Enhanced antibacterial activity of new “composite” biocides with both N-chloramine and quaternary ammonium moieties†
Chenxi Ning‡
a,
Lingdong Li‡b,
Sarvesh Logsettyc,
Sadegh Ghanbard,
Melinda Guoe,
Werner Ensf and
Song Liu*adgh
aDepartment of Textile Sciences, Faculty of Human Ecology, University of Manitoba, Winnipeg, Canada R3T 2N2. E-mail: Song.Liu@umanitoba.ca; Fax: +1-204-474-7593; Tel: +1-204-474-9616
bSchool of Petroleum & Chemical Engineering, Dalian University of Technology (Panjin Campus), No.2 Dagong Road, Liaodongwan New District, Panjin, PR China 124221
cManitoba Firefighters Burn Unit, Department of Surgery, Faculty of Medicine, University of Manitoba, Winnipeg, Canada R3A 1R9
dDepartment of Chemistry, Faculty of Science, University of Manitoba, Winnipeg, Canada R3T 2N2
eDepartment of Chemistry, Faculty of Arts & Science, University of Toronto, Toronto, Ontario, Canada M5S 2J7
fDepartment of Physics, Faculty of Science, University of Manitoba, Winnipeg, Canada R3T 2N2
gDepartment of Biosystems Engineering, Faculty of Agricultural and Food Sciences, University of Manitoba, Winnipeg, Canada R3T 2N2
hDepartment of Medical Microbiology, Faculty of Medicine, University of Manitoba, Winnipeg, Canada R3A 1R9
First published on 23rd October 2015
Abstract
In view of the emerging resistance in bacteria against biocides, this work describes a novel combination of two existing biocides in one molecule to improve the bactericidal activity and overcome bacterial resistance. A new series of “composite” biocides combining an amide based N-chloramine with a quaternary ammonium (QA) moiety in one molecule was synthesized and the antibacterial kinetics of each biocide was tested against two clinically retrieved bacteria: methicillin-resistant S. aureus (MRSA) and multi-drug resistant (MDR) P. aeruginosa. The addition of multiple cationic centers into one N-chloramine molecule did not result in enhanced inactivation of bacteria. The bactericidal activity against both microbes increased dramatically when the length of the alkyl chain of QA moiety in these biocides increased to 12 and 14. Covalently bonding N-chloramine with long-chained QA moieties did result in faster kill of MRSA and MDR P. aeruginosa than the formulation with two separate counterparts (N-chloramine and long-chained QA salts). Uptake isotherm curves of the “composite” biocides with long alkyl chain substitution revealed more uptakes of the “composite” biocides by bacteria than the precursor mono-functional biocides (with only QA moieties). An improved antibacterial activity resulted from covalently bonding the two biocides (N-chloramine and long-chained QA salts) into one molecule.
Introduction
There are growing concerns over Healthcare-Associated Infections (HAIs), particularly those caused by antibiotic-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) and multidrug resistant Pseudomonas aeruginosa.1,2 The overall incidence rates of MRSA in Canadian hospitals have increased 19-fold since 1995 from 0.44 per 1000-patient admission to 8.62 in 2007.3,4 Rising infection rates are leading to unnecessary suffering and death. Each year in Canada, around 8000 deaths occur, with more than 220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 HAI occurrences.5 Strong correlation between hospital environmental hygiene and the rates of HAIs has been reported.6–9 Doorknobs, privacy curtains, bed-frames and lockers, garments of healthcare workers, and surfaces of various shared facilities such as wheelchairs and toilets are all important “media” in transmission of microorganisms from one person to another.10–13 Studies have shown that organisms have a capacity to survive on various environmental surfaces including fabrics, plastics and metals, and some can even stay viable for up to 30 months in the absence of adequate cleaning and disinfection.14–16 These environmental contaminations pose a direct risk to the occupants in the environment, acting as reservoirs for microbial pathogens and causing cross-infections.
000 HAI occurrences.5 Strong correlation between hospital environmental hygiene and the rates of HAIs has been reported.6–9 Doorknobs, privacy curtains, bed-frames and lockers, garments of healthcare workers, and surfaces of various shared facilities such as wheelchairs and toilets are all important “media” in transmission of microorganisms from one person to another.10–13 Studies have shown that organisms have a capacity to survive on various environmental surfaces including fabrics, plastics and metals, and some can even stay viable for up to 30 months in the absence of adequate cleaning and disinfection.14–16 These environmental contaminations pose a direct risk to the occupants in the environment, acting as reservoirs for microbial pathogens and causing cross-infections.
Various disinfection strategies to maintain a good hospital environment have been developed, including use of conventional disinfectants and physical sterilization. Quaternary ammonium (QA) salts are well-known as efficacious biocides against microorganisms including bacteria, and fungi. Recent research has shown that the presence of heteroatoms (N, P, and S) tends to increase the antimicrobial activity of QA salts, achieving 10–17 mm of inhibition zone diameters against various bacterial strains and fungi when compared to 12.3 mm of cetyl trimethyl ammonium bromide.17,18 Activity of those QA salts also increased with increasing length of the hydrophobic alkyl chain which increases the adsorption to microorganisms' membranes and thus increases the relative efficiency of the molecules.17,18 However, there are continuous reports of the emergence of increasing resistance in bacteria against QA salts.19–21 Although the novel, gas-phase disinfection techniques like gas plasma have proven highly effective, they are mainly used for healthcare devices rather than surfaces and they can only be activated in the absence of people.22 Therefore, there is still an urgent need to develop safe and more potent broad-spectrum antimicrobial reagents that are less likely to induce bacterial resistance, in order to achieve a hygiene hospital environment.
Organic N-chloramines, a group of organic compounds bearing N–Cl bonds, are very comparable to hypochlorites in terms of bactericidal efficacy, but are safer, more stable and more resistant to organic matter than hypochlorites.23,24 N-Chloramines function as biocides in different ways: the initial attack results in chlorination of the external protein matrix of bacteria, forming a moderate chlorine cover which does not always result in cellular death; next following penetration of N-chloramines into bacterial cells, N-chloramines attack many vitally important constituents (such as enzymes) containing thiols and thioethers by oxidation, and denature proteins by transchlorination. These reactions cause the eventually death of the bacteria. N-Chloramines are less likely to induce bacterial resistance because they interact with vital proteins in bacteria in a variety of nonspecific ways. N-Chloramines have been used for a long time and no resistance has yet been reported. Nagl (as cited in ref. 25) stated that four strains of bacteria and C. albicans did not develop resistance to N-chlorotaurine after being exposed to 1% N-chlorotaurine for an extended period of time.
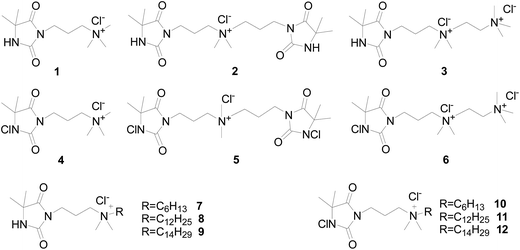
Recently, we introduced a positive charge (n-methyl QA) into an amide based N-chloramine structure and grafted it onto poly(ethylene terephthalate) (PET) and cotton. The introduced positive charge contributes to faster kill of two clinical bacterial strains – MDR-E. coli and MRSA.26 Inspired by the finding, we intended to study the antibacterial behavior of a series of biocides with both N-chloramine and QA moieties (both short- and long-chained) in aqueous solution. In the current study, we synthesized a series of “composite” molecules, as shown in Scheme 1, to study the effects of the ratio of N-chloramine/QA moiety (4–6) and the alkyl chain length of QA moiety (4, 10–12) on the antibacterial activity. Considering the different antibacterial modes of action of N-chloramines and long alkyl chain QA salts, we hypothesized a synergistic bactericidal effect from the two moieties when they are bonded together in one molecule.
