Influence of carbon nanomaterial defects on the formation of protein corona†
Bishwambhar
Sengupta
ab,
Wren E.
Gregory
ab,
Jingyi
Zhu
 a,
Siva
Dasetty
c,
Mehmet
Karakaya
a,
Jared M.
Brown
d,
Apparao M.
Rao
ae,
John K.
Barrows
f,
Sapna
Sarupria
*b and
Ramakrishna
Podila
*abe
a,
Siva
Dasetty
c,
Mehmet
Karakaya
a,
Jared M.
Brown
d,
Apparao M.
Rao
ae,
John K.
Barrows
f,
Sapna
Sarupria
*b and
Ramakrishna
Podila
*abe
aDepartment of Physics and Astronomy, Clemson Nanomaterials Center, Clemson University, Clemson, South Carolina 29634, USA. E-mail: rpodila@g.clemson.edu; Fax: +1-864-656-0805; Tel: +1-864-656-4447
bLaboratory of Nano-biophysics, Clemson University, Clemson, South Carolina 29634, USA. E-mail: ssarupr@g.clemson.edu; Fax: +1-864-656-0784; Tel: +1-864-656-3258
cDepartment of Chemical and Biomolecular Engineering, Clemson University, Clemson, South Carolina 29634, USA
dDepartment of Pharmaceutical Sciences, Skaggs School of Pharmacy and Pharmaceutical Sciences, The University of Colorado Anschutz Medical Campus, Aurora, CO 80045, USA
eCOMSET, Clemson University, Anderson, SC 29625, USA
fDepartment of Genetics and Biochemistry, Clemson University, Clemson, South Carolina 29634, USA
First published on 23rd September 2015
Abstract
In any physiological media, carbon nanomaterials (CNM) strongly interact with biomolecules leading to the formation of biocorona, which subsequently dictate the physiological response and the fate of CNMs. Defects in CNMs play an important role not only in material properties but also in the determination of how materials interact at the nano-bio interface. In this article, we probed the influence of defect-induced hydrophilicity on the biocorona formation using micro-Raman, photoluminescence, infrared spectroscopy, electrochemistry, and molecular dynamics simulations. Our results show that the interaction of proteins (albumin and fibrinogen) with CNMs is strongly influenced by charge-transfer between them, inducing protein unfolding which enhances conformational entropy and higher protein adsorption.
1. Introduction
The increased use of engineered nanomaterials (ENMs) in biomedicine has raised natural concerns regarding their adverse immune response.1,2 Blood is the first physiological environment that ENMs encounter upon intravenous injection for use as a drug delivery vector or a biomedical imaging contrast agent. Previously, it was shown that many different proteins and lipids compete between themselves for adsorbing on to the surface of ENMs to form a bio “corona”.3–5 Interactions between ENMs and the adsorbed proteins in the bio-corona may alter the structural arrangement of proteins leading to changes in their secondary structure through protein unfolding.3–9The formation of biocorona and ensuing protein structural changes play an important role in complement initiation (through C3b protein) and adverse reactions of ENMs.10,11 A detailed understanding of the ENM-biomolecular interactions is therefore necessary to completely understand ENM toxicity and immune response.12–15
Among the wide variety of nanostructures, carbon nanomaterials (CNMs) represent an intriguing set of ENMs from biological and toxicity standpoints because CNMs: (i) possess excellent affinity for proteins through hydrophobic and aromatic π–π stacking interactions,16,17 and (ii) exhibit unique molecular charge-transfer among themselves and with other molecules including proteins.18–20 For instance, C60 is known to display charge-transfer interactions with electron donor molecules including organic amines,20 while graphene and single-walled carbon nanotubes (SWNTs) can act as either electron donors or acceptors with proteins such as streptavidin.19 Indeed, charge transfer interactions between proteins and CNMs have previously been implicated in destabilizing adsorbed proteins/enzymes, which could in turn elicit adverse physiological response.9,21 While the root cause of ENM (particularly CNM) toxicity is still a subject of ongoing research, numerous studies have identified the presence of bioactive defects in ENMs (including Au, Ag, TiO2, SiO2, and CNMs) as the common denominator in their physiological response.1,12–15 In case of CNMS, it has been hypothesized that the presence of neutral and charged defects could elicit an adverse physiological response possibly by generating highly reactive oxygen species and inducing structural changes in proteins through charge transfer reactions.1,9,18 Despite this obvious importance of defects and charge transfer in CNM toxicity, the influence of structural and functional defects in CNMs on biomolecular adsorption and immune response remains poorly understood.21 Although the formation of protein corona on CNMs has been extensively studied in recent years, the underlying physical and chemical processes in protein and defected CNM interactions (e.g., charge transfer between proteins and defects) have not yet been completely understood. To elucidate the role of defects and their associated charge transfer in biological interactions of CNMs, it is imperative to synthesize CNMs with different defect content and study their effects on protein corona.
