Enzyme activity enhancement of chondroitinase ABC I from Proteus vulgaris by site-directed mutagenesis†
Zhenya Chena,
Ye Li*b,
Yue Fenga,
Liang Chenb and
Qipeng Yuan*a
aState Key Laboratory of Chemical Resource Engineering, Beijing University of Chemical Technology, Beijing 100029, China. E-mail: yuanqp@mail.buct.edu.cn; Fax: +86-10-64437610; Tel: +86-10-64437610
bDepartment of Biotechnology, Beijing Polytechnic, Beijing 100029, China. E-mail: liyeyashi@163.com; Fax: +86-10-84649573; Tel: +86-10-84649573
First published on 1st September 2015
Abstract
Chondroitin sulfate (CS) is widely applied in the medical industry, especially CS B which is a kind of CS and widely used in the field of food industry, medicine and scientific research. Because of the high molecular weight of CSs, many functions could not be realized effectively. Chondroitinase ABC I (ChSase ABC I) could degrade CS to low molecular weight CS. In this study, ChSase ABC I was expressed with a maltose binding protein (MBP) tag, and site-directed mutagenesis based on both sequence alignment and molecular docking simulation analysis was conducted. 13 amino acids were selected to be mutated to Ala separately for the first time, 8 out of the 13 single-amino-acid mutants showed decreased activity with CS A as substrate and 11 of them showed decreased activity with CS B as substrate. Mutating Arg660 to Ala caused a total loss of the enzyme activity either with CS A or CS B as substrate, indicating that Arg660 was one of the active sites of ChSase ABC I. The specific activities of Asn795Ala and Trp818Ala were 1.39 and 1.38 times higher than that of wild type enzyme with CS A as substrate, and 1.85 and 1.71 times higher with CS B as substrate. In particular, the specific activity of Asn795Ala in this study was the highest among the reported ones. The kinetic parameters as well as the thermostabilities of the two mutants also showed significant improvement when compared with those of the wild type enzyme.
1. Introduction
Chondroitin sulfate (CS) is a linear polysaccharide of repeated disaccharide units, which are D-glucuronic acid and D-N-acetyl-galactosamine complexes.1 CS A, B, C are commonly used CSs. CS A is also called 4-chondroitin sulfate, in which the sulfuric acid base is on the fourth carbon atom of galactosamine. CS B was an isomer of CS A. CS C, or 6-chondroitin sulfate, has the sulfuric acid base on the sixth carbon atom of galactosamine (Fig. 1).2 CSs are widely used in medical industry, especially CS B. Because of the high molecular weight of CSs, some functions, like regeneration of cartilage and transmission of signal,1 could not be realized effectively. Chondroitinase (ChSase) could degrade high molecular weight CSs into low molecular weight CSs, which show more significant effect and play the role of “spring” in cartilage matrix. According to the types of CSs they react with, the ChSase family could be divided into ChSase ABC, ChSase AC, ChSase B and ChSase C subfamilies. ChSase B and ChSase C could degrade CS B and CS C, respectively, and ChSase ABC could degrade any of CS A, B or C.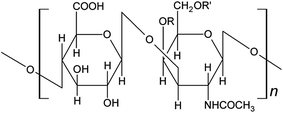 | ||
| Fig. 1 The basic unit of CS. CS A: R = SO3H, R′ = H; CS B: R = SO3H, R′ = H; CS C: R = H, R′ = SO3H. | ||
ChSase could alleviate pain, promote cartilage regeneration and treat arthrosis problems. Makris has used ChSase ABC combined with TGF-β1 and lysyl oxidase to treat fibrocartilage damage. The results showed that the tensile stiffness and strength of the interface were both increased by 730% under treatment, compared to the untreated group.3 ChSase ABC could also stimulate nerve regeneration after brain injury.4 Siebert has studied the effect of ChSase ABC on spinal injury and found that it could improve the axon regeneration significantly.5 Additionally, Lee reported that the adherence of cartilage cell and the surface was enhanced notably under the treatment of ChSase ABC.6
As a tool enzyme to study glycosaminoglycan, a number of ChSase ABCs were identified from Proteus vulgaris (P. vulgaris) and were divided into ChSase ABC I and ChSase ABC II subfamilies, due to their different catalytic sites and kinetic parameters. It has been reported that the catalytic efficiency of ChSase ABC I was higher than that of ChSase ABC II.7,8 ChSase ABC I contained three domains, including the N-terminal domain (residues 25–234), the catalytic domain (residues 235–617) and the C-terminal domain (residues 618–1021). The C-terminal domain showed 21% homology to the C-terminal domain of ChSase AC from Flavobacterium heparinum, and 17–19% similarity to the C-terminal domain of hyaluronidase from Streptococcus pneumonia.9,10 ChSase ABC II also had three domains, including the N-terminal domain (residues 14–170), the catalytic domain (residues 171–593), which was similar to ChSase AC, hyaluronidase and heparinase II, and the C-terminal domain (residues 594–1014).11
ChSase ABC I and ChSase ABC II have been isolated from P. vulgaris and recombinantly expressed in Escherichia coli (E. coli).12,13 His501, Tyr508 and Arg560 of ChSase ABC I were determined as active sites and the catalytic mechanism of the enzyme has been illustrated.9,10,14–16 The substrate first bound ChSase ABC I in a completely open form, and then its β-1,4-galactosaminidic bond linking disaccharide units was cut by the active site amino acids, and the substrate was finally degraded into mainly 4,5-unsaturated uronic acids.17 The active sites of ChSase ABC II were determined to be His454, Tyr461, Arg514 and Glu628.18
Although the enzyme activity of ChSase ABC I has been studied extensively, its activity with CS B as substrate was still relatively low. CS B widely existed in connective tissues and was used to guide many biological processes, such as proliferation of cell, transmission of signal and mediation of inflammation, etc.1,19,20 It was widely used in the field of food industry, medicine and scientific researches.21 Due to its low bioavailability, high molecular weight CS B has to be degraded into low molecular weight ones which could be utilized more efficiently.22,23 Therefore, in this study, we set out to improve the enzyme activity of ChSase ABC I with CS B as substrate.