Experimental section
Reagents and materials
Reagents and solvents were obtained from commercial suppliers such as Sigma, VWR or Fisher. All chemicals were analytical grade and used as received without further purification unless otherwise stated. Synthetic compounds were purified using flash column chromatography on silica gel obtained from Selecto Scientific Georgia USA. NMR spectra were recorded at room temperature in 5 mm NMR tubes on a Bruker Avance 300 MHz NMR spectrometer. Accurate mass measurements were performed using a PerkinElmer Sciex prOTOFTM 2000 MALDI-OTOF Mass Spectrometer. Community-associated (CA)-MRSA #40065 and multi-drug resistant (MDR) P. aeruginosa #73104 were used as the model microorganism to challenge all the biocides. Both were clinical strains were obtained from the CANWARD (Canadian Ward Surveillance) study assessing antimicrobial resistance in Canadian hospitals, http://www.canr.ca.Synthesis of new “composite” compounds
The chemical synthesis of new biocides is depicted in Scheme 2, except for compound 1 and 4 which were prepared according to previously published protocols.5 Codes were created to clearly describe the composition of each biocide. For example, biocide 6 was given the code of “C1 + 2QA + Chlor” since it consists of two methyl QA and one N-chloramine moieties; and biocide 7 was given the code of “C6 + QA + Hyd” since it is composed of one hexyl QA and one hydantoin (precursor of N-chloramine). The biocide numbers and their corresponding codes are listed in Table 1.| Categories | Biocide number | Biocide code | QA alkyl chain length | Ratio of N-chloramine (or precursor) to QA moiety | |
|---|---|---|---|---|---|
| N-Chloramine | With short-chained QA moiety | 4 | C1 + QA + Chlor | Methyl | 1 |
| 5 | C1 + QA + 2Chlor | Methyl | 2 | ||
| 6 | C1 + 2QA + Chlor | Methyl | 0.5 | ||
| With long-chained QA moiety | 10 | C6 + QA + Chlor | Hexyl | 1 | |
| 11 | C12 + QA + Chlor | Dodecyl | |||
| 12 | C14 + QA + Chlor | Tetradecyl | |||
| N-Chloramine precursor with long-chained QA moiety | 7 | C6 + QA + Hyd | Hexyl | ||
| 8 | C12 + QA + Hyd | Dodecyl | |||
| 9 | C14 + QA + Hyd | Tetradecyl | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) to give 15 as a white solid (0.7 g, 51%).
5, v/v) to give 15 as a white solid (0.7 g, 51%).1H NMR (D2O, 300 MHz, δ) 3.55 (t, J = 7.5 Hz, 2H), 2.65 (t, J = 7.5 Hz, 2H), 2.46 (s, 6H; N(CH3)2), 1.88 (m, 2H), 1.44 (s, 6H); 13C NMR (D2O, 75 MHz, δ) 181.0, 157.3, 58.8, 55.6, 43.6, 36.0, 24.3, 23.7; HRMS (MALDI-TOF) m/z: [M + H]+ calculated for C10H20N3O2+, 214.1556; found: 214.1555.
To the solution of 15 (0.25 g, 1.17 mmol) in 10 mL CH3CN was added bromide 13 (0.32 g, 1.1 equiv.). The reaction mixture was allowed to undergo reflux for 24 hours. Solvent was removed and the crude product was applied on chromatography column (MeOH/CH2Cl2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v) to give Br− form product, which was dissolved in a minimum amount of water and slowly passed through an anion-exchange resin (Amberlite R IRA-900, Cl− form) to afford 2 as white solid (0.46 g, 94%).
3, v/v) to give Br− form product, which was dissolved in a minimum amount of water and slowly passed through an anion-exchange resin (Amberlite R IRA-900, Cl− form) to afford 2 as white solid (0.46 g, 94%).
1H NMR (D2O, 300 MHz, δ) 3.6 (t, J = 6 Hz, 2H), 3.37 (t, J = 7.5 Hz, 2H), 3.12 (s, 3H), 2.10 (m, 2H), 1.45 (s, 6H); 13C NMR (D2O, 75 MHz, δ) 180.5, 157, 61.3, 59.2, 50.8, 35.3, 23.5, 21.3; HRMS (MALDI-TOF) m/z: [M − Cl]+ calculated for C18H32N5O4+, 382.2449; found: 382.2454.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v) to give 14 as yellowish oil (1.3 g, 76%).
3, v/v) to give 14 as yellowish oil (1.3 g, 76%).1H NMR (D2O, 300 MHz, δ) 3.61 (t, J = 6.0 Hz, 2H), 3.49 (t, J = 7.5 Hz, 2H), 3.41 (t, J = 6 Hz, 2H), 3.15 (s, 6H), 2.83 (t, J = 7.5 Hz, 2H), 2.30 (s, 6H), 2.09–2.18 (m, 2H), 1.45 (s, 6H); 13C NMR (CDCl3, 75 MHz, δ) 180.57, 157.04, 61.8, 60.7, 59.2, 53.5, 44.4, 43.7, 35.4, 23.6, 21.4; HRMS (MALDI-TOF) m/z: [M − Br]+ calculated for C14H29N4O2+, 285.2285; found: 285.2290.
To the solution of 14 (0.9 g, 3.15 mmol) in 30 mL mixed solvent CH3CN/CH3OH (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) was added excess methyl iodide (2 mL, 10 equiv.). The resulting solution was continuously stirred at room temperature for 22 hours before solvent and excess of methyl iodide were removed under vacuum. The oily residue was purified by column chromatography (MeOH/CH2Cl2, 1
1, v/v) was added excess methyl iodide (2 mL, 10 equiv.). The resulting solution was continuously stirred at room temperature for 22 hours before solvent and excess of methyl iodide were removed under vacuum. The oily residue was purified by column chromatography (MeOH/CH2Cl2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3–1
3–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) to give I form product as yellow solid. Then product was dissolved in minimum amount of water and slowly passed through an anion-exchange resin (Amberlite R IRA-900, Cl−) to afford 3 as white solid (0.87 g, 74%).
2, v/v) to give I form product as yellow solid. Then product was dissolved in minimum amount of water and slowly passed through an anion-exchange resin (Amberlite R IRA-900, Cl−) to afford 3 as white solid (0.87 g, 74%).
1H NMR (D2O, 300 MHz, δ) 4.05 (s, 4H), 3.64 (t, J = 7.5 Hz, 2H), 3.56 (t, J = 7.5 Hz, 2H), 3.33 (s, 9H), 3.28 (s, 6H), 2.22 (m, 2H), 1.46 (s, 6H); 13C NMR (CDCl3, 75 MHz, δ) 180.6, 157.1, 63.1, 59.2, 57.5, 56.3, 53.8, 35.2, 23.4, 21.4.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) to afford the corresponding Br form product (7, 90%; 8, 92% and 9, 88%), which was dissolved in a minimum amount of water and slowly passed through an anion-exchange resin (Amberlite R IRA-900, Cl−) to afford Cl− form product as white solid.
3) to afford the corresponding Br form product (7, 90%; 8, 92% and 9, 88%), which was dissolved in a minimum amount of water and slowly passed through an anion-exchange resin (Amberlite R IRA-900, Cl−) to afford Cl− form product as white solid.7 (N-(3-(4,4-dimethyl-2,5-dioxoimidazolidin-1-yl)propyl)-N,N-dimethylhexan-1-aminium) 1H NMR (D2O, 300 MHz, δ) 3.62 (t, J = 6.6 Hz, 2H), 3.28–3.37 (m, 4H), 3.09 (s, 6H), 2.09–2.17 (m, 2H), 1.70–1.75 (m, 2H), 1.45 (s, 6H), 1.35–1.40 (m, 6H), 0.90 (t, J = 6.4 Hz, 3H); 13C NMR (D2O, 75 MHz, δ) 180.6, 157.1, 64.1, 60.7, 59.2, 50.9, 35.4, 30.4, 25.0, 23.5, 21.8, 21.6, 21.2, 13.2; HRMS (MALDI-TOF) m/z: [M − Cl]+ calculated for C16H32N3O2+, 298.2489; found: 298.2494.
8 (N-(3-(4,4-dimethyl-2,5-dioxoimidazolidin-1-yl)propyl)-N,N-dimethyldodecan-1-aminium) 1H NMR (D2O, 300 MHz, δ) 3.61 (t, J = 6.2 Hz, 2H), 3.41–3.43 (m, 4H), 3.09 (s, 6H), 2.14–2.17 (m, 2H), 1.76–1.77 (m, 2H), 1.47 (s, 6H), 1.32–1.40 (m, 18H), 0.92 (t, J = 6.3 Hz, 3H); 13C NMR (D2O, 75 MHz, δ) 179.7, 156.8, 63.6, 60.5, 58.9, 51.3, 35.4, 31.9, 29.7, 29.6, 29.3, 29.0, 26.0, 23.8, 22.6, 22.2, 21.4, 13.9; HRMS (MALDI-TOF) m/z: [M − Cl]+ calculated for C22H44N3O2+, 382.3428; found: 382.3433.