In this article, we investigated the interaction of bovine serum albumin (BSA) and fibrinogen with defected CNMs such as multi-walled carbon nanotubes (MWNTs), graphene, and graphene oxide nanoribbons (GNRs and GONRs) using micro-Raman spectroscopy, photoluminescence (PL), infrared absorption, electrochemistry and molecular dynamics (MD) simulations. The defects in the CNMs used in this study include edges (e.g., edges coming from finiteness of GNRs), functional groups (e.g., hydroxyl and carboxyl groups on GONRs), and other topological defects (e.g., vacancies and Stone–Wales defects in MWNTs). While MWNTs are seamless cylinders with some topological defects and only a few available edges, GNRs and GONRs provide more edges with different functional groups and are well suited for investigating the influence of defects on protein adsorption. Furthermore, the lack of functional group-type defects on MWNTs makes them more hydrophobic than GNRs (weakly hydrophilic) and GONRs (strongly hydrophilic), allowing us to understand the interplay between shape and defect-induced hydrophilicity of CNMs on their biomolecular interactions. We have identified fibrinogen (tubular structure with high-internal stability) and albumin (globular with relatively low internal stability) as the model proteins of interest due to their contrasting properties and binding affinities. We observed: (i) that BSA exhibited similar adsorption on all the CNMs, whereas fibrinogen showed better binding to GNRs and GONRs, (ii) the charge transfer for the case of BSA (/fibrinogen) adsorbed on MWNTs (/GONR) to be the highest, and (iii) an enhanced relaxation of α-helices in proteins with highest charge transfer/adsorption (viz., MWNTs in case of BSA and GONRs for fibrinogen) suggesting that the charge transfer reactions between proteins and CNMs may be critical to control ENM-biomolecular interactions.
2. Experimental methods and materials
2.1 Sample preparation
In this study, M-grade MWNTs (diameter: 50 nm and length > 5 μm) were obtained from NanoTechLabs, Yadkinville, NC. The unzipping and refluxing methods were used to prepare GNRs and GONRs from MWNTs. In the unzipping process, 500 mg of MWNTs were mixed with 100 mL of concentrated H2SO4 (Sigma Aldrich, 95–98% purity) and bath-sonicated for 3 hours (Aquasonic P250HT). Subsequently, 2 g of KMnO4 (Sigma Aldrich, >98% purity) was added and stirred for 3 hours at 70 °C. Thereafter 3 mL of 30% H2O2 (VWR international, 30% w/w) was added to finish the reaction. The unzipped GNRs were collected by centrifugation (Heraeus Instruments, Labofuge 400, at 2900 rpm for 15 min). The pellet was resuspended 3 times in de-ionized (DI) water and re-centrifuged to remove the residual acid and inorganic salts. All pellets were air dried overnight to remove any remaining impurities. The obtained GNRs were further refluxed with 100 mL 30% H2O2 and stirred for 2 hours at 70 °C. After the completion of reflux, 200 mL of concentrated H2SO4 (VWR international, 95–98%) was slowly added and the mixture was left to stir for another 1 hour at 70 °C. The resulting suspension was diluted and filtered through 0.45 μm polyamide filter, dried, and resuspended in pure DI water. This procedure was repeated at least 3 times to wash away residual chemicals and obtain GONRs. For the protein binding, BSA (Spectrum Chemical Mfg. Corp, CA) and fibrinogen (Alfa Aesar) were incubated at 37 °C for 12 hours with CNMs. We used physiologically relevant concentrations of BSA 5–60 g L−1 and fibrinogen 0.5–6 g L−1. All dilutions for BSA (/fibrinogen) were done in DI water (/0.9% NaCl). After incubation, suspensions were centrifuged for 15 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (eppendorf minispin) and the obtained pellets were resuspended in DI water. The procedure was repeated at least 3 times to remove any unabsorbed protein.
000 rpm (eppendorf minispin) and the obtained pellets were resuspended in DI water. The procedure was repeated at least 3 times to remove any unabsorbed protein.