Currently, two main genetic manipulation approaches have been used to enhance the activity of an enzyme. One approach is directed evolution, which combined random mutagenesis and high-throughput screening.24,25 The other approach is rational design or semi-rational design, which depends on the knowledge about the protein structure, catalytic mechanism, and the sequence alignment among the related homologues.26 In this study, 13 amino acids of ChSase ABC I were selected for site-directed mutagenesis through sequence alignment and molecular docking simulation analysis for the first time. Among these sites, we identified a new active site Arg660, mutation of which caused the total loss of the enzyme activity. We also revealed that mutating Asn795 to Ala or Trp818 to Ala could improve the specific activity both with CS A and CS B. In addition, the kinetic parameters Vmax, kcat/Km and the thermostabilities of the Asn795Ala and Trp818Ala mutants all showed improvement to different extent, compared with those of the wild type enzyme.
2. Materials and methods
2.1 Strains, plasmids, and reagents
P. vulgaris (KCTC 2579) was purchased from Korea KCTC storage. E. coli DH5α and E. coli BL21(DE3) were purchased from Beijing Biomed Biological Technology Co., Ltd. The plasmid pMAL-c2x was stored in our laboratory. Q5™ High-Fidelity 2× Master Mix, T4 DNA ligase and the restriction enzyme were obtained from New England Biolabs. The substrates CS A and CS B (MW: 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were purchased from Nanjing Oddo's Biological Technology Co., Ltd. All the reagents used in this study were analytical grade.
000) were purchased from Nanjing Oddo's Biological Technology Co., Ltd. All the reagents used in this study were analytical grade.
2.2 Construction of the plasmid pMAL-c2x-ChSase ABC I
Genomic DNA extracted from P. vulgaris was amplified by PCR using the upstream primer P1: 5′-CG![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ATGGCCACCAGCAATCCTGCATT-3′ (underlined sequence was the position of BamH I site) and the downstream primer P2: 5′-AA
ATGGCCACCAGCAATCCTGCATT-3′ (underlined sequence was the position of BamH I site) and the downstream primer P2: 5′-AA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TTATCAAGGGAGTGGCGAGAGTTTG-3′ (underlined sequence was the position of a Pst I site). The PCR program was 98 °C for 3 min, followed by 30 cycles of 98 °C for 7 s, 60 °C for 30 s, 72 °C for 2 min, followed by an elongation at 72 °C for 2 min.
TTATCAAGGGAGTGGCGAGAGTTTG-3′ (underlined sequence was the position of a Pst I site). The PCR program was 98 °C for 3 min, followed by 30 cycles of 98 °C for 7 s, 60 °C for 30 s, 72 °C for 2 min, followed by an elongation at 72 °C for 2 min.
The amplified ChSase ABC I fragment was digested by BamH I and Pst I, and ligated with T4 DNA ligase into plasmid pMAL-c2x which was digested by the same restriction enzymes, to construct plasmid pMAL-c2x-ChSase ABC I. The constructed plasmid was sequenced by Beijing Genomics Institute and proved to be 100% identical to the sequence in the NCBI website (GQ996964.1).