9 (N-(3-(4,4-dimethyl-2,5-dioxoimidazolidin-1-yl)propyl)-N,N-dimethyltetradecan-1-aminium) 1H-NMR (D2O, 300 Hz) 3.61 (t, J = 6.7 Hz, 2H), 3.31–3.38 (m, 4H), 3.15 (s, 6H), 2.09–2.14 (m, 2H), 1.71–1.73 (m, 2H), 1.45 (s, 6H), 1.31–1.37 (m, 22H), 0.91 (t, J = 6.7 Hz, 3H); 13C-NMR (CDCl3, 75 Hz) 179.6, 156.8, 63.6, 60.5, 58.9, 51.3, 35.4, 31.9, 29.9, 29.8, 29.7, 29.5, 29.4, 29.1, 26.0, 23.8, 22.6, 22.2, 21.4, 13.9; HRMS (MALDI-TOF) m/z: [M − Cl]+ calculated for C24H48N3O2+, 410.3741; found: 410.3746.
Chlorination of the N-chloramine precursors
To the solution of non-chlorinated compound (2, 3, 7–9) in mixed solvent (t-BuOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 4
H2O, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), 3 equivalent excess t-butyl hypochlorite was added. The reaction was allowed to stir vigorously for 22–24 h (except 3). Due to poor solubility in this mixed solvent, the chlorination of 3 was prolonged to three days to reach satisfactory conversion. Excess t-butyl hypochlorite and solvent were removed under vacuum and the corresponding chlorinated forms (5, 6, 10–12) were thus obtained as white solid.
1, v/v), 3 equivalent excess t-butyl hypochlorite was added. The reaction was allowed to stir vigorously for 22–24 h (except 3). Due to poor solubility in this mixed solvent, the chlorination of 3 was prolonged to three days to reach satisfactory conversion. Excess t-butyl hypochlorite and solvent were removed under vacuum and the corresponding chlorinated forms (5, 6, 10–12) were thus obtained as white solid.
5: 1H NMR (D2O, 300 MHz, δ) 3.7 (t, J = 7.5 Hz, 2H), 3.38 (t, J = 4.5 Hz, 2H), 3.12 (s, 3H), 2.13 (m, 2H), 1.51 (s, 6H); 13C NMR (D2O, 75 MHz, δ) 176.7, 155.4, 66.3, 61.3, 50.9, 36.5, 21.4, 20.9; HRMS (MALDI-TOF) m/z: [M − Cl]+ calculated for C18H30Cl2N5O4+, 450.1669; found: 450.1675.
6: 1H NMR (D2O, 300 MHz, δ) 4.03 (m, 4H), 3.72 (t, J = 6.8 Hz 2H), 3.56 (t, J = 7.4 Hz, 2H), 3.32 (s, 9H), 3.27 (s, 6H), 2.21–2.26 (m, 2H), 1.52 (s, 6H); 13C NMR (D2O, 75 MHz, δ) 176.8, 155.4, 66.4, 63.3, 57.5, 56.3, 53.9, 51.1, 36.4, 21.2, 20.9; HRMS (MALDI-TOF) m/z: [M − 3Cl-N(CH3)3]+ calculated for C12H22N3O2+, 450.1669; found: 450.1675.
10: 1H NMR (D2O, 300 MHz, δ) 3.7 (t, J = 6.4 Hz, 2H), 3.29–3.38 (m, 4H), 3.09 (s, 6H), 2.09–2.18 (m, 2H), 1.71–1.76 (m, 2H), 1.52 (s, 6H), 1.35–1.41 (m, 6H), 0.9 (t, J = 6.5 Hz, 3H); 13C NMR (D2O, 75 MHz, δ) 176.8, 155.4, 66.3, 64.2, 60.6, 50.8, 36.6, 30.4, 25.0, 21.8, 21.1, 21.0, 13.2; HRMS (MALDI-TOF) m/z: [M − Cl]+ calculated for C16H31ClN3O2+, 332.2099; found: 332.2105.
11: 1H NMR (D2O, 300 MHz, δ) 3.73 (t, J = 6.0 Hz, 2H), 3.33–3.37 (m, 4H), 3.15 (s, 6H), 2.15–2.17 (m, 2H), 1.76–1.77 (m, 2H), 1.52 (s, 6H), 1.32–1.38 (m, 18H), 0.92 (t, J = 6.0 Hz, 3H);13C NMR (D2O, 75 MHz, δ) 175.7, 155.0, 66.1, 63, 59.7, 51.7, 36.7, 32, 29.8, 29.6, 29.4, 29.1, 25.9, 22.7, 22.2, 21.5, 21.3, 13.9; HRMS (MALDI-TOF) m/z: [M − Cl]+ calculated for C22H43ClN3O2+, 416.3038; found: 416.3044.
12: 1H-NMR (D2O, 300 Hz) 3.73 (t, J = 6.7 Hz, 2H), 3.26–3.40 (m, 4H), 3.17 (s, 6H), 2.12–2.17 (m, 2H), 1.66–1.70 (m, 2H), 1.52 (s, 6H), 1.31–1.37 (m, 22H), 0.92 (t, J = 6.7 Hz, 3H); 13C-NMR (CDCl3, 75 Hz) 175.4, 155.0, 66.0, 63, 59.7, 51.8, 36.8, 32.0, 30.0, 29.9, 29.6, 29.5, 29.2, 26, 22.7, 22.3, 21.6, 21.3, 13.9; HRMS (MALDI-TOF) m/z: [M − Cl]+ calculated for C24H47ClN3O2+, 444.3351; found: 444.3357.
Purities of all the compounds 1–12 were checked by HPLC-MS or absolute quantitative 1H NMR (qHNMR) and found to be higher than 95%. Analytic HPLC was run on a Varian 212 HPLC instrument, equipped with X-Bridge™ BEH C18 2.5 μm and Inertsil C8 3.3 μm columns and interfaced with Varian 500 MS-Ion trap detector. Eluent system was gradient: acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5
water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95, v/v, 0.1% of formic acid), a linear gradient was applied for 20 min, up to an acetonitrile
95, v/v, 0.1% of formic acid), a linear gradient was applied for 20 min, up to an acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water ratio of 90
water ratio of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v), after which elution with a acetonitrile
10 (v/v), after which elution with a acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (5
water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95, v/v) gradient was used. Flow: 0.4 mL min−1. Maleic acid (99.94% pure) was used as internal calibrant in absolute quantitative 1H NMR (qHNMR) analysis.
95, v/v) gradient was used. Flow: 0.4 mL min−1. Maleic acid (99.94% pure) was used as internal calibrant in absolute quantitative 1H NMR (qHNMR) analysis.
Quantitative killing assays
For the antibacterial studies, logarithmic-phase cultures were prepared by initially suspending several colonies in phosphate-buffered saline (PBS, 0.1 M, pH 7.4) at a density equivalent to a 0.5 McFarland standard of 1 × 108 colony forming units (CFU) mL−1 and then diluted 100 times to 1 × 106 CFU mL−1. 20 μL of the diluted MDR P. aeruginosa and MRSA suspension was further diluted into 60 mL cation-supplemented Mueller-Hinton (MH) broth and Tryptone Soya broth, respectively. After culturing in the incubator at 37 °C for overnight, the concentration of bacteria went up to 108 CFU mL−1 again and suspensions were diluted 100 or 1000 times in PBS (pH 7.4, 0.1 M), yielding a starting inoculum of 106 CFU mL−1 or 105 CFU mL−1 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ppm (based on the concentration of Cl+) stock solution of each biocide was prepared in PBS. 30 μL of each biocide solution was added into 20 mL diluted bacteria suspension, mixing thoroughly by vortex to give a final concentration of Cl+ of 15 ppm. Following different contact time, 1 mL samples were removed and transferred to 1 mL neutralizer solution (N-chlorine was quenched by 0.02 M sodium thiosulfate (Na2S2O4) solution and long alkyl chain was quenched by PBS buffer consisting of 1.4% [w/v] lecithin and 10% [w/v] Tween 80). Then 100 μL of bacterial suspension was taken out and diluted to 1 × 101, 1 × 102 and 1 × 103 times in sequence. Finally, 100 μL each of the bacterial suspension as well as the three diluted solutions were placed onto four zones of a Tryptone Soya agar plate (CM 0131, OXOID) and incubated at 37 °C for 18–20 h. The same procedure was also applied to the blanks (bacterial solution only) as controls but it was diluted to 1 × 102, 1 × 103, 1 × 104 and 1 × 105 times in sequence before plating on the agar plate.