2.2 Characterization
Transmission electron microscopy (TEM; Hitachi 7600) was performed to investigate changes in CNM structure upon chemical treatment. For TEM, the samples were dispersed in water for 10 min using 1/8′′ tip sonicator and the sample was drop casted on a holey carbon (400 mesh; Ted Pella) and was dried overnight. Contact angle measurements were performed on freestanding CNM buckypapers using a custom-built setup equipped with Celestron's 44302 digital USB microscope. We prepared freestanding buckypapers for all different types of CNMs by vacuum filtering CNM suspensions in water (1 mg mL−1, sonicated using 1/8′′ tip sonicator for 30 minutes) through a 0.45 mm nylon filter paper. Subsequent to filtration, the CNM materials were peeled off from the filter paper by heat treating the samples at 60 °C overnight. All the Raman spectra were obtained on dry powders using a 514.5 nm Ar+ excitation coupled to a Renishaw InVia micro-Raman spectrometer.PL spectra was measured at 280 nm excitation wavelength and 300–450 nm emission range with 5 nm slit width using a Horiba iHR 550 spectrometer equipped with a TRIAX 550 liquid N2 cooled CCD. For the PL measurements, 3 mL of CNM–protein suspension was used in a 10 mm wide quartz cuvette. Fourier Transform Infrared (FTIR) spectroscopy was performed on a Thermo Scientific Nicolet 6700 ATR-FTIR and Bruker IFS 66v/S. For FTIR measurements, the samples were drop casted onto a monolithic diamond crystal and the signal from DI water was used as the background.
Cyclic voltammetry was carried out to measure the charge transfer properties between the MWNTs and proteins solutions. The experiments were performed with a Reference 3000 Potentiostats electrochemical measurement system (Gamry Instruments, Inc.). A platinum mesh was used as the counter electrode and Ag/AgCl as the reference electrode. The electrolyte solution consisted 40 g L−1 of BSA or 4 g L−1 of fibrinogen. All the data was obtained at a low scan rate (5 mV s−1) to avoid any diffusion limitations.
2.3 Molecular dynamics simulation
We performed large-scale MD simulations of BSA (4F5S.pdb)22 in explicit water in the presence of two different CNMs – GNR and GONR. The surface area of both the CNMs was kept the same in the simulations. GONR was generated using the methodology described in DeFever et al.23 and was 30% oxidized. The Amberff99SB-ILDN force field was used to describe the protein and GNR.24 TIP3P model was used for water.25 GONR was described using the OPLS force field24 because this eliminates the necessity of determining the partial charges on the GONR atoms using ab initio calculations.The simulation systems comprised of the CNM, 9220 protein atoms and approximately 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 water molecules. Ions were added to neutralize the system. To enhance the sampling of protein adsorption in the MD simulations we performed 10 simulations for each system. In these 10 simulations, we used 10 different protein orientations in the starting configurations. This enabled us to sample different regions of the protein that could adsorb to the CNMs. We note that this does not capture all the regions of the protein that could adsorb to the CNMs and also does not specify the affinity of various regions to the CNMs – neither of which are the goals of these simulations. The protein was placed at a distance such that no two heavy atoms of the protein and CNM were closer than 8 Å in the starting configurations of the simulation. Each simulation was run for 100 ns, resulting in effectively 1 μs (10 orientations × 100 ns) simulation per CNM–BSA–water system.
000 water molecules. Ions were added to neutralize the system. To enhance the sampling of protein adsorption in the MD simulations we performed 10 simulations for each system. In these 10 simulations, we used 10 different protein orientations in the starting configurations. This enabled us to sample different regions of the protein that could adsorb to the CNMs. We note that this does not capture all the regions of the protein that could adsorb to the CNMs and also does not specify the affinity of various regions to the CNMs – neither of which are the goals of these simulations. The protein was placed at a distance such that no two heavy atoms of the protein and CNM were closer than 8 Å in the starting configurations of the simulation. Each simulation was run for 100 ns, resulting in effectively 1 μs (10 orientations × 100 ns) simulation per CNM–BSA–water system.
The long-range electrostatic interactions were calculated using PME as implemented in GROMACS v5.0.2.26 The velocity-rescaling thermostat27 and Berendsen barostat28 were used to maintain the temperature at 300 K and pressure at 1 bar, respectively. The bonds involving hydrogen atoms were constrained using LINCS algorithm.29 The time step used was 2 fs. The simulations were performed on 20 CPUs + 2 K20 GPUs and each 100 ns long simulation took ∼6 days to complete. Configurations were stored every 20 ps for further analysis.