2.3 Mutagenesis of ChSase ABC I
Site-directed mutagenesis was performed using the Quik-Change Site-Directed Mutagenesis Kit (Stratagene, USA) following the manufacturer's instructions. After the PCR process, the products were subjected to a treatment with Dpn I (2 μL) for 1 h at 37 °C to ensure complete digestion of the original templates (cloned ChSase ABC I in pMAL-c2x). The primers for mutating the thirteen amino acids to Ala were shown in Table 1. Mutations were verified through DNA sequencing.| Mutants | CS A (IU mg−1 protein) | CS B (IU mg−1 protein) |
|---|---|---|
| Wild type | 22.52 ± 0.91 | 14.60 ± 0.06 |
| Asp433Ala | 7.13 ± 0.03 | 5.86 ± 0.02 |
| Ser441Ala | 22.86 ± 0.97 | 13.39 ± 0.61 |
| Asn468Ala | 23.59 ± 1.01 | 14.49 ± 0.63 |
| Ser474Ala | 21.98 ± 1.21 | 12.64 ± 0.56 |
| Asn515Ala | 17.36 ± 0.88 | 10.03 ± 0.45 |
| Asn564Ala | 26.16 ± 1.34 | 11.76 ± 0.55 |
| Tyr575Ala | 28.82 ± 1.36 | 13.90 ± 0.56 |
| Tyr594Ala | 28.12 ± 0.88 | 13.26 ± 0.64 |
| Phe609Ala | 28.97 ± 0.97 | 14.07 ± 0.78 |
| Tyr623Ala | 24.88 ± 0.67 | 11.57 ± 0.52 |
| Arg660Ala | 0 | 0 |
| Asn795Ala | 31.32 ± 1.45 | 27.07 ± 1.21 |
| Trp818Ala | 31.14 ± 1.22 | 24.90 ± 1.25 |
2.4 Protein expression and purification
The wild-type and mutated plasmids were transformed into E. coli BL21 (DE3) for protein expression. The transformants were cultured with shaking at 200 rpm at 37 °C for 12 h as seed culture. 500 μL of seed culture was transferred into 50 mL fresh medium which was in 250 mL triangle bottle and cultured until OD600 reached around 0.6 at 37 °C, and then IPTG (final concentration 0.5 mmol L−1) was added for induction. The bacterial cells were cultured with shaking at 180 rpm at 16 °C for 20 h.After induction, the cells were centrifuged for 6 min at 8000 rpm at 4 °C. The supernatant was discarded, and then 15 mL buffer A (20 mmol L−1 Tris–HCl, pH 7.4) was added to resuspend the cells, which were then disrupted by ultrasonic treatment with SCIENTZ (JY 92-IIN) instrument. The conditions were described as follows: ultrasonic sound 3 s, intermittent 5 s, ultrasonic power 60%, total time 15 min. After another centrifugation, the supernatant was obtained as crude enzyme solution. Then the supernatant was loaded onto MBPTrap HP affinity column (USA GE Healthcare), and eluted with buffer A containing 10 mmol L−1 maltose. The purity of MBP–ChSase ABC I was detected by SDS-PAGE analysis. The concentrations of acrylamide of separation gel and upper gel were 12% and 5%. Separation gel was made up of 1.75 mL ddH2O, 1.25 mL 1.5 mol L−1 Tris–HCl, pH 8.8, 2 mL 30% acrylamide, 0.05 mL 10% ammonium persulfate and 0.008 mL TEMED. Upper gel was made up of 1.45 mL ddH2O, 0.625 mL 0.5 mol L−1 Tris–HCl, pH 6.8, 0.42 mL 30% acrylamide, 0.03 mL 10% ammonium persulfate and 0.005 mL TEMED. The condition of electrophoresis was 220 V, 30 min.
2.5 Activity assay
MBP–ChSase ABC I activity was measured according to the UV 232 nm method.27 The enzymatic reaction was carried out at 37 °C using CS A and CS B as substrates in the same buffer (20 mmol L−1 Tris–HCl, pH 7.4). CS A and CS B degradation was monitored by UV absorbance at 232 nm and the activity was calculated using a molar extinction coefficient of 3800 L mol−1 cm−1. Protein concentration was detected by Bradford Protein Assay Kit (Bio-rad). One international unit was defined as the amount of protein that could form 1 μmol L−1 4,5-unsaturated uronic acid per minute at 37 °C.282.6 Determination of kinetic parameters and thermostability of mutated enzymes
