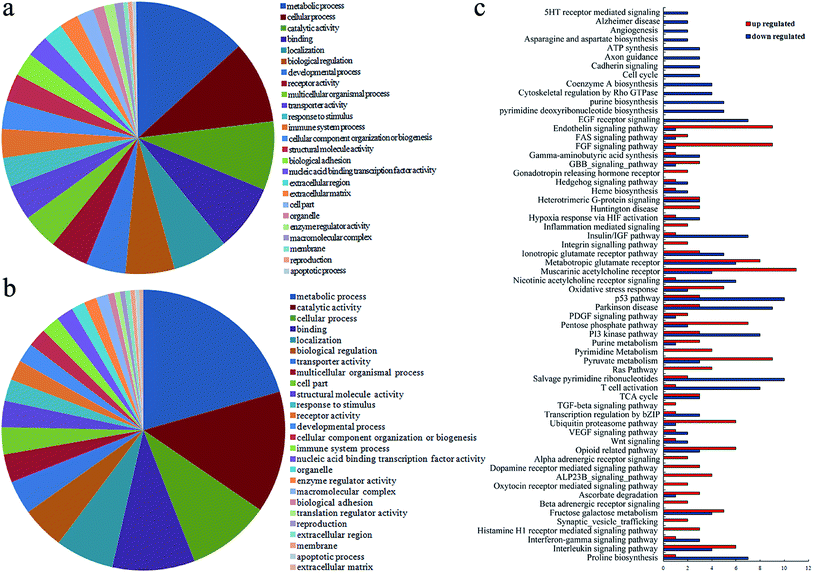
Gene ontology analysis can be used to provide the ontology of defined terms and gene product properties in terms of their associated biological processes, cellular components, and molecular functions.32 Based on the dysregulated mRNAs, the biological processes involved in the in vivo GO toxicity were evaluated in the present study (Tables S2 and S3†). The significantly affected gene ontology terms were classified into several categories, which were at least involved in the biological processes of development, reproduction, cell adhesion, apoptosis, enzyme activity, cellular component, cellular localization and transportation, response to stimulus, immune response, and cell metabolism (Fig. 4a and b). This information is helpful for understanding the function of the dysregulated genes in GO-exposed nematodes.The signaling pathways mediated by the dysregulated genes in GO-exposed nematodes based on KEGG analysis
KEGG pathway mapping, a bioinformatics resource, can be used to map the molecular data sets in genomics,33 and the related signaling pathways were extracted by the pathway mining tool. To further examine the molecular basis of mRNAs differentially expressed in GO-exposed nematodes, we used the KEGG pathway database to identify the related signal pathways mediated by the dysregulated mRNAs. We identified 50 signaling pathways for up-regulated miRNAs and 45 signaling pathways for down-regulated mRNAs in GO-exposed nematodes (Tables S4 and S5†). The signaling pathways possibly influenced by in vivo GO exposure mainly contained the signaling pathways related to development, cell cycle, cell death, neuronal degeneration, transcription regulation, stress response, cellular component, vesicle transportation, immune response, neuronal development, and cell metabolism (Fig. 4c). Signaling pathway-based annotation and analysis will be further helpful for understanding the roles of the dysregulated genes in regulating GO toxicity in nematodes.The mutations of the gene encoding JNK signaling pathways altered by GO toxicity in nematodes
In nematodes, GO exposure altered the expression patterns of the genes (mek-1, jkk-1, and jnk-1) in the JNK signaling pathway as indicated above. To examine whether the gene encoding JNK signaling pathways are involved in the control of GO toxicity in nematodes, we employed some endpoints to investigate the effects of mutations in the gene encoding JNK signaling pathways on GO toxicity. In C. elegans, both reproductive organs and neurons are the important secondary targeted organs for toxicants, including ENMs.34–36 We selected the endpoints of brood size, head thrash, and body bend to investigate the effects of mutations in the gene encoding JNK signaling pathways on the function of secondary targeted organs in GO-exposed nematodes. After GO exposure, the mek-1, jkk-1, and jnk-1 mutants showed more severe decreases in both brood size and locomotion behavior compared with wild-type N2 (Fig. 5).In C. elegans, the intestine is the primary targeted organ for toxicants, including ENMs.34,37 We next selected the endpoint of intestinal ROS production to investigate the effects of mutations in the gene encoding JNK signaling pathways on functions of primary targeted organs in GO-exposed nematodes. After GO exposure, the mek-1, jkk-1, and jnk-1 mutants showed a more severe induction of intestinal ROS production compared with wild-type N2 (Fig. 6). These results suggest that the gene encoding JNK signaling pathways are involved in the control of GO toxicity in nematodes.