3. Results and discussions
3.1 Electron microscopy and Raman spectroscopy
As shown in Fig. 1a–c, we synthesized GNRs and GONRs by unzipping and subsequently oxidizing pristine MWNTs. FTIR measurements (see ESI Fig. S1†) revealed the absence of functional group-type defects on MWNTs, while showed the presence of hydroxyl groups on GNRs, which possibly formed on the edges of unzipped MWNTs during chemical reflux and the unzipping process. In contrast to MWNTs and GNRs, GONRs exhibited more functional groups such as epoxide, hydroxyl, and carboxyl functionalities (see Fig. S1†). The presence of polar functional groups on GNRs and GONRs indicate that they are more hydrophilic than MWNTs and could be ranked as MWNTs > GNRs > GONRs in terms of hydrophobicity. Concurrent with the observations from FTIR, our contact angle measurements confirmed the non-wettable nature of MWNTs and GNRs (Fig. S2†). As expected from the presence of polar groups on GONRs, water was found to wet GONRs confirming the hydrophilic nature.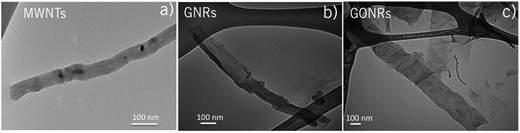 | ||
| Fig. 1 Transmission electron microscope images of multi-walled carbon nanotubes (MWNTs) (a), graphene nanoribbons (GNRs) (b), and graphene oxide nanoribbons (GONRs) (c). The scale bars are 100 nm. | ||
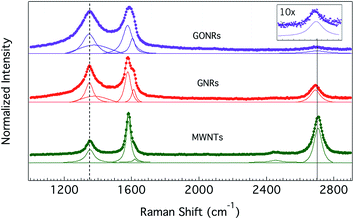
The Raman spectra of all CNMs used in this study (Fig. 2) showed the graphitic band (or G-band), arising from longitudinal and transverse optical phonons ∼1585 cm−1, along with the so-called disorder or D-band at 1350 cm−1. The integrated intensity of the D-band to that of the G-band (ID/IG ratio) is often used as a measure of average defect–defect spacing in CNMs.30 Clearly, as seen from Fig. 2, the ID/IG ratio is higher for GONR and GNRs than the pristine MWNTs, which could be attributed to the harsh chemical reactions in the unzipping and oxidation processes. While GNRs are unzipped through only one-time exposure to sulphuric acid and KMnO4, GONRs undergo a two-step chemical process (i.e., unzipping and subsequent oxidation) leading to an increased D-band intensity in their Raman spectrum. Furthermore, the overtone of D-band (called 2D-band ∼2700 cm−1) was found to significantly decrease in intensity (at least by 10 times, as shown in Fig. 2 inset) in GONRs due to oxidation. The full-width at half-maximum of G-band in GNRs (∼28 cm−1) and GONRs (∼40 cm−1) also increased significantly relative to pristine MWNTs (∼20 cm−1) indicating a decrease in phonon lifetime due to the presence of different type of defects (particularly, edges and functional group-type defects in GNRs and GONRs). Additionally, a defect-induced Raman feature ∼1620 cm−1, often activated at high defect concentrations, is clearly observed for both GNRs and GONRs. Juxtaposing Raman, FTIR, and contact angle measurements, it could be concluded that MWNTs are less-defective (ID/IG ∼ 0.35) and more hydrophobic (no presence of functional groups), while GNRs (/GONRs) are slightly (/highly) defective (ID/IG ∼ 0.83 for GNRs and 0.94 for GONRs) and weakly (/strongly) hydrophilic due to the presence of polar functional groups.
3.2 Adsorption isotherms
The protein adsorption on different CNMs was characterized using adsorption isotherms obtained through photoluminescence studies. Previously, we showed that the intrinsic emission of proteins (∼345 nm upon 280 nm excitation) from aromatic amino acids such as tryptophan, tyrosine and phenylalanine could be used to quantify the unknown concentration of a protein through the use of a standard curve.9 We obtained a standard calibration curves for BSA and fibrinogen (see Fig. S3†) by measuring their intrinsic photoluminescence at various concentrations in an aqueous medium. Subsequently, the adsorption isotherms for BSA and fibrinogen on CNMs were obtained by measuring the PL signal from adsorbed proteins and acquiring the protein concentration from the standard curves in Fig. S3.† Although the shape of experimental isotherms for protein adsorption often appears strikingly similar to the Langmuir isotherm, the use of the Langmuir isotherm is inappropriate in our case because of its assumptions such as: (i) the equivalence of all adsorption sites, (ii) one-to-one binding between each adsorption site and the adsorbed molecules, (iii) the absence of interaction between adsorbed solutes, and (iv) the dynamic reversibility of the adsorption process.31 In reality, adsorbed proteins tend to rapidly undergo surface-induced unfolding and reorientation to increase their contact area to irreversibly stick to the material surface.As shown in Fig. 3, the adsorption of BSA and fibrinogen on CNMs was observed to follow a Freundlich isotherm. Unlike Langmuir model, the Freundlich isotherm is more appropriate to describe the adsorption processes, as it does not assume uniformity, one-to-one binding, and the absence of protein–protein interactions. The Freundlich adsorption isotherm is mathematically expressed as:
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a = (1/n)log a = (1/n)log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + log C + log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K K | (1) |
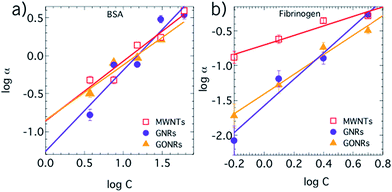 | ||
| Fig. 3 Adsorption isotherms for BSA (a) and fibrinogen (b) on to various carbon nanomaterials (CNMs) obtained using the intrinsic photoluminescence of aromatic acids in BSA and fibrinogen. The trends match a Freundlich isotherm model suggesting non-uniform and multivalent adsorption sites on carbon nanomaterials. BSA exhibits similar adsorption on all CNMs with slightly lower adsorption on GONRs (cf.Table 1). Fibrinogen exhibits a relatively strong adsorption of GNRs and GONRs, relative to MWNTs, possibly due to their flat-sheet like structure. | ||
| Nanomaterial | 1/n | k | ||
|---|---|---|---|---|
| BSA | Fibrinogen | BSA | Fibrinogen | |
| MWNTs | 0.79 | 0.68 | 0.42 | 0.49 |
| GNR | 0.9 | 1.9 | 0.30 | 0.2 |
| GONR | 0.73 | 1.4 | 0.42 | 0.24 |
We observed that 1/n and K values are similar for the adsorption of BSA on all CNMs (Table 1). In the case of MWNTs (hydrophobic) and GNRs (which are only weakly hydrophilic), a “shell” of interacting water molecules plausibly forms around the hydrophobic surface decreasing the entropy. The disruption of this shell upon protein adsorption is more energetically favourable because the release of otherwise constrained water molecules leads to an increase in the entropy. Additionally, the energy of BSA–MWNT/GNR system could be reduced by the relaxation of protein secondary structures to increase the protein entropy. Thus, the adsorption of BSA on to MWNTs and GNRs could be expected to increase with increasing protein concentration (Fig. 3). The low 1/n values (cf.Table 1) for GONRs showed that GONR exhibited relatively weaker interaction with BSA. Surprisingly, the difference in 1/n values for the adsorption of BSA on GONRs was only slightly different from MWNTs and GNRs despite their hydrophilicity. The hydrophilic nature of GONRs does not constrain water molecules, unlike MWNTs and GNRs, and thus the adsorption of BSA on GONRs is not accompanied by significant entropy increase from the release of water molecules. Nevertheless, a comparable 1/n value for the adsorption of BSA on GONRs could be attributed to the formation of energetically favorable hydrogen bonds between BSA and functional group-type defects on GONRs (particularly, hydroxyl and carboxyl groups shown in Fig. S1†).
3.3 Molecular dynamics simulations
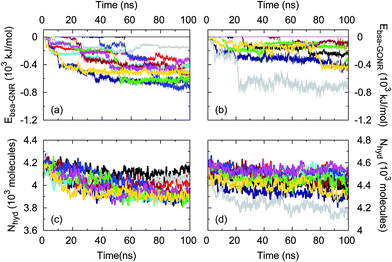
To further understand these unexpected CNM–BSA interactions, we performed large-scale molecular dynamics (MD) simulations of BSA–GNR/GONR–water systems. As shown in Fig. 4a and b, we observed a decrease in the BSA–GNR/GONR interaction energy indicating that BSA associates with GNRs and GONRs consistent with our experiments. Furthermore, the adsorption process was found to occur in steps (as seen in the series of plateaus and steep drops in Fig. 4a and b) primarily because BSA undergoes conformational changes after initial contact (in accordance with our experimental observations, discussed later in Fig. 5) leading to some protein regions collapsing on to the CNM surface rather than gradually spreading from the initial region of contact.32 In the case of GNRs, most simulations resulted in interaction energies <−380 kJ mol−1 within 100 ns of the simulation unlike BSA–GONR–water simulations. This suggests that BSA displays more affinity to GNR compared to GONR, consistent with the experimental findings that BSA prefers more hydrophobic surfaces (as seen by higher 1/n values for adsorption of BSA on GNR compared to GONR in Table 1). We observed that water molecules are released from the hydration shells of BSA, GNR and GONR as BSA adsorbs to the CNMs (Fig. 4c and d). The total number of water molecules in the hydration shell of the protein and GNR is lower (i.e., the number of released water molecules is higher) than that in the case of the BSA–GONR system. As discussed before (cf.Fig. 3), such a result is expected because relatively more water molecules interact with GONR due to its hydrophilicity. Nonetheless, in both cases of GNRs and GONRs, BSA adsorbs well on to the CNM due to favourable interaction energy and a decrease in the total number of water molecules in the hydration shell of the BSA–CNM complex.Returning to Fig. 3, in case of fibrinogen, 1/n and K values for MWNTs were found to be lower when compared to GNRs and GONRs. It may be rationalized that fibrinogen, which is a hard protein due to its excellent internal stability, has a lower tendency than BSA (a soft protein) to relax its secondary structure for adsorbing on to a tubular structure as in the case of MWNTs. GNRs and GONRs, unlike MWNTs, provide a flat-sheet like structure (cf.Fig. 1) which is more energetically favourable for the adsorption of fibrinogen even without unfolding. Furthermore, similar to the case of BSA, it could be expected that the functional groups on GONRs facilitate hydrogen bonds with fibrinogen.