The kinetic parameters, Vmax, Km and kcat of recombinant MBP–ChSase ABC I were determined by substrates CS A and CS B in concentration range of 0–80 μmol L−1 (0, 10, 20, 40, 60, 80 μmol L−1) at 50 °C in 20 mmol L−1 Tris–HCl pH 7.73 buffer.29 The thermostabilities of wild type and mutants were investigated at pH 7.4 after incubation of the enzyme solutions in the absence of substrate at various temperatures (30–50 °C) for 30, 60, 90, 120, 150, 180, 210 min, respectively.2.7 Molecular docking simulation
Molecular docking simulation of ChSase ABC I with sulfated glycosaminoglycan molecule was performed using Discovery Studio (version 2.5). The tertiary structures of ChSase ABC I (PDB code: 1HN0) and sulfated glycosaminoglycan molecule (PDB code: 1C4S) were all downloaded from the Protein Data Bank (http://www.rcsb.org). For the molecular docking of the ChSase ABC I protein, His501, Tyr508 and Arg560 (ref. 9,16) were used as the active sites and binding sites. The molecular docking simulations were performed with the default parameters of the CDOCKER module. The binding free energy of the complexes was calculated using a Discovery Studio protocol with default parameters. The complexes between the sulfated glycosaminoglycan molecule and the wild type or mutants with the lowest binding free energy were used as the modeled docking structures.303. Results
3.1 Selection of promising sites of ChSase ABC I for site-directed mutagenesis
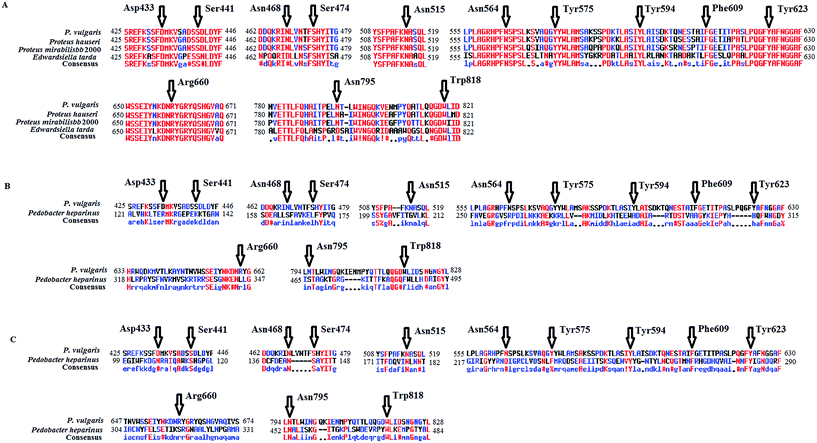
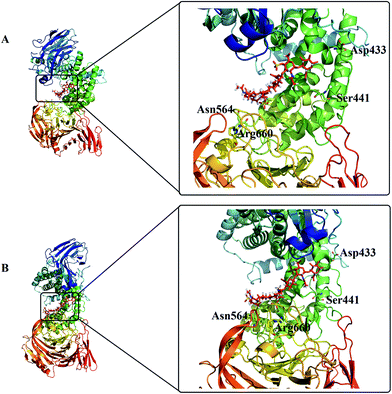
To select promising sites to be mutated for enhancing activity, first the amino acid sequence of ChSase ABC I from P. vulgaris was aligned with those of three homologs from other organisms. We also did a comparison between the sequences of ChSase ABC I and ChSase AC from Pedobacter heparinus (Fig. 2B), and an alignment between the sequences of ChSase ABC I and ChSase B from Pedobacter heparinus (Fig. 2C). The alignments resulted in stretches of amino acids which were conserved among the homologs (Fig. 2A). And then, these residues were subjected to a more detailed screening. Based on the solved crystal structure, the possibility of the conserved residues in stabilizing the enzyme, facilitating the catalysis, and maintaining the structural integrity were assessed. The possible active sites were determined to be Asp433, Ser441, Asn468, Ser474, Asn515, Asn564, Tyr575, Tyr594, Phe609, Tyr623, Arg660, Asn795 and Trp818, which were highly conserved amino acids among different sources of ChSase ABC I enzymes. Specially, the alignment of ChSase ABC I and ChSase AC showed that these amino acids were all different from the corresponding amino acids of ChSase AC, however, the alignment of ChSase ABC I and ChSase B showed that those amino acids was same with the amino acids of ChSase B except Arg660.Molecular docking simulation of ChSase ABC I with sulfated glycosaminoglycan molecule (total ligand–receptor interaction energy was −6.31 kcal mol−1) showed that most of the amino acids obtained from sequence alignment (Asp433, Ser441, Asn468, Ser474, Asn515, Asn564, Tyr575, Tyr594, Phe609, Tyr623 and Arg660) were around the active pocket of the enzyme (Fig. 3). Two amino acids, Asn795 and Trp818 were both away from the previously suggested active sites. Combining the results of sequence alignment and molecular docking simulation analysis, we finally selected the 13 amino acids for site-directed mutagenesis. The residues were mutated to Ala separately, and the impact of each mutation on enzyme activity was studied respectively.