Genetic interactions of the gene encoding JNK signaling pathways in regulating GO toxicity in nematodes
In C. elegans, the mek-1 and jkk-1 genes encode MAP kinase kinases, homolog of human MKK-7a, act as an activator of JNK. The jnk-1 gene encodes a serine/threonine kinase, homolog of human JNK, serves as the sole member of the JNK subgroup of MAP kinase. We first investigated whether mek-1 and jnk-1 may function in the same genetic pathway for regulating GO toxicity in nematodes. With the aid of brood size, locomotion behavior, and intestinal ROS production as the endpoints, after GO exposure, we found that the jnk-1(gk7);mek-1(ks54) double mutants showed similar brood size, locomotion behavior, or intestinal ROS production to that found in the mek-1(ks54) or jnk-1(gk7) single mutants (Fig. 5 and 6). Similarly, after GO exposure, we found that the jnk-1(gk7);jkk-1(km2) double mutants showed similar brood size, locomotion behavior, or intestinal ROS production to that found in the jkk-1(km2) or jnk-1(gk7) single mutants (Fig. 5 and 6). Therefore, the MEK-1 and JKK-1 genes may act genetically in the same pathway with JNK-1 to regulate GO toxicity in nematodes.The miRNAs–mRNAs network involved in the control of GO toxicity
In C. elegans, miRNAs regulate biological processes through their multiple targeted genes.28,38,39 Our previous study has suggested that GO induced some dysregulated miRNAs in exposed nematodes.27 The dysregulated miRNAs induced by GO exposure contained mir-259, mir-1820, mir-36, mir-82, mir-239, mir-246, mir-247, mir-392, mir-4806, mir-2217, mir-360, mir-4810, mir-4807, mir-1822, mir-4805, mir-800, mir-1830, mir-236, mir-4806, mir-244, mir-235, mir-4937, mir-4812, mir-43, mir-1834, mir-231, mir-5546, mir-42, mir-2214, mir-2210, and mir-73.27 Based on the bioinformatics analysis, among these miRNAs, mir-259, mir-1820, mir-36, mir-82, mir-239, mir-246, mir-392, mir-2217, mir-360, mir-4810, mir-4807, mir-4805, mir-800, mir-1830, mir-236, mir-4806, mir-244, mir-235, mir-4812, mir-43, mir-231, mir-42, mir-2210, and mir-73 may be involved in the control of GO toxicity through the function of dysregulated genes in GO-exposed nematodes (Table 1). More interestingly, we found that the gas-1 gene may serve as the molecular target for mir-4810 to regulate oxidative stress in GO-exposed nematodes (Table 1). The par-6 gene may serve as the molecular target for mir-1820 to regulate intestinal development and function in GO-exposed nematodes (Table 1). In addition, mir-231, mir-236, mir-259, mir-36, mir-42, mir-43, mir-73, mir-82, and mir-4805 may act through the function of the unc-44, mig-2, itr-1, ced-10, unc-93, fat-2, smp-1, and/or cab-1 gene(s) to regulate defecation behavior in GO-exposed nematodes (Table 1). These results suggest that a miRNAs–mRNAs network may exist and function to regulate GO toxicity in nematodes. This miRNAs–mRNAs network may regulate GO toxicity by influencing the induction of oxidative stress, intestinal development and function, and defecation behavior in nematodes.| a The blue color indicates genes associated with the control of oxidative stress, the purple color indicates genes associated with the control of intestinal development, and the red color indicates genes associated with the control of defecation behavior. The underlines indicate the genes encoding the JNK signaling pathway. |
|---|
 |
The genetic interaction of mir-73 and sek-1 in regulating GO toxicity in nematodes
To confirm that the miRNAs–mRNAs network was involved in the control of GO toxicity, we selected mir-73 and sek-1 to investigate the possible genetic interaction of mir-73 and sek-1 in regulating GO toxicity in nematodes. The data mentioned above imply that the sek-1 gene may be the targeted gene for mir-37 in GO-exposed nematodes (Table 1). In C. elegans, the sek-1 gene encodes a mitogen-activated protein kinase kinase (MAPKK), a member of the p38 MAPK signaling pathway. With the aid of locomotion behavior as the endpoint, we found that the mir-73 mutants were resistant to GO toxicity. However, the sek-1 mutants were susceptible to GO toxicity (Fig. 7). Moreover, we observed that the locomotion behavior in the GO-exposed double mutant of mir-73;sek-1 was similar to that found in the GO-exposed sek-1 mutant (Fig. 7), i.e. the sek-1 mutation may suppress the phenotype of the GO-exposed mir-73 mutant in nematodes, which suggests the important role of sek-1 in acting as the molecular target for mir-73 in GO-exposed nematodes.In the present study, we first employed the HiSeq 2000 sequencing technique to investigate the possible dysregulated mRNAs in GO-exposed nematodes. Based on the HiSeq 2000 sequencing data, we identified 970 up-regulated and 995 down-regulated mRNAs in the GO-exposed nematodes (Fig. 3b, Table S1†). Gene ontology analysis suggested that these dysregulated mRNAs may be at least involved in the biological processes