000 ppm (based on the concentration of Cl+) stock solution of each biocide was prepared in PBS. 30 μL of each biocide solution was added into 20 mL diluted bacteria suspension, mixing thoroughly by vortex to give a final concentration of Cl+ of 15 ppm. Following different contact time, 1 mL samples were removed and transferred to 1 mL neutralizer solution (N-chlorine was quenched by 0.02 M sodium thiosulfate (Na2S2O4) solution and long alkyl chain was quenched by PBS buffer consisting of 1.4% [w/v] lecithin and 10% [w/v] Tween 80). Then 100 μL of bacterial suspension was taken out and diluted to 1 × 101, 1 × 102 and 1 × 103 times in sequence. Finally, 100 μL each of the bacterial suspension as well as the three diluted solutions were placed onto four zones of a Tryptone Soya agar plate (CM 0131, OXOID) and incubated at 37 °C for 18–20 h. The same procedure was also applied to the blanks (bacterial solution only) as controls but it was diluted to 1 × 102, 1 × 103, 1 × 104 and 1 × 105 times in sequence before plating on the agar plate.
The number of viable bacteria on 4 zones of the agar plates for the controls (A, CFU mL−1) and for the biocides treated samples (B, CFU mL−1) were counted with valid counts in the range of 25–250 colonies, and the total number of bacteria was calculated using the number of viable bacteria multiplied by the dilution factor. The percentage reduction of bacteria (%) = (A − B)/A × 100; and logarithm reduction = log(A/B). The antibacterial test of each biocide was repeated at least three times.
Uptake isotherm measurements
Final concentrations of 4.5, 9, 12, 15, 25, and 35 μg mL−1 of each compound were prepared in PBS buffer, and 4 × 10−4 M orange II dye (Sigma-Aldrich) was prepared in 0.1 M NaCl solution. 4 mL of each concentration of each compound was taken out and mixed with 1 mL prepared orange II dye solution for 5 min, followed by the addition of 5 mL chloroform to extract the dye–compound complex. The mixture was vortexed for 30 s or even longer to ensure that the chloroform and dye were mixed thoroughly. 700 μL of the chloroform phase (the bottom layer) was removed into a UV silica cuvette (VWR Spectrophotometer Cell, 10 mm light path), and the absorbance was measured at 485 nm. A PBS buffer control extracted in the same way was used to blank the spectrophotometer. Then, a standard calibration curve of each compound was achieved by plotting the absorbance at 485 nm (A485 nm) vs. the compound concentrations.Subsequently, 1 mL of the stock suspension (CA-MRSA/MDR P. aeruginosa, 109 CFU mL−1) was added into 9 mL of each concentration of each compound, giving a final bacterial concentration of 108 CFU mL−1. The mixture was vortexted every 20 min for a total contact time of 1 h, followed by 15 min centrifugation at 2500 RPM. 4 mL of the supernatant liquid was then removed and mixed with 1 mL orange II dye solution, and the following procedure was the same as described in the preceding paragraph. The equilibrium (unbound) concentration of compounds was calculated directly using the established calibration curves, and then the uptake concentration was obtained by subtracting the equilibrium concentration from the initial concentrations. Isotherm profiles comparing the equilibrium concentration (μg mL−1) with the uptake concentration (μg per 108 cells) were plotted.
Statistical analysis
Two-sample t-test was used to check for statistical differences among the test results of the antibacterial activity. JMP software was used to determine the significant difference between two curves in the time-kill profiles of the biocides. Statistical significance was considered at p ≤ 0.05.Results and discussion
Chemistry
We previously reported the synthesis of a 5,5-dimethyl hydantoin (DMH) analog (compound 1 in Scheme 1) that contains one DMH moiety and one short-chained QA salt moiety. In this paper, we synthesized a series of new DMH analogs with multiple QA cationic moieties or with long-chained QAs to explore the contribution of positive charge, length of alkyl chain and possible biocidal synergism from the combination of the N-chloramine and antibacterial long-chained QA moieties. The chemical synthesis is depicted in Scheme 2. DMH bromide 13 was prepared according to our previously published procedure.24 Reacting the large excess commercially available N,N,N′,N′-tetramethylethylenediamine with bromide 13 under reflux conditions gave the QA-tertiary amine 14 as white solid. We completed the quaternization of the structural terminal tertiary amine by treating 14 with excess MeI at room temperature. By passing the double quaternized 14 through an ion exchange resin, we then obtained compound 3 (with chloride anion, Cl−) which could be chlorinated using t-BuOCl to give 6. DMH abutted with tertiary amine (15) was produced by reacting bromide 13 with excess dimethyl amine. Following quaternization of 15 with 13 smoothly afforded the product 2. Similar quaternization with different alkyl bromides yield QA products 7–9 containing long alkyl chains that may render them antibacterial activity. All these synthetic QA compounds, including 2, 7–9, were transformed into corresponding Cl− form before the final chlorination step to give the new “composite” biocides with both N-chloramine and QA moieties: 5, 10–12. The ratios of N-chloramine versus QA moiety in compounds 4, 5 and 6 are 1/1, 2/1 and 1/2, respectively. All the biocides are listed in the summary table (Table 1) with corresponding numbers and codes.Impact of the ratio of N-chloramine versus short-chained QAs on the antibacterial efficacy of the “composite” biocides
MRSA and MDR P. aeruginosa have been two of the most frequently isolated organisms in burn wounds,27 thus, they were selected as the test microbes to evaluate the antibacterial performance of the synthesized compounds. According to our previous study,26 compound 4 of dimethyl-hydantoin (DMH) based N-chloramine covalently linked with a positively charged, short-chained QA moiety (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) exhibited more efficient antibacterial activity, when compared to N-chloramine bonded with a negative charge moiety (phosphorus ion). Herein we sought to study the effect of the ratio of N-chloramine/QA moiety on the bactericidal efficacy. Prior to evaluating the antibacterial activity of the final products, the efficacy of the non-chlorinated precursors (compound 1–3, 7–9) was evaluated to fully understand the benefit of designing a combination approach.
1) exhibited more efficient antibacterial activity, when compared to N-chloramine bonded with a negative charge moiety (phosphorus ion). Herein we sought to study the effect of the ratio of N-chloramine/QA moiety on the bactericidal efficacy. Prior to evaluating the antibacterial activity of the final products, the efficacy of the non-chlorinated precursors (compound 1–3, 7–9) was evaluated to fully understand the benefit of designing a combination approach.
Compounds 1–6 with short-chained QA moieties at the concentrations of 0.423 mM (or 15 ppm of [Cl+] for N-chloramines) were challenged with 106 CFU mL−1 of MRSA and MDR P. aeruginosa, providing time-kill profiles for each compound. As it can be seen from Table 2, there was no antibacterial activity from the non-chlorinated precursors 1–3 with short alkyl chain (methyl). This is not a surprising finding since QA salts with n-alkyl chain shorter than 4 carbons have been reported to be inactive against bacteria.28 The chlorinated counterparts 4–6 exerted considerable killing efficiency against MRSA, achieving more than 99.55% (2.35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log) bacterial reduction after 60 min of contact. However, there was no significant activity against MDR P. aeruginosa within the studied period of contact time.
log) bacterial reduction after 60 min of contact. However, there was no significant activity against MDR P. aeruginosa within the studied period of contact time.