3.4 FTIR studies on secondary structure changes
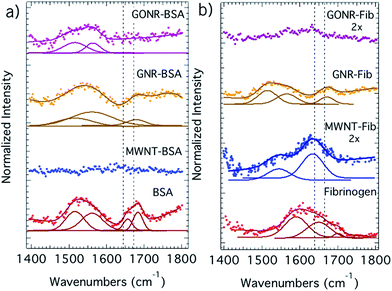
In the formation of the protein corona, the protein molecules initially adsorb on to the CNMs while largely retaining their native-state structure. Subsequently, the adsorbed protein can begin to relax its secondary structure, unfold, and spread out on the CNM surface and transition from an end-on to a side-on orientation.33,34 The degree of protein unfolding on a surface is influenced by the strength of the protein–surface interactions relative to the internal stability of the protein.35–37 Accordingly, to elucidate the adsorption-induced structural changes in proteins, we obtained the FTIR spectra of adsorbed proteins as shown in Fig. 5. It should be noted that CNMs exhibit strong absorption <240 nm due to their π-electron system precluding the use of traditional tools such as circular dichroism for the evaluation of protein secondary structure. As evident from Fig. 5a and b, the α-helical content in BSA leads to strong adsorption ∼1640–1660 cm−1 (shown in dashed lines) while the lower frequency component at ∼1620–1640 cm−1 and the peak ∼1555 cm−1 arise from β-sheets.36,37 Clearly, the rich secondary structure of BSA (particularly, the peak relating to α-helical content) significantly disappears upon its adsorption on to all CNMs, as expected from its low internal stability. Indeed, the changes in secondary structure are higher in the case of MWNTs (i.e., complete disappearance of secondary structure) suggesting that BSA unfolds much more, relative to GNRs and GONRs, in order to adhere to the tubular MWNTs. GNR and GONR retain BSA secondary structure to certain extent, as shown by the presence of ∼1555 cm−1 for β-sheets. In the case of fibrinogen, the secondary structural changes are found to be higher for GONRs compared to MWNTs and GNRs plausibly due to the formation of hydrogen bonds. The α-helix peak was found to partially disappear for fibrinogen adsorbed on MWNTs and GNRs. Lastly, the structural changes for fibrinogen on GNRs seemed to be less pronounced than MWNTs possibly due to its shape. It could be rationalized that fibrinogen must unfold more to adhere to MWNTs due to their higher curvature than GNRs.3.5 Cyclic voltammetry
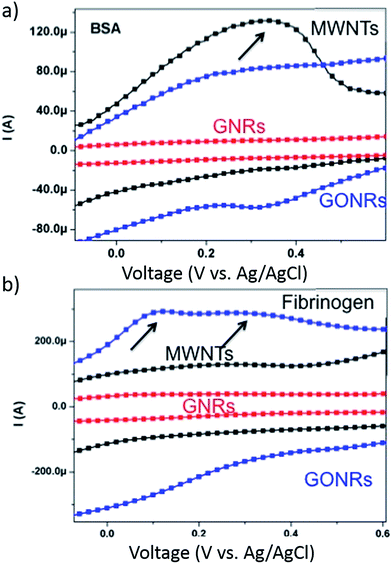
The chemisorption of proteins on bulk material surfaces has been known to occur through charge transfer processes.34,38–40 It may be expected that a surface facilitating higher charge transfer at the nanoscale may lead to stronger surface–protein interactions and a subsequent increase in protein adsorption. To validate such a hypothesis, we performed cyclic voltammetry (CV) measurements with CNMs as a working electrode in protein electrolyte solution (Fig. 6). In CV characterization, the application of gate voltage on the working electrode modulates its electronic energy levels, which when above (/below) the LUMO (/HOMO) levels of the protein can result in a charge transfer. Although the proximity of electronic levels make charge transfer between protein in electrolyte solution and the working electrode (i.e., CNMs) thermodynamically favourable, the probability of charge transfer depends upon density of electronic states at the Fermi level (DOS(EF)) in the working electrode (CNMs in this case). In defect-free CNMs (e.g., perfect sheet of graphene with no defects), the DOS(EF) is very low (almost zero for graphene) and therefore charge transfer is often not observable in the experiments.9,41 However, in our case, the presence of defects (introduced through unzipping in GNRs and oxidation in GONRs) induces new electronic states in CNMs which could increase the DOS(EF), and thereby lead to strong interactions between the protein and CNMs through charge transfer. In a typical CV plot, a