of development, reproduction, cell adhesion, apoptosis, enzyme activity, cellular component, cellular localization and transportation, response to stimulus, immune response, and cell metabolism in nematodes (Fig. 4a and b). KEGG pathway analysis further indicated that these dysregulated mRNAs may at least mediate the signaling pathways related to development, cell cycle, cell death, neuronal degeneration, transcription regulation, stress response, cellular component, vesicle transportation, immune response, neuronal development, and cell metabolism in nematodes (Fig. 4c). Based on the KEGG pathway analysis, the influenced signaling pathways associated with the development contain Wnt, integrin, EGF, TGF-beta, Ras, heterotrimeric G-protein, cadherin, BMP, PI3 kinase, insulin/IGF, oxytocin receptor, and toll receptor signaling pathways; the influenced signaling pathways associated with the cell death or cell cycle contain ubiquitin proteasome, p53, and apoptosis signaling pathways; the influenced signaling pathways associated with the metabolism contain pyruvate metabolism, asparagine and aspartate biosynthesis, TCA cycle, purine metabolism, pentose phosphate metabolism, heme biosynthesis, pyrimidine deoxyribonucleotide biosynthesis, co-enzyme A biosynthesis, ATP synthesis, and glycolysis signaling pathways; the influenced signaling pathways associated with the neuronal development, degeneration, or function contain Huntington disease, Parkinson's disease, Alzheimer disease, axon guidance, synaptic vesicle trafficking, nicotinic acetylcholine receptor, muscarinic acetylcholine receptor, glutamate receptor, 5HT receptor, and dopamine receptor signaling pathways; and the influenced signaling pathways associated with the stress response or immunity contain hypoxia response, oxidative stress response, inflammation mediated by chemokine and cytokine, and interleukin signaling pathways (Tables S4 and S5†). The obtained data on dysregulated mRNAs induced by GO exposure in nematodes are largely consistent with the data on dysregulated proteins or mRNAs induced by GO in other biological assay systems.12,13,40 Based on the micro-array data, the KEGG analysis suggested that the dysregulated signaling pathways caused by GO exposure contained ribosome, metabolism of xenobiotics by cytochrome P450, drug metabolism, Alzheimer's disease, Huntington's disease, TGF-beta, cytokine–cytokine receptor interaction, cell adhesion molecules, ECM–receptor interaction, proteasome, hormone recombination, and non-hormone end-joining in HepG2 cells.13 Based on the proteome analysis data, 30 differentially expressed proteins involved in metabolic pathways, redox regulation, cytoskeleton formation, and cell growth were identified in GO-exposed HepG2 cells.12
Our previous studies demonstrated that miRNAs may also play an important role in regulating the toxicity and translocation of ENMs in nematodes.27,41 In organisms, because miRNAs function through their targeted genes to regulate the biological processes,28,38,39 we hypothesize that the dysregulated miRNAs may act through the function of some dysregulated mRNAs to regulate the toxicity and translocation of GO in exposed nematodes. In C. elegans, we have suggested a miRNAs–mRNAs network, which is potentially involved in the control of GO toxicity and translocation in nematodes. Among the dysregulated miRNAs induced by GO exposure, our bioinformatics analysis suggests that most of them may regulate the GO toxicity with the dysregulated mRNAs as their targeted genes in GO-exposed nematodes (Table 1). Our study establishes an important link between miRNAs and mRNAs for the molecular mechanism of nanotoxicity formation in organisms. More importantly, our data imply that the identified miRNAs–mRNAs network may further regulate GO toxicity through influencing the signaling pathways associated with the development, cell death or cell cycle, metabolism, neuronal development, degeneration and function, stress response, or immunity as mentioned above. In other words, the dysregulated miRNAs induced by GO exposure may result in the dysregulated expression of targeted genes, which will in turn further induce the dysregulated signaling pathways and lead to the formation of abnormal development, cell death or cell cycle, metabolism, neuronal development, degeneration and function, stress response, or immunity in animals. In addition, considering the fact that the GO exposure was performed from L1-larvae to adult day-1, transcriptomic fluctuations may also reflect the adaptation of GO to a certain degree. However, because the concentration for GO examined was 10 mg L−1, a relatively high dose for nematodes,24 most of the transcriptomic fluctuations may reflect the direct response to GO toxicity. Our results provide an important basis for further elucidating the molecular mechanisms of toxicity and translocation of GO in organisms.