3.2 Specific activities of wild type and ChSase ABC I mutants
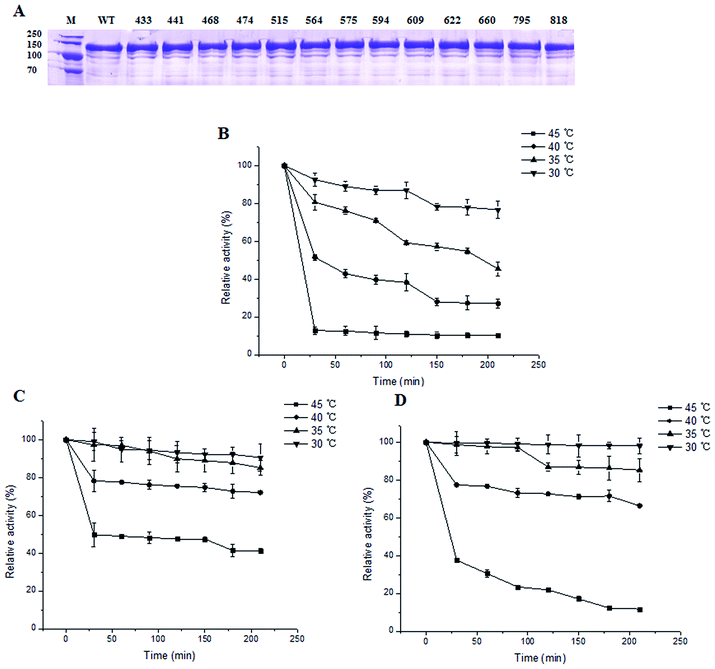
The results of the specific activity assay were shown in Table 1. Among the 13 mutants, 8 mutants showed decreased activity towards CS A and 11 mutants showed decreased activity towards CS B. Specifically, mutating Arg660 to Ala caused a total loss of the enzyme activity either with CS A or CS B as substrate. We further mutated Arg660 to other amino acids including Gly, Leu, Asp, Cys, Gln, Glu, His, Ile, Asn, Lys, Met, Pro, Phe, Ser, Thr, Trp, Tyr, and Val, however, none of the mutants showed any activity to CS A or B (data not shown). The results illustrated that Arg660 was an active sites of the enzyme.On the other hand, the specific activities of Asn795Ala and Trp818Ala were 31.32 ± 1.45 and 31.14 ± 1.22 IU mg−1 protein when CS A was used as substrate. The specific activities of the two mutants were 1.39 and 1.38 times higher than that of the wild type enzyme. With CS B as substrate, the specific activities of Asn795Ala and Trp818Ala mutants were 1.85 and 1.71 times higher than that of the wild type enzyme respectively. None of the 13 mutations disrupted the expression and purity of the MBP–ChSase ABC I according to the SDS-PAGE analysis (Fig. 4A).
3.3 Kinetic studies of the wild type and mutants
Since both Asn795Ala and Trp818Ala mutants showed higher activities than the wild type either with CS A or CS B as substrate, we moved on to characterize the kinetic parameters of the mutants and the wild type. The kinetic parameters, Vmax, Km and kcat were determined by substrate CS A and CS B and calculated according to Michaelis–Menten (1/v = Km/(Vmax × [S])+1/Vmax). The results showed that when CS A was used as substrate, Vmax and kcat/Km of the Asn795Ala mutant were 1.20 and 6.28 times higher than those of the wild type, respectively (Table 2, Fig. S1†). The catalytic efficiency was increased with substrate CS A. On the other hand, however, Vmax and kcat/Km were 1.10 and 1.57 times lower than those of wild type with CS B as substrate. The catalytic efficiency was decreased with substrate CS B.| Mutants | CS A | CS B | ||||
|---|---|---|---|---|---|---|
| Vmax (μmol L−1 s−1) | Km (μmol L−1) | kcat/Km (L μmol−1 s−1) | Vmax (μmol L−1 s−1) | Km (μmol L−1) | kcat/Km (L μmol−1 s−1) | |
| Wild type | 18.70 ± 0.52 | 73.10 ± 4.64 | 8.00 ± 0.07 | 5.04 ± 0.05 | 8.17 ± 0.01 | 19.16 ± 0.45 |
| Asn795Ala | 22.52 ± 1.02 | 7.25 ± 0.04 | 50.25 ± 3.01 | 4.59 ± 0.04 | 13.65 ± 0.31 | 12.22 ± 0.57 |
| Trp818Ala | 19.84 ± 0.87 | 16.92 ± 0.32 | 14.83 ± 0.56 | 2.94 ± 0.01 | 2.29 ± 0.01 | 58.44 ± 4.51 |
For the Trp818Ala mutant, Vmax and kcat/Km were 1.06 and 1.85 times higher than those of wild type with CS A as substrate. When CS B was used as substrate, the Vmax was 2.94 ± 0.01 μmol L−1 s−1, which was lower than that of wild type, and kcat/Km was 3.05 times higher than that of wild type. Those illustrated that the catalytic efficiency of the Trp818Ala mutant was also improved either CS A or CS B used as substrate.