peak in current (in a current vs. voltage plot such as the one shown in Fig. 6) indicates the presence of charge transfer between the electrolyte (i.e., protein solution in our case) and the working electrode (i.e., CNMs). Interestingly, the amount of charge transfer (Fig. 6) does not concomitantly increase with increasing ID/IG (cf.Fig. 2). Such an observation may be rationalized by the fact that the electronic structure (i.e., both energy levels and DOS(EF)), which is dependent on shape, size, and defect density of CNMs, is different for MWNTs, GNRs, and GONRs. Nonetheless, in the case of BSA (/fibrinogen), the charge transfer was found to be the highest for MWNTs (/GONRs), which exhibited highest secondary structural changes (cf.Fig. 5) for BSA (/fibrinogen) suggesting that charge-transfer may induce protein unfolding or vice versa.When taken together, photoluminescence, Raman, infrared, and electrochemistry measurements suggest that: (i) the defect-induced hydrophilicity can alter the formation of protein corona in CNMs, and (ii) the net charge transfer between protein and CNM and the change in secondary structures may be correlated, indicating that protein corona formation is accompanied by both charge-transfer and protein unfolding.
4. Conclusions
In summary, we investigated the binding of bovine serum albumin (BSA) and fibrinogen with different carbon nanomaterials. Raman spectroscopy measurements showed a higher amount of defects in graphene and graphene oxide nanoribbons (GNRs and GONRs) compared to pristine multi-walled nanotubes (MWNTs). The binding experiments showed that the BSA adsorbs equally on all MWNTs, GNRs, and GONRs while fibrinogen showed a significantly lower adsorption on MWNTs. The lower adsorption of fibrinogen on MWNTs was attributed to its hardness relative to BSA. Furthermore, it is observed that the net conformational changes (gleaned from infrared spectroscopy) in protein structure were highest for the cases of highest charge transfer (observed in cyclic voltammetry) between protein and CNMs. Our results show that the formation of protein corona is sensitive to the defects on CNMs and is accompanied by both charge-transfer and protein unfolding.Acknowledgements
J. B. and R. P. gratefully acknowledge funding support from NIH NIEHS R03-ES023036. R. P. and S. S. acknowledge their Clemson University Start-Up funds and resources from Palmetto Supercomputer maintained by Clemson Computing and Information Technology group. R. P and A. M. R thank Aleksandr Kakinen, Estonia for his initial contributions to the project.References
- R. Podila and J. M. Brown, J. Biochem. Mol. Toxicol., 2013, 27(1), 50–55 CrossRef CAS PubMed.
- P. Wick, M. J. D. Clift, M. Roesslein and B. Rothen-Rutishauser, ChemSusChem, 2011, 4(7), 905–911 CrossRef CAS PubMed.
- J. H. Shannahan, X. Lai, P. C. Ke, R. Podila, J. M. Brown and F. A. Witzmann, PLoS One, 2013, 8(9), e74001 CAS.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2050–2055 CrossRef CAS PubMed.
- A. E. Nel, L. Maedler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8(7), 543–557 CrossRef CAS PubMed.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3(1–2), 40–47 CrossRef CAS.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132(16), 5761–5768 CrossRef CAS PubMed.
- R. Podila, R. Chen, P. C. Ke, J. M. Brown and A. M. Rao, Appl. Phys. Lett., 2012, 101(26), 263701 CrossRef PubMed.
- R. Podila, P. Vedantam, P. C. Ke, J. M. Brown and A. M. Rao, J. Phys. Chem. C, 2012, 116(41), 22098–22103 CAS.
- A. A. Aldossari, J. H. Shannahana, R. Podila and J. M. Brown, Toxicol. in Vitro, 2015, 29(1), 195–203 CrossRef CAS PubMed.
- J. H. Shannahan, R. Podila, A. A. Aldossari, H. Emerson, B. A. Powell, P. C. Ke, A. M. Rao and J. M. Brown, Toxicol. Sci., 2015, 143(1), 136–146 CrossRef CAS PubMed.