Our previous studies have suggested that the formation of GO toxicity may be largely due to the combined effects of oxidative stress, increased intestinal permeability, and abnormal defecation behavior in nematodes.24–26 We further found that some dysregulated miRNAs, including mir-4810, mir-1820, mir-231, mir-236, mir-259, mir-36, mir-42, mir-43, mir-73, mir-82, and mir-4805, can function with the dysregulated genes associated with the control of oxidative stress, intestinal development and function, or defecation behavior as targeted genes to regulate the GO toxicity in exposed nematodes. Therefore, the miRNAs–mRNAs network suggested here explains the molecular basis for the crucial roles of oxidative stress, intestinal development and function, and defecation behavior in regulating GO toxicity in nematodes.
Previous studies have implied the important role of the JNK signaling pathway in regulating stress response in nematodes.29,30,42 For example, the mutants of genes encoding the JNK signaling pathway are hypersensitive to copper and cadmium.29,42 In the present study, we provide evidence to show the possible function of the JNK signaling pathway in regulating the formation of toxicity due to ENMs. Our results demonstrate that mutation of the jkk-1, mek-1, or jnk-1 gene caused the hypersensitive property to GO toxicity in nematodes as reflected by the endpoints of reproduction, locomotion behavior, and intestinal ROS production (Fig. 5 and 6). Previous studies have implied that at least two different JNK signaling pathways mediated by JKK-1 and MEK-1 exist in C. elegans.42 Our genetic data further imply that JKK-1 and JNK-1, as well as MEK-1 and JNK-1, function in the same genetic pathway to regulate GO toxicity in nematodes as reflected by the endpoints of reproduction, locomotion behavior, and intestinal ROS production (Fig. 5 and 6). Furthermore, we also found that mutation of the sek-1 gene caused a susceptible property of nematodes to GO toxicity (Fig. 7). Moreover, GO exposure down-regulated the expression level of pmk-1 gene encoding MAPK (Table S1†). sek-1 and pmk-1 are key components for the p38 MAPK signaling pathway in nematodes. Therefore, our data indicate the crucial in vivo role of the JNK and p38 MAPK signaling pathways in regulating GO toxicity in nematodes.
Conclusions
With the aid of a HiSeq 2000 sequencing technique, we first examined the dysregulated mRNAs in GO-exposed nematodes and identified 970 up-regulated and 995 down-regulated mRNAs induced by GO exposure. Both gene ontology analysis and KEGG pathway analysis implied that the dysregulated mRNAs may mediate many important biological processes. Some dysregulated genes encoding the JNK signaling pathway were proven to be involved in the control of GO toxicity. Moreover, we have suggested a miRNAs–mRNAs network, which explains the molecular basis for roles of oxidative stress, intestinal development and function, and defecation behavior in regulating in vivo GO toxicity. The established link between miRNAs and mRNAs provides the key basis for further elucidating the molecular mechanisms of GO toxicity in organisms.Experimental
Preparation and characterizations of GO
GO was prepared as described previously.43,44 After addition of graphite (2 g) and sodium nitrate (1 g), concentrated H2SO4 (50 mL) was slowly added with stirring for 30 min on ice into a 250 mL flask. Then, KMnO4 (7 g) was added to the mixture over 1 h. After the temperature of the mixture was warmed to 35 °C, 90 mL of H2O was slowly added dropwise to cause an increase in temperature to 70 °C. The diluted suspension was stirred at 70 °C for 15 min and further treated with a mixture of 7 mL of 30% H2O2 and 55 mL of H2O. The resulting warm suspension was filtered to obtain a yellow-brown filter cake. The yellow-brown filter cake was washed with a solution of 3% HCl, followed by drying at 40 °C for 24 h. GO was finally obtained by ultrasonication of the as-made graphite oxide in water for 1 h. All the other chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA).The prepared GO was characterized by TEM (JEM-200CX, JEOL, Japan) and AFM (SPM-9600, Shimadzu, Japan). The zeta potential was analyzed with a Nano Zetasizer using a dynamic light scattering (DLS) technique.