3.4 Thermostability of wild type and ChSase ABC I mutants
The thermostabilities of Asn795Ala and Trp818Ala mutants were also investigated. Generally, the enzyme activities of the wild type, Asn795Ala and Trp818Ala mutants all decreased gradually with the increase of temperature and time. However, the thermostabilities of both Asn795Ala (Fig. 4C) and Trp818Ala (Fig. 4D) mutants were improved when compared with that of wild type (Fig. 4B). The absolute activities for 100% relative activity of wile type, Asn795Ala and Trp8185Ala mutants which was purified were 76.00 ± 1.71 IU mg−1 protein, 123.77 ± 0.13 IU mg−1 protein and 112.07 ± 0.71 IU mg−1 protein with CS A as substrate.The relative activity of Asn795Ala mutant could retain 90% after 210 min at 30 °C and 35 °C. The relative activity of Trp818Ala mutant was 98% after 210 min at 30 °C and 90% at 35 °C, compared with 78% at 30 °C and 45% in 35 °C of the wild type enzyme. The Asn795Ala mutant retained 72% activity after 210 min at 40 °C, which was 2.7 times higher than wild type. Furthermore, the relative activity of Asn795Ala mutant could keep 41% after 210 min at 45 °C, compared with 10% and 12% of the wild type protein and the Trp818Ala mutant, respectively. Taken together, these results indicated that the thermostabilities of Asn795Ala and Trp818Ala mutants were both improved to different extent.
4. Discussion
Heterologous expression was used widely in the process of industrial production to obtain large amounts of products. E. coli was well recognized as host for recombinant protein expression, due to its ease of genetic manipulation, low cost of cultivation, and capability for fast and large-scale production.31 However, many problems also emerged, such as low level of expression, misfolding/unfolding proteins and insoluble expression of proteins like inclusion body.32 Expression of a soluble protein was hampered by the size of the gene, the signal sequence and the effect of some amino acids. So far, the csl ABC was expressed in E. coli and obtained through complicated steps of separation and purification; however, most of the enzyme was in the inclusion body form.12,13,33 Several methods were used to solve those problems, such as using different tags like MBP, His and GST,4,29,34 and mutating specific amino acids.35–38In this study, ChSase ABC I was expressed with an MBP tag and site-directed mutations were conducted. We identified 8 residues whose mutations caused decreased enzyme activities with CS A as substrate and 11 residues whose mutations caused decreased enzyme activities with CS B as substrate, suggesting that these residues either constitute part of the active site or their mutations influence the structural integrity of the enzyme. Of these mutants, the Arg660Ala mutant completely lost its activity, revealing that Arg660 was a new active site, together with previously identified His501, Tyr508 and Arg560.9,10,14–16 Sequence alignment revealed that Arg660 is highly conserved among ChSase ABC I enzymes, but is not conserved in ChSase AC and ChSase B enzymes (Fig. 2). Investigating the molecular docking simulation suggested that Arg660 sits adjacent to the proposed substrate binding site, indicating that this residue might participate in the degradation process of the substrate (Fig. 3).
In our study, the enzyme activities of the Asn795Ala and Trp818Ala mutants were higher than that of wild type either with CS A or CS B as substrate. In Tkalec et al.'s studies, the enzyme activity of ChSase AC with CS A as substrate and that of ChSase B with CS B as substrate were 79.5 mU per mL A600 unit and 29.4 mU per mL A600 unit,39 both of which were lower than those of the two mutants in this study. Site-directed mutagenesis of ChSase B from Flavobacterium heparinum had also been investigated and the kcat/Km were also determined to be 41.3 μmol L−1 s−1,40 lower than that of the Trp818Ala mutant. Sequence alignment showed that Asn795 and Trp818 of ChSase ABC I were different from the corresponding amino acids of ChSase AC, but were same with the corresponding amino acids of ChSase B. Structural analysis revealed that Asn795 and Trp818 are both away from the previously suggested active site, and the surfaces of Asn795Ala and Trp818Ala mutants sunk more than that of wild type (Fig. 5). The mutations of the two amino acids might not directly influence the catalytic process. However, both of the two amino acids are located in the C-terminal domain of the enzyme, and the mutations of them might slightly change the conformation of the C-terminal domain, thus influencing the active site in the catalytic domain through domain–domain interaction. This might result in the more closely binding of the substrate in the enzyme. However, the detailed variations of the active sites in the two mutants awaited the characterization of their crystal structures.
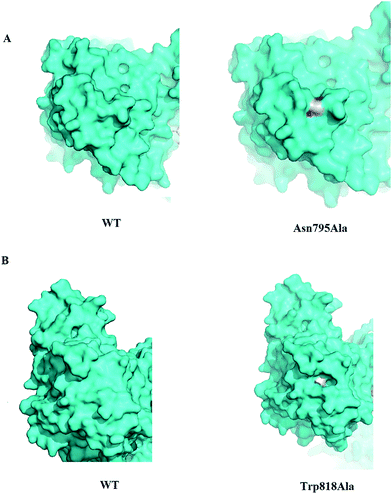 | ||
| Fig. 5 The surface of mutants. (A) The surfaces of wild type and Asn795Ala mutant. (B) The surfaces of wild type and Trp818Ala mutant. | ||
Gln140 was chosen previously as an important site, which influenced the thermostability of ChSase ABC I because of its improper value of φ and ψ in the Ramachandran plot. The Gln140Gly and Gln140Ala mutants were able to improve both the activity and thermostability of the enzyme, but Gln140Asn reduced the enzyme activity and destabilized it. This investigation demonstrated that relief of conformational tension could be considered as a possible approach to increase the stability of the protein.35 Nazari-Robati et al. also used glycerin, sorbitol and trehalose to improve the thermostability of ChSase ABC I. Results showed that trehalose improved both enzyme activity and thermostability.41 In this study, the increase of the thermostability indicated that these two mutants might improve the conformational stability of the enzyme and delay the loss of activity of the enzyme.