| Synthetic compounds | Bacterial reduction at various contact time (min) | |||||
|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 10 | 60 | ||
| a Inoculum concentration of MRSA and MDR P. aeruginosa was 1.62 × 106 CFU mL−1 and 2.06 × 106 CFU mL−1, respectively. | ||||||
| Gram-positive MRSA (106 CFU mL−1)a | ||||||
| 1–3, 7 | % | NO antibacterial activity | ||||
| 4 | % | 82.94 ± 0.20 | 91.86 ± 0.90 | 86.12 ± 0.50 | 93.03 ± 1.16 | 99.88 ± 2.93 |
| log10 | 1.09 ± 0.05 | 0.86 ± 0.02 | 1.16 ± 0.02 | 2.93 ± 0.02 | ||
| 5 | % | 91.54 ± 1.35 | 91.43 ± 1.10 | 87.22 ± 0.95 | 89.13 ± 0.65 | 99.55 ± 0.04 |
| log10 | 1.08 ± 0.07 | 1.07 ± 0.06 | 0.89 ± 0.03 | 0.96 ± 0.03 | 2.35 ± 0.04 | |
| 6 | % | 93.66 ± 0.25 | 93.06 ± 0.30 | 90.97 ± 0.25 | 91.75 ± 0.35 | 99.56 ± 0.06 |
| log10 | 1.20 ± 0.02 | 1.16 ± 0.02 | 1.04 ± 0.01 | 1.08 ± 0.02 | 2.36 ± 0.06 | |
| 10 | % | 78.26 ± 1.48 | 78.86 ± 0.04 | 82.81 ± 1.90 | 89.27 ± 0.47 | 98.80 ± 0.34 |
| log10 | 1.93 ± 0.12 | |||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Gram-negative MDR P. aeruginosa (106 CFU mL−1)a | ||||||
| 1–3, 7 | % | NO antibacterial activity | ||||
| 4 | % | 21.84 ± 6.18 | 26.21 ± 10.98 | 21.36 ± 6.87 | 41.26 ± 4.81 | 10.68 ± 0.05 |
| 5 | % | 0.00 ± 32.27 | 5.83 ± 6.87 | 8.74 ± 0.00 | 10.19 ± 0.69 | 14.08 ± 8.92 |
| 6 | % | 38.83 ± 1.37 | 37.86 ± 4.12 | 31.55 ± 3.43 | 25.73 ± 2.06 | 26.21 ± 1.37 |
For these “composite” compounds 4–6 with short chain QA moieties, N-chloramine, rather than the QA cationic center, was considered as the primary cause of bacterial death within the studied time frame. The first action of N-chloramine on bacteria was reported to be the formation of chlorine cover, i.e. formation of N–Cl bonds on the superficial surface of bacterial cell wall or membrane. This chlorine cover does not necessarily impair the viability of bacteria below a critical concentration (3.3 × 10−16 mole Cl+ per CFU).29 At the same time, N-chloramine molecule penetrates into the intact bacteria, oxidizing mainly S–H and S–S groups in proteins which comprise the vital sites of bacteria. Penetration of N-chloramines into bacteria can cause faster inactivation. Worley and his coworkers had reported that the bacterial inactivation by N-chloramines is caused by the entire molecular structure, not the limited amount of free chlorine generated in hydrolysis equilibrium.30 Therefore, the bactericidal efficacy of N-chloramines is strongly relevant to the cell accessibility of the integrated compounds. Gram-negative P. aeruginosa proved to be less sensitive to compound 4–6 than Gram-positive MRSA, which might be attributed to the rigid outer membrane. The lipopolysaccharide outer membrane of P. aeruginosa is likely to offer a hindrance for the penetration of N-chloramines into cells, contributing to a higher intrinsic resistance to this mechanism of antibacterial activity. Thus, in order to obtain useable data on the effect of number of QA moieties in these bacteria, the inoculum concentration of P. aeruginosa was decreased to 105 CFU mL−1 and different contact times were chosen (Table 3).
| Gram-negative MDR P. aeruginosaa (105 CFU mL−1) | ||||||
|---|---|---|---|---|---|---|
| Synthetic compounds | Bacterial reduction at various contact time (min) | |||||
| 3 | 10 | 20 | 30 | 60 | ||
| a Inoculum concentration of MDR P. aeruginosa was 2.26 × 105 CFU mL−1 for 1–6 and 2.98 × 105 CFU mL−1 for 7, 10. | ||||||
| 1–3, 7 | % | NO antibacterial activity | ||||
| 4 | % | 37.61 ± 1.88 | 61.50 ± 1.88 | 99.65 ± 0.00 | 99.99 ± 0.00 | 100 ± 0.00 |
| log10 | 2.46 ± 0.02 | 4.02 ± 0.21 | 5.35 ± 0.00 | |||
| 5 | % | 22.57 ± 4.38 | 26.55 ± 12.52 | 70.81 ± 5.22 | 99.96 ± 0.02 | 100 ± 0.00 |
| log10 | 3.36 ± 0.25 | 5.35 ± 0.00 | ||||
| 6 | % | 39.82 ± 10.01 | 62.83 ± 1.25 | 99.67 ± 0.12 | 100 | 100 ± 0.00 |
| log10 | 2.50 ± 0.17 | 4.82 ± 0.92 | 5.35 ± 0.00 | |||
| 10 | % | 42.68 ± 0.67 | 49.34 ± 16.39 | 68.56 ± 3.59 | 97.00 ± 0.61 | 100 ± 0.00 |
| log10 | 1.52 ± 0.09 | 5.80 ± 0.00 | ||||
No significant difference existed in the time-kill profile (P > 0.05) of compound 4, 5, and 6 with different ratios of N-chloramine/QA moiety against MRSA (Table 2). However, as shown in Table 3, compound 5 with the highest ratio (the ratio of N-chloramine/QA moiety: 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) exerted much slower killing kinetics against 105 CFU mL−1 of MDR P. aeruginosa. Only 70.81% (0.54
1) exerted much slower killing kinetics against 105 CFU mL−1 of MDR P. aeruginosa. Only 70.81% (0.54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log) bacterial reduction was achieved within 20 minutes of contact, which was significantly (P < 0.05) lower than 99.65% (2.46
log) bacterial reduction was achieved within 20 minutes of contact, which was significantly (P < 0.05) lower than 99.65% (2.46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log) and 99.67% (2.50
log) and 99.67% (2.50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log) reduction achieved by 4 and 6, respectively. The higher bulk and hydrophobicity of compound 5 might have resulted in a slower diffusion rate into intact cells through the water-filled porins in the lipidic outer membrane of P. aeruginosa. There was no further acceleration in the killing kinetics of compound 6 with an additional cationic QA center (the ratio of N-chloramine/QA moiety: 1
log) reduction achieved by 4 and 6, respectively. The higher bulk and hydrophobicity of compound 5 might have resulted in a slower diffusion rate into intact cells through the water-filled porins in the lipidic outer membrane of P. aeruginosa. There was no further acceleration in the killing kinetics of compound 6 with an additional cationic QA center (the ratio of N-chloramine/QA moiety: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). The cell accessibility for the penetration of biocides, rather than the ionic interaction, seems to play a more important role in the antibacterial activity. The structural difference of compounds #4–6 was not manifested in the time kill profile against MRSA probably because the size difference of the three compounds is not big enough to cause any difference in their diffusion across the loose cell wall of S. aureus. Even polymers with molecular weight up to 9 × 104 Dalton have been reported to diffuse through the cell wall of S. aureus.31
2). The cell accessibility for the penetration of biocides, rather than the ionic interaction, seems to play a more important role in the antibacterial activity. The structural difference of compounds #4–6 was not manifested in the time kill profile against MRSA probably because the size difference of the three compounds is not big enough to cause any difference in their diffusion across the loose cell wall of S. aureus. Even polymers with molecular weight up to 9 × 104 Dalton have been reported to diffuse through the cell wall of S. aureus.31
Impact of chain length of QA on the antibacterial efficacy of the “composite” biocide
The effect of alkyl chain length of QA moiety (compounds 10–12) on the antibacterial activity was also investigated in this study, hypothesizing a synergistic effect between N-chloramine and long-chained QA moiety.The bactericidal efficacy of QA salts is reported to exhibit a strong dependence on the alkyl chain length, however there was no bacterial reduction when increasing the length from methyl group (compound 1) to hexyl group (compound 7). Hexamer has been reported to show characteristically slower disinfection rates since its mode of action primarily involves inhibition of DNA activity instead of membrane disruption.32 The killing kinetics of compound 10 (C6 + QA + Chlor), the chloramine counterpart of compound 7 (C6 + QA + Hyd), was even slower than compound 4 (C1 + QA + Chlor), especially against MRD P. aeruginosa, arriving at merely 1.52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction after 30 min in comparison with 4.02
log reduction after 30 min in comparison with 4.02![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction of 4 as listed in Table 3. It can be attributed to that penetration into pathogens is favored for N-chloramines with low bulk, giving the fastest killing kinetics of methyl 4. Thus, the combination of N-chloramine with hexyl QA has no synergistic, but perhaps antagonistic effect on the antibacterial activity. Further increasing the alkyl chain length to dodecyl 11 (C12 + QA + Chlor) and tetradecyl 12 (C14 + QA + Chlor) resulted in an instantaneous total kill of 106 CFU mL−1 of both microbes (data not shown). In order to detect the difference of the antibacterial efficacy between of 8 & 11, and 9 & 12, the inoculum concentration was raised up to 107 CFU mL−1 in repeat experiments (Table 4).
log reduction of 4 as listed in Table 3. It can be attributed to that penetration into pathogens is favored for N-chloramines with low bulk, giving the fastest killing kinetics of methyl 4. Thus, the combination of N-chloramine with hexyl QA has no synergistic, but perhaps antagonistic effect on the antibacterial activity. Further increasing the alkyl chain length to dodecyl 11 (C12 + QA + Chlor) and tetradecyl 12 (C14 + QA + Chlor) resulted in an instantaneous total kill of 106 CFU mL−1 of both microbes (data not shown). In order to detect the difference of the antibacterial efficacy between of 8 & 11, and 9 & 12, the inoculum concentration was raised up to 107 CFU mL−1 in repeat experiments (Table 4).