C. elegans maintenance
The nematodes used were wild-type N2 and mutants of jnk-1(gk7), jkk-1(km2), mek-1(ks54), sek-1(km4), mir-73(nDf47), and mir-73(nDf47);sek-1(km4), which were maintained on nematode growth medium (NGM) plates seeded with Escherichia coli OP50 at 20 °C as described previously.14 Gravid nematodes were washed off the plates into centrifuge tubes, and further lysed with a bleaching mixture (0.45 mol L−1 NaOH, 2% HOCl). Age synchronous populations of the L1-larvae were obtained as described previously.45GO exposure
GO exposure was performed from L1-larvae to adult day-1 at 20 °C in 12-well sterile tissue culture plates. The GO exposure concentration was 10 mg L−1.27 After exposure, nematodes were used for the toxicity assessment with lifespan, reproduction, locomotion behavior, and intestinal ROS production as the endpoints.RNA-seq library preparation and HiSeq 2000 sequencing
Total RNA was isolated from wild-type N2 nematodes exposed to 10 mg L−1 of GO or without GO exposure using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). Contaminating DNA was removed with Ambion TURBO DNA-free kit (Applied Biosystems, Austin, TX, USA). RNA quality was determined with a Nano Photometer P-Class. mRNA libraries were established using a RNA-seq sample preparation kit (Illumina, Inc., San Diego, CA, USA). An Illumina HiSeq™ 2000 sequencing platform was used to obtain 22.1 million and 20.7 million 100-nucleotide paired-end reads for the control and GO exposure group mRNAs, respectively. Three replicates were performed.RNA-seq data analysis
The quality of the reads was checked with fast QC. The draft genome of C. elegans (version WS220, Release 62, Ensembl, fttp://ftp.ensembl.org/pub/release-62/fasta/caenorhabditis_elegans/dna/) was used for reference-guided mapping of the transcriptome sequencing reads with TopHat 1.3.1 that uses Bowtie and SAMtools 0.1.16. The total read numbers of the control and GO exposure group data sets were normalized to equal levels and the relative gene abundance was defined by log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 of the normalized read number. Transcripts with false discovery rate-corrected p-values < 0.05 and fold change > 2 were defined as differentially expressed.
10 of the normalized read number. Transcripts with false discovery rate-corrected p-values < 0.05 and fold change > 2 were defined as differentially expressed.
Pathway analysis
Differentially expressed genes (DEGs) were performed using the default parameters to adjust the p value with the whole genome set as the background. Enriched gene ontology terms were generated using the gene ontology database (http://www.Geneontology.org/) based on blast with C. elegans database as the reference background. Gene ontology analysis and enrichment were found separately for the up-regulated and down-regulated genes with the DAVID functional annotation tool. The KEGG orthology database (http://www.genome.jp/kegg/ko.html) was used for pathway mapping. The pathway analysis was performed using the KEGG pathway exploiting the DAVID database.Bioinformatics analysis for the miRNAs–mRNAs network
The corresponding miRNA(s) for the dysregulated genes were predicted using TargetScan version 6.2 (http://www.targetscan.org/worm_52/). TargetScan can be used to predict the biological targets of miRNAs by searching for the presence of conserved sites that match the seed region of each miRNA. We searched whether the dysregulated genes are the possible biological targets of dysregulated miRNAs caused by GO exposure.27Reverse-transcription and qRT-PCR
Total RNA was isolated from nematodes using Trizol (Invitrogen, UK). RNA purities and concentrations were assessed by OD260/280 in a spectrophotometer. cDNA synthesis was finished in a 12.5 μL reaction volume containing 625 ng total RNA, 0.5 mmol L−1 reverse-transcript primers, 10 mmol L−1 dithiothreitol, 50 mmol L−1 Tris–HCl, 75 mmol L−1 KCl, 3 mmol L−1 MgCl2, 20 units ribonuclease inhibitor, and 100 U reverse transcriptase (Takara, China). Relative expression levels were determined by real-time PCR in an ABI 7500 real-time PCR system with Evagreen (Biotium, USA). The 20 μL PCR reaction system included cDNA (1 μL), SYBR Green I Master Mix (8 μL), SYBR Green I Master Plus (2 μL), forward primer (1.2 μL), reverse primer (1.2 μL), and ddH2O (6.6 μL). The reactions were incubated at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, annealing temperature for 30 s, and 72 °C for 30 s. After amplification, the fluorescent data were converted to threshold cycle values (Ct). All reactions were performed in triplicate with the aid of the same cDNA samples. Targeted gene transcript levels were normalized against the reference gene (tba-1) transcript levels. The final results were expressed as the relative expression ratio between targeted gene and reference gene. The primer information is shown in Table S6.†Toxicity assessment
Lifespan assay was performed as described previously.46,47 Hermaphrodite nematodes were transferred daily for the first 4 days of adulthood. Nematodes were checked everyday and scored as dead if they did not move after repeated taps with a pick. Forty nematodes were examined per treatment. The graphs are representative of three trials.Reproduction was evaluated by the endpoint of brood size. The method was performed as previously described.48,49 The off-spring number at all stages beyond the egg was counted. Twenty nematodes were examined per treatment. Three replicates were performed.