- R. Landsiedel, L. Ma-Hock, A. Kroll, D. Hahn, J. Schnekenburger, K. Wiench and W. Wohlleben, Adv. Mater., 2010, 22(24), 2601–2627 CrossRef CAS PubMed.
- F. Watari, N. Takashi, A. Yokoyama, M. Uo, T. Akasaka, Y. Sato, S. Abe, Y. Totsuka and K. Tohji, J. R. Soc., Interface, 2009, 6, S371–S388 CrossRef CAS PubMed.
- B. Fubini, I. Fenoglio, M. Tomatis and F. Turci, Nanomedicine, 2011, 6(5), 899–920 CrossRef CAS PubMed.
- X. Zhu, Y. Chang and Y. Chen, Chemosphere, 2010, 78(3), 209–215 CrossRef CAS PubMed.
- Z. Yang, Z. Wang, X. Tian, P. Xiu and R. Zhou, J. Chem. Phys., 2012, 136(2), 025103 CrossRef PubMed.
- R. Haddad, S. Cosnier, A. Maaref and M. Holzinger, Analyst, 2009, 134(12), 2412–2418 RSC.
- C. P. Firme III and P. R. Bandaru, Nanomedicine: Nanotechnology, Biology and Medicine, 2010, 6(2), 245–256 CrossRef PubMed.
- K. Bradley, M. Briman, A. Star and G. Grüner, Nano Lett., 2004, 4(2), 253–256 CrossRef CAS.
- C. N. R. Rao and R. Voggu, Mater. Today, 2010, 13(9), 34–40 CrossRef CAS.
- R. Podila, J. M. Brown, A. Kahru and A. M. Rao, MRS Bull., 2014, 39(11), 990–995 CrossRef CAS.
- A. Bujacz, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68(10), 1278–1289 CrossRef CAS PubMed.
- R. S. DeFever, N. K. Geitner, P. Bhattacharya, F. Ding, P. C. Ke and S. Sarupria, Environ. Sci. Technol., 2015, 49(7), 4490–4497 CrossRef CAS PubMed.
- K. Lindorff-Larsen, S. Piana, K. Palmo, P. Maragakis, J. L. Klepeis, R. O. Dror and D. E. Shaw, Proteins: Struct., Funct., Bioinf., 2010, 78(8), 1950–1958 CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79(2), 926–935 CrossRef CAS PubMed.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4(3), 435–447 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126(1), 014101 CrossRef PubMed.
- H. J. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. R. H. J. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81(8), 3684–3690 CrossRef CAS PubMed.
- B. Hess, H. Bekker, H. J. Berendsen and J. G. Fraaije, J. Comput. Chem., 1997, 18(12), 1463–1472 CrossRef CAS.
- L. M. Malard, M. A. Pimenta, G. Dresselhaus and M. S. Dresselhaus, Phys. Rep., 2009, 473(5), 51–87 CrossRef CAS PubMed.
- R. A. Latour, J. Biomed. Mater. Res., Part A, 2015, 103(3), 949–958 CrossRef PubMed.
- J. W. Shen, T. Wu, Q. Wang and Y. Kang, Biomaterials, 2008, 29(28), 3847–3855 CrossRef CAS PubMed.
- C. M. Alves, R. L. Reis and J. A. Hunt, J. R. Soc., Interface, 2010, 7(50), 1367–1377 CrossRef CAS PubMed.
- M. Rabe, D. Verdes and S. Seeger, Adv. Colloid Interface Sci., 2011, 162(1–2), 87–106 CrossRef CAS PubMed.
- S. Gupta, M. Camargo, J. Stellbrink, J. Allgaier, A. Radulescu, P. Lindner, E. Zaccarelli, C. N. Likos and D. Richter, Nanoscale, 2015, 7(33), 13924–13934 RSC.
- M. Jackson and H. H. Mantsch, Crit. Rev. Biochem. Mol. Biol., 1995, 30(2), 95–120 CrossRef CAS PubMed.
- J. Kong and S. Yu, Acta Biochim. Biophys. Sin., 2007, 39(8), 549–559 CrossRef CAS PubMed.
- W. Norde, Croat. Chem. Acta, 1983, 56(4), 705–720 CAS.
- N. R. Cabilio, S. Omanovic and S. G. Roscoe, Langmuir, 2000, 16(22), 8480–8488 CrossRef CAS.
- A. Carre, J. Adhes. Sci. Technol., 2010, 24(5), 813–814 CrossRef CAS PubMed.
- R. Podila, T. Moore, F. Alexis and A. M. Rao, RSC Adv., 2013, 3(6), 1660–1665 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra15007h |
| This journal is © The Royal Society of Chemistry 2015 |