A benzimidazolium-based organic trication: a selective fluorescent sensor for detecting cysteine in water†
Abstract
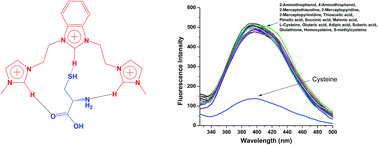
A benzimidazolium- and imidazolium-based trication was developed as a fluorescence probe for cysteine (Cys). The probe exhibits fluorescence responses to Cys in water with high selectivity and a nano-molar detection limit. Such specificity towards Cys is based on differences in the hydrogen bonding of R1 with Cys, and it provides a method for detecting Cys in the presence of other analytes in a real biological system such as human serum. Density functional theory (DFT) calculations and 1H NMR titration confirmed the cavity-based recognition of Cys.

 Please wait while we load your content...
Please wait while we load your content...