Locomotion behavior was assessed by the endpoints of head thrash and body bend. The methods were performed as previously described.50,51 A head thrash was defined as a change in direction of bending at the mid-body. A body bend was defined as a change in direction of the part of the nematode corresponding to the posterior bulb of the pharynx along the y-axis, assuming that the nematode was traveling along the x-axis. Twenty nematodes were examined per treatment. Three replicates were performed.
The method for ROS production was performed as previously described.29 Nematodes were transferred to a solution of 1 μmol L−1 5′,6′-chloromethyl-2′,7′-dichlorodihydro-fluorescein diacetate (CM-H2DCFDA) and incubated for 3 h in 12-well sterile tissue culture plates at 20 °C. Nematodes mounted on 2% agar pads were examined with a laser scanning confocal microscope (Leica, TCS SP2, Bensheim, Germany) with an excitation wavelength of 488 nm and emission filter at 510 nm. The relative fluorescent intensity of the intestine was semi-quantified. Fifty nematodes were examined per treatment. Three replicates were performed.
Statistical analysis
All data were expressed as the mean ± standard error of the mean (S.E.M.). Statistical analysis was performed using SPSS 12.0 software (SPSS Inc., Chicago, USA). The differences between the groups were analyzed using the analysis of variance (ANOVA). Probability levels of 0.05 and 0.01 were considered to be statistically significant. The graphs were generated using Microsoft Excel software (Microsoft Corp., Redmond, WA).Acknowledgements
This study was supported by grants from the National Basic Research Program of China (No. 2011CB933404), the National Natural Science Foundation of China (No. 81202233, 81172698), the Jiangsu Province Ordinary University Graduate Research and Innovation Program (No. CXZZ13_0136), and the Fundamental Research Funds for the Central University.References
- K. Yang, L. Feng, X. Shi and Z. Liu, Chem. Soc. Rev., 2013, 42, 530–547 RSC.
- D. Bitounis, H. Ali-Boucetta, B. H. Hong, D. Min and K. Kostarelos, Adv. Mater., 2013, 25, 2258–2268 CrossRef CAS PubMed.
- J. Liu, S. Yang, H. Wang, Y. Chang, A. Cao and Y. Liu, Nanomedicine, 2012, 7, 1801–1812 CrossRef CAS PubMed.
- B. Li, J. Yang, Q. Huang, Y. Zhang, C. Peng, Y. Zhang, Y. He, J. Shi, W. Li, J. Hu and C. Fan, NPG Asia Mater., 2013, 5, e44 CrossRef CAS.
- X. Zhi, H. Fang, C. Bao, G. Shen, J. Zhang, K. Wang, S. Guo, T. Wan and D. Cui, Biomaterials, 2013, 34, 5254–5261 CrossRef CAS PubMed.
- X. Hu and Q. Zhou, Chem. Rev., 2013, 113, 3815–3835 CrossRef CAS PubMed.
- Y. P. Li, Q. L. Wu, Y. L. Zhao, Y. F. Bai, P. S. Chen, T. Xia and D. Y. Wang, ACS Nano, 2014, 8, 2100–2110 CrossRef CAS PubMed.