| Synthetic compounds | Bacterial reduction at various contact time (min) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 10 | 30 | 60 | 90 | ||
| a Inoculum concentration of MRSA and MDR P. aeruginosa was in the range of 1.07–5.29 × 107 CFU mL−1 and 1.55–7.90 × 107 CFU mL−1, respectively.b 4 + 8 and 4 + 9 represents the mixture of separate biocides 4 and 8 or 9 with the same dose of 0.423 mM for each. | ||||||||
| Gram-positive MRSA (107 CFU mL−1)a | ||||||||
| 8 | % | 26.79 ± 5.50 | 22.32 ± 5.68 | 56.25 ± 3.79 | 55.36 ± 5.41 | 58.93 ± 8.84 | 77.23 ± 1.89 | 88.21 ± 0.25 |
| 11 | log10 | 3.52 ± 0.01 | 5.44 ± 0.00 | 7.05 ± 0.00 | 7.05 ± 0.00 | 7.05 ± 0.00 | 7.05 ± 0.00 | 7.05 ± 0.00 |
| 4 + 8 | % | 16.31 ± 4.68 | 23.60 ± 0.00 | 68.11 ± 2.80 | 99.97 ± 0.00 | 100 ± 0.00 | 100 ± 0.00 | 100 ± 0.00 |
| log10 | 3.54 ± 0.04 | 5.20 ± 0.34 | 7.05 ± 0.00 | 7.05 ± 0.00 | ||||
| 9 | log10 | 3.53 ± 0.02 | 4.79 ± 0.11 | 5.19 ± 0.44 | 6.42 ± 0.59 | 7.55 ± 0.22 | 7.55 ± 0.22 | 7.55 ± 0.22 |
| 12 | log10 | 4.93 ± 0.91 | 7.11 ± 0.29 | 7.55 ± 0.22 | 7.55 ± 0.22 | 7.55 ± 0.22 | 7.55 ± 0.22 | 7.55 ± 0.22 |
| 4 + 9 | log10 | 3.54 ± 0.05 | 6.42 ± 0.31 | 6.95 ± 0.41 | 7.55 ± 0.22 | 7.55 ± 0.22 | 7.55 ± 0.22 | 7.55 ± 0.22 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Gram-negative MDR P. aeruginosa (107 CFU mL−1)a | ||||||||
| 8 | % | 74.27 ± 8.47 | 96.70 ± 1.99 | 98.60 ± 0.73 | 98.61 ± 0.81 | 99.19 ± 0.29 | 99.33 ± 0.22 | 99.33 ± 0.20 |
| log10 | 1.73 ± 0.29 | 2.07 ± 0.29 | 2.10 ± 0.29 | 2.21 ± 0.22 | 2.28 ± 0.21 | 2.26 ± 0.19 | ||
| 11 | % | 34.54 ± 3.81 | 80.36 ± 5.59 | 89.37 ± 1.76 | 93.91 ± 0.39 | 99.90 ± 0.05 | 99.99 ± 0.01 | 100 ± 0.00 |
| log10 | 0.99 ± 0.07 | 1.88 ± 0.12 | 3.17 ± 0.25 | 4.13 ± 0.18 | 5.57 ± 0.75 | |||
| 4 + 8 | % | 16.31 ± 4.68 | 23.60 ± 0.00 | 68.11 ± 2.80 | 93.91 ± 0.38 | 98.10 ± 0.19 | 98.74 ± 0.15 | 98.84 ± 0.40 |
| log10 | 1.22 ± 0.03 | 1.73 ± 0.05 | 1.91 ± 0.05 | 2.00 ± 0.14 | ||||
| 9 | log10 | 2.11 ± 0.19 | 4.30 ± 0.53 | 5.79 ± 0.86 | 6.43 ± 0.41 | 7.50 ± 0.17 | 7.50 ± 0.17 | 7.50 ± 0.17 |
| 12 | log10 | 1.75 ± 0.16 | 3.82 ± 0.45 | 4.60 ± 0.52 | 5.39 ± 0.31 | 6.63 ± 0.33 | 7.50 ± 0.17 | 7.50 ± 0.17 |
| 4 + 9 | log10 | 1.69 ± 0.24 | 3.29 ± 0.27 | 4.08 ± 0.26 | 4.66 ± 0.12 | 4.91 ± 0.34 | 5.58 ± 0.24 | 6.13 ± 0.55 |
7.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log and 7.55
log and 7.55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction of MRSA was achieved by compound 11 and 12 at 5 min, respectively, while 5.57
log reduction of MRSA was achieved by compound 11 and 12 at 5 min, respectively, while 5.57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log and 7.50
log and 7.50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction of MDR P. aeruginosa within 90 min. Long-chained QA salts endow the “composite” biocides with an additional killing mechanism by inserting the long alkyl chains into membrane lipid domains of bacteria, inducing physical disruption and rapid inactivation. Longer alkyl chains show more potent bactericidal efficacy due to the formation of bi-polar dimmers, enabling stronger interaction with bacterial cytoplasmic membranes.33 This finding is consistent with previous report of dramatic increase of biocidal efficacy of QA with >10 n-alkyl chain length.34 Even though the long-chained QA moieties seems to outperform the N-chloramine moieties in the “composite” biocides in terms of the antibacterial activity, compounds 11 and 12 presented significantly faster inactivation of MRSA than their mono-functional QA counterparts 8 and 9 (C12 + QA + Hyd and C14 + QA + Hyd, dodecyl and tetradecyl QAs).
log reduction of MDR P. aeruginosa within 90 min. Long-chained QA salts endow the “composite” biocides with an additional killing mechanism by inserting the long alkyl chains into membrane lipid domains of bacteria, inducing physical disruption and rapid inactivation. Longer alkyl chains show more potent bactericidal efficacy due to the formation of bi-polar dimmers, enabling stronger interaction with bacterial cytoplasmic membranes.33 This finding is consistent with previous report of dramatic increase of biocidal efficacy of QA with >10 n-alkyl chain length.34 Even though the long-chained QA moieties seems to outperform the N-chloramine moieties in the “composite” biocides in terms of the antibacterial activity, compounds 11 and 12 presented significantly faster inactivation of MRSA than their mono-functional QA counterparts 8 and 9 (C12 + QA + Hyd and C14 + QA + Hyd, dodecyl and tetradecyl QAs).