5. Conclusions
In all, we identified a new active site residue Arg660 in ChSase ABC I. Additionally, the specific activities of Asn795Ala and Trp818Ala with CS A and CS B as substrates were the highest ever reported. The two mutants also showed enhanced thermostabilities compared with the wild type enzyme. Our studies shed light on the further manipulation of the ChSase ABC I for better enzyme activity and thermostability. This study can also guide the industrial process of producing ChSase ABC I and low molecular weight CS B and the application of ChSase ABC I and low molecular weight CS B in food industry and clinical therapies.Acknowledgements
The authors would like to acknowledge financial support of the National Natural Science Foundation of China (No. 21306002 and 21176018), the Youth Elite Project of Higher Education of Beijing (YETP1801), the Science-Technology Foundation of Beijing Municipal Commission of Education (KM201410858002), the Science and Technology Innovation Platform (PXM2014 014306 000064).References
- M. Bernfield, M. Gotte, P. W. Park, O. Reizes, M. L. Fitzgerald and J. Lincecum, Annu. Rev. Biochem., 1999, 68, 729–777 CrossRef CAS PubMed.
- R. M. Lauder, Complement Ther. Med., 2009, 17, 56–62 CrossRef PubMed.
- E. A. Makris, R. F. MacBarb, N. K. Paschosa, J. C. Hua and K. A. Athanasioua, Biomaterials, 2014, 35, 6787–6796 CrossRef CAS PubMed.
- X. R. Chen, S. J. Liao, L. X. Ye, Q. Gong, Q. Ding, J. S. Zeng and J. Yu, Brain Res., 2014, 1543, 324–333 CrossRef CAS PubMed.
- J. R. Siebert, D. J. Stelzner and D. J. Osterhout, Exp. Neurol., 2011, 231, 19–29 CrossRef CAS PubMed.
- M. C. Lee, K. L. Sung and M. S. Kurtis, Clin. Orthop. Relat. Res., 2000, 370, 286–294 CrossRef.
- T. Yamagata, H. Saito, O. Habuchi and S. Suzuki, J. Biol. Chem., 1968, 243, 1523–1535 CAS.
- A. Hamai, N. Hashimoto, H. Mochizuki, F. Kato, Y. Makiguchi, K. Horie and S. Suzuki, J. Biol. Chem., 1997, 272, 9123–9130 CrossRef CAS PubMed.
- W. J. Huang, V. V. Lunin, Y. Li, S. Suzuki, N. Sugiura, H. Miyazono and M. Cygler, J. Mol. Biol., 2003, 328, 623–634 CrossRef CAS.
- J. Fethiere, B. Eggimann and M. Cygler, J. Mol. Biol., 1999, 288, 635–647 CrossRef CAS PubMed.
- D. Shaya, B. S. Hahn, N. Y. Park, J. S. Sim, Y. S. Kim and M. Cygler, Biochemistry, 2008, 47, 6650–6661 CrossRef CAS PubMed.
- N. Sato, M. Shimada, H. Nakajima, H. Oda and S. Kimura, Appl. Microbiol. Biotechnol., 1994, 41, 39–46 CrossRef CAS.
- V. Prabhakar, I. Capila, C. J. Bosques, K. Pojasek and R. Sasisekharan, Biochem. J., 2005, 386, 103–112 CrossRef CAS PubMed.
- V. Prabhakar, I. Capila, R. Raman, A. Srinivasan, C. J. Bosques, K. Pojasek, M. A. Wrick and R. Sasisekharan, Biochemistry, 2006, 45, 11130–11139 CrossRef CAS PubMed.
- W. J. Huang, A. Matte, S. Suzuki, N. Sugiura, H. Miyazono and M. Cygler, Acta Crystallogr., Sect. A: Found. Crystallogr., 2000, 56, 904–906 CrossRef CAS PubMed.
- V. Prabhakar, I. Capila, C. J. Bosques, K. Pojasek and R. Sasisekharan, Biochem. J., 2005, 390, 395–405 CrossRef CAS PubMed.
- C. S. Rye and S. G. Withers, J. Am. Chem. Soc., 2002, 124, 9756–9767 CrossRef CAS PubMed.