- A. B. Seabra, A. J. Paula, R. de Lima, O. L. Alves and N. Duran, Chem. Res. Toxicol., 2014, 27, 159–168 CrossRef CAS PubMed.
- Y. Li, L. Feng, X. Shi, X. Wang, Y. Yang, K. Yang, T. Liu, G. Yang and Z. Liu, Small, 2014, 8, 1544–1554 CrossRef PubMed.
- Q. Q. Wang, S. Q. Zhao, Y. L. Zhao, Q. Rui and D. Y. Wang, RSC Adv., 2014, 4, 60891–60901 RSC.
- G. Qu, S. Liu, S. Zhang, L. Wang, X. Wang, B. Sun, N. Yin, X. Gao, T. Xia, J. Chen and G. Jiang, ACS Nano, 2013, 7, 5732–5745 CrossRef CAS PubMed.
- J. Yuan, H. Gao, J. Sui, H. Duan, W. N. Chen and C. B. Ching, Toxicol. Sci., 2012, 126, 149–161 CrossRef CAS PubMed.
- N. Chatterjee, H. Eom and J. Choi, Biomaterials, 2014, 35, 1109–1127 CrossRef CAS PubMed.
- S. Brenner, Genetics, 1974, 77, 71–94 CAS.
- M. C. Leung, P. L. Williams, A. Benedetto, C. Au, K. J. Helmcke, M. Aschner and J. N. Meyer, Toxicol. Sci., 2008, 106, 5–28 CrossRef CAS PubMed.
- Y.-L. Zhao, Q.-L. Wu, Y.-P. Li and D.-Y. Wang, RSC Adv., 2013, 3, 5741–5757 RSC.
- E. Zanni, G. de Bellis, M. P. Bracciale, A. Broggi, M. Santarelli, M. S. Sarto, C. Palleschi and D. Uccelletti, Nano Lett., 2012, 12, 2740–2744 CrossRef CAS PubMed.
- P. Chen, K. Hsiao and C. Chou, Biomaterials, 2013, 34, 5661–5669 CrossRef CAS PubMed.
- A. Nouara, Q.-L. Wu, Y.-X. Li, M. Tang, H.-F. Wang, Y.-L. Zhao and D.-Y. Wang, Nanoscale, 2013, 5, 6088–6096 RSC.
- C. M. Goodwin, G. G. Lewis, A. Fiorella, M. Ellison and R. Kohn, RSC Adv., 2014, 4, 5893–5900 RSC.
- C. J. Shu, X. M. Yu, Q. L. Wu, Z. H. Zhuang, W. M. Zhang and D. Y. Wang, RSC Adv., 2015, 5, 8942–8951 RSC.
- Y.-L. Zhao, Q. Liu, S. Shakoor, J. R. Gong and D.-Y. Wang, Toxicol. Res., 2015, 4, 270–280 RSC.
- W. Zhang, C. Wang, Z. Li, Z. Lu, Y. Li, J. Yin, Y. Zhou, X. Gao, Y. Fang, G. Nie and Y. Zhao, Adv. Mater., 2012, 24, 5391–5397 CrossRef CAS PubMed.
- Q.-L. Wu, L. Yin, X. Li, M. Tang, T. Zhang and D.-Y. Wang, Nanoscale, 2013, 5, 9934–9943 RSC.
- Q.-L. Wu, Y.-L. Zhao, J.-P. Fang and D.-Y. Wang, Nanoscale, 2014, 6, 5894–5906 RSC.
- Q.-L. Wu, Y.-L. Zhao, Y.-P. Li and D.-Y. Wang, Nanoscale, 2014, 6, 11204–11212 RSC.
- Q.-L. Wu, Y.-L. Zhao, G. Zhao and D.-Y. Wang, Nanomedicine: Nanotechnology, Biology and Medicine, 2014, 10, 1401–1410 CrossRef CAS PubMed.
- D. P. Bartel, Cell, 2004, 116, 281–297 CrossRef CAS.
- M. Koga, R. Zwaal, K. Guan, L. Avery and Y. Ohshima, EMBO J., 2000, 19, 5148–5166 CrossRef CAS PubMed.
- T. Mizuno, N. Hisamoto, T. Terada, T. Kondo, M. Adachi, E. Nishida, D. H. Kim, F. M. Ausubel and K. Matsumoto, EMBO J., 2004, 23, 2226–2234 CrossRef CAS PubMed.