Boosted antibacterial effect by covalently joining long-chained QA with N-chloramine
In order to study the possible synergistic effect of the “composite” biocide (11, C12 + QA + Chlor or 12, C14 + QA + Chlor), the mixture of separate N-chloramine (4, C1 + QA + Chlor) and long-chained QA salt (8, C12 + QA + Hyd or 9, C14 + QA + Hyd) was also formulated at equal dose of 0.423 mM for each, marked as 4 + 8 or 4 + 9. Compound 4 was chosen as “mono-functional” N-chloramine to ensure that both components in this formulation were endowed with a cationic center, consistent with the “composite” compound. Time-kill profiles in Fig. 1 were plotted using the data in Table 4 for better comparative purposes. Before chlorination, there was only 0.93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log (88.21%) reduction against 107 CFU mL−1 of MRSA achieved by dodecyl 8 within 90 min. The formulation of 4 + 8 provided a much more favorable killing efficiency, arriving at 7.05
log (88.21%) reduction against 107 CFU mL−1 of MRSA achieved by dodecyl 8 within 90 min. The formulation of 4 + 8 provided a much more favorable killing efficiency, arriving at 7.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log (100%) reduction at 60 min, and there was a further enhancement of “composite” compound 11 with a complete elimination (7.05
log (100%) reduction at 60 min, and there was a further enhancement of “composite” compound 11 with a complete elimination (7.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction) at 5 min. Smaller differences were noted in the batch of tetradecyl 9, 12 and 4 + 9, but the trend in the antibacterial activity was accordant. Compound 9 with long chain alone exerted the least potent bacterial efficacy. Biocide 12 achieved a total killing of 7.11
log reduction) at 5 min. Smaller differences were noted in the batch of tetradecyl 9, 12 and 4 + 9, but the trend in the antibacterial activity was accordant. Compound 9 with long chain alone exerted the least potent bacterial efficacy. Biocide 12 achieved a total killing of 7.11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log bacterial reduction at 3 min, as compared to 6.42
log bacterial reduction at 3 min, as compared to 6.42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log of compound 4 + 9 (0.05 < p < 0.1). Covalently combining N-chloramine and long-chained QA moiety proved advantageous with the fastest disinfection kinetics, indicating a synergistic effect of these two components against MRSA. But unlike MRSA, MDR P. aeruginosa appeared to be most sensitive to C12 + QA + Hyd (biocide 8) during the first 10 min of contact, achieving the maximum of approximately 2
log of compound 4 + 9 (0.05 < p < 0.1). Covalently combining N-chloramine and long-chained QA moiety proved advantageous with the fastest disinfection kinetics, indicating a synergistic effect of these two components against MRSA. But unlike MRSA, MDR P. aeruginosa appeared to be most sensitive to C12 + QA + Hyd (biocide 8) during the first 10 min of contact, achieving the maximum of approximately 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction. The bacterial reduction of chlorinated 11 slowly increased to 1.88
log reduction. The bacterial reduction of chlorinated 11 slowly increased to 1.88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction at 10 min and exceeded that of compound 8, arriving at 5.57
log reduction at 10 min and exceeded that of compound 8, arriving at 5.57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction after 90 min. The longer alkyl chains make the compounds work better against P. aeruginosa as evidenced by 7.5
log reduction after 90 min. The longer alkyl chains make the compounds work better against P. aeruginosa as evidenced by 7.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction by both C14 + QA + Hyd (9) and C14 + QA + Chlor (12) with 30 min. No significant difference existed in the time-kill profile of C14 + QA + Hyd (9) and C14 + QA + Chlor (12). The mixture of 4 + 8 and 4 + 9 exhibited the worst antibacterial performance against P. aeruginosa. The different ranking of the time-kill kinetics of the biocides implies different modes of action against these two microbes which is currently under study in our lab. Long-chained QAs manifests fast inactivation of P. aeruginosa since they directly act on the cell membrane. Inactivation of P. aeruginosa by C12 + QA + Hyd (8) plateaued after 30 min of contact whereas C12 + QA + Chlor (12) continued to act on bacterial cells to reach 5.57
log reduction by both C14 + QA + Hyd (9) and C14 + QA + Chlor (12) with 30 min. No significant difference existed in the time-kill profile of C14 + QA + Hyd (9) and C14 + QA + Chlor (12). The mixture of 4 + 8 and 4 + 9 exhibited the worst antibacterial performance against P. aeruginosa. The different ranking of the time-kill kinetics of the biocides implies different modes of action against these two microbes which is currently under study in our lab. Long-chained QAs manifests fast inactivation of P. aeruginosa since they directly act on the cell membrane. Inactivation of P. aeruginosa by C12 + QA + Hyd (8) plateaued after 30 min of contact whereas C12 + QA + Chlor (12) continued to act on bacterial cells to reach 5.57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction after 90 min, indicating more effective inactivation of the bacterial cells by the “composite” biocide. The antibacterial activity of C12 + QA + Hyd (9) and C12 + QA + Chlor (12) was compared with two benchmark benzalkonium chlorides: benzyldodecyldimethyl-ammonium bromide (C12) and benzyltetradecyldimethylammonium chloride (C14), as shown in Table S7.† It was found that C12 + QA + Hyd (9) and C12 + QA + Chlor (12) present similar killing kinetics with C14 against both MRSA and P. aeruginosa, all of which are faster than C12. With the knowledge of the synergistic antibacterial activity of N-chloramines and long-chained QA salts, one can design and synthesize new “composite” biocides based on the more effective QA salts developed by the recent studies.17,18
log reduction after 90 min, indicating more effective inactivation of the bacterial cells by the “composite” biocide. The antibacterial activity of C12 + QA + Hyd (9) and C12 + QA + Chlor (12) was compared with two benchmark benzalkonium chlorides: benzyldodecyldimethyl-ammonium bromide (C12) and benzyltetradecyldimethylammonium chloride (C14), as shown in Table S7.† It was found that C12 + QA + Hyd (9) and C12 + QA + Chlor (12) present similar killing kinetics with C14 against both MRSA and P. aeruginosa, all of which are faster than C12. With the knowledge of the synergistic antibacterial activity of N-chloramines and long-chained QA salts, one can design and synthesize new “composite” biocides based on the more effective QA salts developed by the recent studies.17,18
Uptake isotherms
The interaction of a biocide with a given cell is conventionally measured by determining its adsorption, providing estimable information on the availability of target sites. To identify the differences in the mode of action of compound 8–12 against MRSA and MDR P. aeruginosa, the uptake isotherms of each compound by both bacteria were obtained via a spectrophotometric method.35 To avoid multiple antibacterial effects at high concentrations of biocides, low concentrations were used to single out the prime lesion of biocide adsorption to cell membrane. According to the uptake classification scheme developed by Giles, all the uptake isotherms in Fig. 2 followed the Langmuir (L) pattern but different subclass groups, where the adsorption of biocides become more difficult due to the decreasing vacant binding sites with the progressive covering of the bacterial membrane. Overall, the uptake of C14 + QA + Hyd (tetradecyl 9) was almost two to three times that of C12 + QA + Hyd (dodecyl 8), that is, the uptake of biocides by bacterial cells increased with the increasing length of alkyl chain of QA moiety. The longer chain has a higher binding affinity to the cell membrane due to its higher lipophilicity, thus giving much faster inactivation kinetics. The isotherm profiles in Fig. 2(A) against MRSA belonged to L3 pattern. “Monofunctional” C12 + QA + Hyd (dodecyl 8) had a lower ultimate uptake of 6.4 μg per 108 cells, when compared to 9.2 μg per 108 cells of “composite” C12 + QA + Chlor (11). The more uptake of compound 11 by MRSA can be interpreted as higher affinity of 11 towards MRSA cell wall and membrane. The increase uptake may result from additional binding sites on the cell wall and membrane arising from N-chloramine moieties. The increased affinity of “composite” biocide 11 towards MRSA, as compared with mono-functional QA biocide 8, gave rise to better bactericidal efficacy of 11 against MRSA. The difference in uptake isotherms of tetradecyl 9 and 12 was much smaller, which is concordant with the antibacterial results. | ||
| Fig. 2 Uptake isotherms of compound 8–12 against (A) 2.70 × 108 CFU mL−1 of MRSA and (B) 1.54 × 108 CFU mL−1 of MDR P. aeruginosa in PBS (0.1 M, pH 7.4). | ||
The uptake isotherms of compound 8 and 9 against MDR P. aeruginosa in Fig. 2(B) followed an L2 pattern, arriving at the final plateau with a maximum of 2.9 μg per 108 cells and 9.3 μg per 108 cells, respectively. Generally, the uptakes by P. aeruginosa were generally lower than MRSA, owing to the outer membrane which acted as a barrier to prevent the access of biocides.36–38 Tischer39 et al. demonstrated that long-chained QA salts must first pass the outer membrane by lysis of this layer and then interact with the inner membrane. There appeared to be a limit to the amount of adsorption of long-chained QA salts, especially compound 8, possibly due to the exhaustion of binding sites. This observation could be linked to the killing kinetics where a plateau effect was observed, suggesting no further killing of cells at the given ratio of biocide to bacteria. However, the uptake profiles of “composite” compound 11 and 12 turned to follow L3 pattern, continuing to increase with the rising concentration to 8.8 μg per 108 cells and 12.6 μg per 108 cells, respectively. Higher uptake of compound 12 as compared with compound 9 led us to the prediction that compound 12 might demonstrate higher log reduction of MDR P. aeruginosa than compound 9 eventually as in the case of compounds 8 and 11 if both compounds (9 and 12) are challenged with higher bacterial concentration. This study is ongoing and will be reported in another paper.
Conclusions
New “composite” biocides combining N-chloramine and QA moieties were synthesized and challenged with both MRSA and MDR P. aeruginosa. Changing the ratio of N-chloramine to n-methyl QA moiety didn't result in enhanced biocidal efficacy against either bacteria. By keeping the ratio of N-chloramine to QA moiety at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the n-alkyl chain length was increased from 1 to 6, 12 and 14. Antibacterial synergism against both bacteria was observed when the n-alkyl chain length reached 12. This might be due to the different modes of action of N-chloramine and long-chained QA. The dissolution of long-chained QA in the bacterial membrane may facilitate the penetration of the whole molecule into the bacteria, allowing N-chloramine component to exert oxidative stress inside cells causing faster inactivation of the cells. Covalently joining N-chloramine with long-chained QA not only results in new biocides with enhanced biocidal activity, but also reduces the risk for potential bacterial resistance associated with QA as shown in another study recently published by us.40 These “composite” biocides possess the potential to be used as surface disinfectants. This is the first time that the antibacterial synergism is demonstrated by covalently combining N-chloramine and long-chained QA. This opens a door for the creation of more new broad-spectrum biocides.