- D. Shaya, B. S. Hahn, T. M. Bjerkan, W. S. Kim, N. Y. Park, J. S. Sim, Y. S. Kim and M. Cygler, Glycobiology, 2008, 18, 270–277 CrossRef CAS PubMed.
- X. Bao, S. Nishimura, T. Mikami, S. Yamada, N. Itoh and K. Sugahara, J. Biol. Chem., 2004, 279, 9765–9776 CrossRef CAS PubMed.
- K. Sugahara, T. Mikami, T. Uyama, S. Mizuguchi, K. Nomura and H. Kitagawa, Curr. Opin. Struct. Biol., 2003, 13, 612–620 CrossRef CAS PubMed.
- S. F. Penc, B. Pomahac, E. Eriksson, M. Detmar and R. L. Gallo, J. Clin. Invest., 1999, 103, 1329–1335 CrossRef CAS PubMed.
- U. Aich, Z. Shriver, K. Tharakaraman, R. Raman and R. Sasisekharan, Anal. Chem., 2011, 83, 7815–7822 CrossRef CAS PubMed.
- M. L. Garron and M. Cygler, Glycobiology, 2010, 20, 1547–1573 CrossRef CAS PubMed.
- S. Molloy, J. Nikodinovic-Runic, L. B. Martin, H. Hartmann, F. Solano, H. Decker and K. E. O’Connor, Biotechnol. Bioeng., 2013, 110, 1849–1857 CrossRef CAS PubMed.
- K. Iinoya, T. Kotani, Y. Sasano and H. Takagi, Biotechnol. Bioeng., 2009, 103, 341–352 CrossRef CAS PubMed.
- C. Q. Zhong, S. Song, N. Fang, X. Liang, H. Zhu, X. F. Tang and B. Tang, Biotechnol. Bioeng., 2009, 104, 862–870 CrossRef CAS PubMed.
- L. B. Shields, Y. P. Zhang, D. A. Burke, R. Gray and C. B. Shields, Surg. Neurol., 2008, 69, 568–577 CrossRef PubMed.
- S. Chen, F. C. Ye, Y. Chen, Y. Chen, H. X. Zhao, R. Yatsunami, S. Nakamura, F. Arisaka and X. H. Xing, Biotechnol. Bioeng., 2011, 108, 1841–1851 CrossRef CAS PubMed.
- Z. Y. Chen, Y. Li and Q. P. Yuan, Int. J. Biol. Macromol., 2015, 72, 6–10 CrossRef CAS PubMed.
- S. J. Kim, J. C. Joo, B. K. Song, Y. J. Yoo and Y. H. Kim, Biotechnol. Bioeng., 2015, 112, 668–676 CrossRef CAS PubMed.
- S. Chen, Z. L. Huang, J. J. Wu, Y. Chen, F. C. Ye, C. Zhang, R. Yatsunami, S. Nakamura and X. H. Xing, Appl. Microbiol. Biotechnol., 2013, 97, 2907–2916 CrossRef CAS PubMed.
- D. M. Francis and R. Page, Strategies to optimize protein expression in E. coli, John Wiley & Sons, Inc., 2010, vol. 5.24, pp. 1–29 Search PubMed.
- V. Prabhakar, I. Capila, V. Soundararajan, R. Raman and R. Sasisekharan, J. Biol. Chem., 2009, 284, 974–982 CrossRef CAS PubMed.
- Z. Y. Chen, Y. Li and Q. P. Yuan, Int. J. Biol. Macromol., 2015, 78, 96–101 CrossRef CAS PubMed.
- M. Nazari-Robati, K. Khajeh, M. Aminian, N. Mollania and A. Golestani, Biochim. Biophys. Acta, 2013, 1834, 479–486 CrossRef CAS PubMed.
- C. Zhang, J. Li, B. Yang, F. He, S. Y. Yang, X. Q. Yu and Q. Wang, RSC Adv., 2014, 4, 27526–27531 RSC.
- Z. A. Sánchez-Quitian, V. Rodrigues-Junior, J. G. Rehm, P. Eichler, D. B. B. Trivella, C. V. Bizarro, L. A. Basso and D. S. Santos, RSC Adv., 2015, 5, 830–840 RSC.
- N. Minovski, M. Novic and T. Solmajer, RSC Adv., 2015, 5, 16162–16172 RSC.
- L. A. Tkalec, D. Fink and F. Blain, Appl. Environ. Microbiol., 2000, 66, 29–35 CrossRef.
- K. Pojasek, Z. Shriver, P. Kiley, G. Venkataraman and R. Sasisekharan, Biochem. Biophys. Res. Commun., 2001, 286, 343–351 CrossRef CAS PubMed.
- M. Nazari-Robati, K. Khajeh, M. Aminian, M. Fathi-Roudsari and A. Golestani, Int. J. Biol. Macromol., 2012, 50, 487–492 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra15220h |
| This journal is © The Royal Society of Chemistry 2015 |