- Y.-L. Zhao, Z.-Q. Lin, R.-H. Jia, G.-J. Li, Z.-G. Xi and D.-Y. Wang, J. Hazard. Mater., 2014, 274, 106–114 CrossRef CAS PubMed.
- E. Camon, M. Magrane, D. Barrell, D. Binns, W. Fleischmann, P. Kersey, N. Mulder, T. Oinn, J. Maslen, A. Cox and R. Apweiler, Genome Res., 2003, 13, 662–672 CrossRef CAS PubMed.
- M. Kanehisa, M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu and Y. Yamanishi, Nucleic Acids Res., 2008, 36, D480–D484 CrossRef CAS PubMed.
- Q.-L. Wu, Y.-X. Li, Y.-P. Li, Y.-L. Zhao, L. Ge, H.-F. Wang and D.-Y. Wang, Nanoscale, 2013, 5, 11166–11178 RSC.
- Y.-L. Zhao, X. Wang, Q.-L. Wu, Y.-P. Li and D.-Y. Wang, J. Hazard. Mater., 2015, 283, 480–489 CrossRef CAS PubMed.
- Y.-L. Zhao, X. Wang, Q.-L. Wu, Y.-P. Li, M. Tang and D.-Y. Wang, Toxicol. Res., 2015, 4, 399–408 RSC.
- R.-L. Yang, Y.-L. Zhao, X.-M. Yu, Z.-Q. Lin, Z.-G. Xi, Q. Rui and D.-Y. Wang, Toxicol. Res., 2015, 4, 333–343 RSC.
- Y.-O. Hu, Y. Sun, B.-P. Ye and D.-Y. Wang, Neurosci. Bull., 2007, 23, 9–20 CrossRef CAS PubMed.
- Y. Sun, Y.-N. Zhao and D.-Y. Wang, Neurosci. Bull., 2006, 22, 339–349 CAS.
- T. Nezakati, B. G. Cousins and A. M. Seifalian, Arch. Toxicol., 2014, 88, 1987–2012 CrossRef CAS PubMed.
- Y.-L. Zhao, Q.-L. Wu, Y.-P. Li, A. Nouara, R.-H. Jia and D.-Y. Wang, Nanoscale, 2014, 6, 4275–4284 RSC.
- A. Villanueva, J. Lozano, A. Morales, X. Lin, X. Deng, M. O. Hengartner and R. N. Kolesnick, EMBO J., 2001, 20, 5114–5128 CrossRef CAS PubMed.
- W. S. Hummers Jr and R. E. Offerman, J. Am. Chem. Soc., 1958, 80, 1339 CrossRef.
- N. I. Kovtyukhova, P. J. Olivier, B. R. Martin, T. E. Mallouk, S. A. Chizhik, E. V. Buzaneva and A. D. Gorchinskiy, Chem. Mater., 1999, 11, 771–778 CrossRef CAS.
- S. G. Donkin and D. B. Dusenbery, Arch. Environ. Contam. Toxicol., 1993, 25, 145–151 CrossRef CAS.
- D. Y. Wang, P. D. Liu, Y. C. Yang and L. L. Shen, Ecotoxicol. Environ. Saf., 2010, 73, 1221–1230 CrossRef CAS PubMed.
- W.-M. Zhang, T. Lv, M. Li, Q.-L. Wu, L.-S. Yang, H. Liu, D.-F. Sun, L.-M. Sun, Z.-H. Zhuang and D.-Y. Wang, PLoS One, 2013, 8, e74553 CAS.
- D. Y. Wang and X. J. Xing, Ecotoxicol. Environ. Saf., 2010, 73, 423–429 CrossRef CAS PubMed.
- Q. Rui, Y.-L. Zhao, Q.-L. Wu, M. Tang and D.-Y. Wang, Chemosphere, 2013, 93, 2289–2296 CrossRef CAS PubMed.
- Y. Qiao, Y.-L. Zhao, Q.-L. Wu, L.-M. Sun, Q.-L. Ruan, Y.-Y. Chen, M. Wang, J.-A. Duan and D.-Y. Wang, PLoS One, 2014, 9, e91825 Search PubMed.
- Q.-L. Wu, Y.-L. Zhao, Y.-P. Li and D.-Y. Wang, Nanomedicine: Nanotechnology, Biology and Medicine, 2014, 10, 1263–1271 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra16142h |
| This journal is © The Royal Society of Chemistry 2015 |