1, the n-alkyl chain length was increased from 1 to 6, 12 and 14. Antibacterial synergism against both bacteria was observed when the n-alkyl chain length reached 12. This might be due to the different modes of action of N-chloramine and long-chained QA. The dissolution of long-chained QA in the bacterial membrane may facilitate the penetration of the whole molecule into the bacteria, allowing N-chloramine component to exert oxidative stress inside cells causing faster inactivation of the cells. Covalently joining N-chloramine with long-chained QA not only results in new biocides with enhanced biocidal activity, but also reduces the risk for potential bacterial resistance associated with QA as shown in another study recently published by us.40 These “composite” biocides possess the potential to be used as surface disinfectants. This is the first time that the antibacterial synergism is demonstrated by covalently combining N-chloramine and long-chained QA. This opens a door for the creation of more new broad-spectrum biocides.
Acknowledgements
This project was supported by Collaborative Health Research Project (CHRP) operating grant (Grant no. CHRP 413713-2012), and the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant (Grant no. RGPIN/04922-2014).References
- S. Oie, A. Yanagi, H. Matsui, T. Nishida, M. Tomita and A. Kamiya, Biol. Pharm. Bull., 2005, 28, 120–123 CAS.
- S. Hota, Z. Hirji, K. Stockton, C. Lemieux, H. Dedier, G. Wolfaardt and M. A. Gardam, Infect. Contr. Hosp. Epidemiol., 2009, 30, 25–33 CrossRef PubMed.
- Canadian Nosocomial Infection Surveillance Program (CNISP). Surveillance for Methicillin-resistant Staphylococcus aureus (MRSA) in Patients Hospitalized in Canadian Acute-Care Hospitals Participating in CNISP: 2006–2007 preliminary results [Internet]. Ottawa: Public Health Agency of Canada. Available from: http://www.phac-aspc.gc.ca/nois-sinp/pdf/mrsa-sarm-eng.pdf (accessed September 2015).
- Canadian Nosocomial Infection Surveillance Program (CNISP), Antimicrobial resistant organism presentation, CNISP Annual Meeting, Montreal, 2005 Search PubMed.
- D. E. Zoutman, B. D. Ford, E. Bryce, M. Gourdeau, G. Hébert, E. Henderson and S. Paton, Am. J. Infect. Control, 2003, 31, 266–272 CrossRef PubMed.
- S. J. Dancer, J. Hosp. Infect., 1999, 43, 85–100 CrossRef CAS PubMed.
- M. Drees, D. R. Snydman, C. H. Schmid, L. Barefoot, K. Hansjosten, P. M. Vue, M. Cronin, S. A. Nasraway and Y. Golan, Clin. Infect. Dis., 2008, 46, 678–685 CrossRef CAS PubMed.
- D. J. Weber, W. A. Rutala, M. B. Miller, K. Huslage and E. Sickbert-Bennett, Am. J. Infect. Control, 2010, 38, 25–33 CrossRef PubMed.
- K. L. Cheng, M. V. Boost and J. W. Y. Chung, Am. J. Infect. Control, 2011, 39, 577–580 CrossRef PubMed.
- D. Talon, J. Hosp. Infect., 1999, 43, 13–17 CrossRef CAS PubMed.
- M. Schweizer, M. Graham, M. Ohl, K. Heilmann, L. Boyken and D. Diekema, Infect. Contr. Hosp. Epidemiol., 2012, 33(11), 1081–1085 CrossRef PubMed.
- S. J. Dancer, J. Hosp. Infect., 2009, 73, 378–385 CrossRef CAS PubMed.
- C. Zollfrank, K. Gutbrod, P. Wechsler and J. P. Guggenbichler, Mater. Sci. Eng., C, 2012, 32, 47–54 CrossRef CAS PubMed.
- A. Kramer, I. Schwebke and G. Kampf, BMC Infect. Dis., 2006, 6, 1–8 CrossRef PubMed.
- B. Hota, Clin. Infect. Dis., 2004, 39, 1182–1189 CrossRef PubMed.
- S. A. Wilks, H. Michels and C. W. Keevil, Int. J. Food Microbiol., 2005, 105, 445–454 CrossRef CAS PubMed.
- N. A. Negm, Y. M. Elkholy, F. M. Ghuiba, M. K. Zahran, S. A. Mahmoud and S. M. Tawfik, J. Dispersion Sci. Technol., 2011, 32, 512–518 CrossRef CAS.
- N. A. Negm and S. M. Tawfik, Chem. Today, 2012, 30, 5–8 CAS.
- S. Buffet-Bataillon, P. Tattevin, M. Bonnaure-Mallet and A. Jolivet-Gougeon, Int. J. Antimicrob. Agents, 2012, 39, 381–389 CrossRef CAS PubMed.
- A. Muller, K. Rychli, A. Zaiser, C. Wieser, M. Wagner and S. Schmits-Esser, FEMS Microbiol. Lett., 2014, 311, 166–173 CrossRef PubMed.
- T. M. Wassenaar, D. Ussery, L. N. Nielsen and H. Ingmer, Eur. J. Microbiol. Immunol., 2015, 5, 44–61 CrossRef PubMed.
- M. L. Burts, I. Alexeff, E. T. Mekk and J. A. McCullers, Am. J. Infect. Control, 2009, 37, 729–733 CrossRef PubMed.
- S. D. Worley, D. E. Williams and R. A. Crawford, Crit. Rev. Environ. Control, 1988, 18, 133–175 CrossRef CAS.
- S. B. Barnela, S. D. Worley and D. E. Williams, J. Pharm. Sci., 2006, 76, 245–247 CrossRef.
- W. Gottardi and M. Nagl, J. Antimicrob. Chemother., 2010, 65, 399–409 CrossRef CAS PubMed.
- L. Li, T. Pu, G. Zhanel, N. Zhao, W. Ens and S. Liu, Adv. Healthcare Mater., 2012, 1, 609–620 CrossRef CAS PubMed.
- J. L. S. Macedo and J. B. Santos, Mem. Inst. Oswaldo Cruz, 2005, 100, 535–539 CrossRef PubMed.
- P. Gilbert and L. E. Moore, J. Appl. Microbiol., 2005, 99, 703–715 CrossRef CAS PubMed.
- W. Gottardi and M. Nagl, J. Antimicrob. Chemother., 2005, 55, 475–482 CrossRef CAS PubMed.
- D. E. Williams, E. D. Elder and S. D. Worley, Appl. Environ. Microbiol., 1998, 54, 2583–2585 Search PubMed.
- E. R. Kenawy, S. D. Worley and R. Broughton, Biomacromolecules, 2007, 8, 1359–1384 CrossRef CAS PubMed.
- S. Rotem, I. S. Radzishevsky, D. Bourdetsky, S. Navon-Venezia, Y. Carmeli and A. Mor, FASEB J., 2008, 22, 2652–2661 CrossRef CAS PubMed.
- N. N. D. Daoud, N. A. Dickinson and P. Gilbert, Microbios, 1983, 37, 75–85 Search PubMed.
- P. Gilbert and A. N. A. Al-Taae, Lett. Appl. Microbiol., 1985, 1, 101–105 CrossRef CAS.
- C. J. Ioannou, G. W. Hanlon and S. P. Denyer, Antimicrob. Agents Chemother., 2007, 51, 296–306 CrossRef CAS PubMed.
- S. M. Tawfik, A. A. Abd-Elaal, S. M. Shaban and A. A. Roshdy, J. Ind. Eng. Chem., 2015, 30, 112–119 CrossRef.
- S. M. Tawfik, J. Ind. Eng. Chem., 2015, 28, 171–183 CrossRef CAS.
- S. M. Tawfik, J. Mol. Liq., 2015, 209, 320–326 CrossRef CAS.
- M. Tischer, G. Pradel, K. Ohlsen and U. Holzgrabe, ChemMedChem, 2012, 7, 22–31 CrossRef CAS PubMed.
- M. D. Silva, C. Ning, S. Ghanbar, G. Zhanel, S. Logsetty, S. Liu and A. Kumar, J. Hosp. Infect., 2015, 91, 53–58 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra15714e |
| ‡ Same contribution. |
| This journal is © The Royal Society of Chemistry 2015 |